Introduction
Haemosporidian parasites (phylum Apicomplexa) infect a wide range of mammals including primates, rodents and bats (Garnham, Reference Garnham1966). The human-infecting Plasmodium species belong to a large group of haemosporidian parasites of about 500 closely related species (Martinsen and Perkins, Reference Martinsen, Perkins, Carlton, Perkins and Deitsch2013; Galen et al., Reference Galen, Borner, Martinsen, Schaer, Austin, West and Perkins2018). These parasites use a diverse array of dipteran and vertebrate hosts to complete their life cycle, the latter comprising birds, saurian reptiles and mammals (Garnham, Reference Garnham1966; Levine, Reference Levine1988). Mammalian haemosporidian parasites are classified in ten different genera, including Plasmodium, Hepatocystis and Nycteria (Perkins and Schaer, Reference Perkins and Schaer2016). Previous studies led to the discovery of several parasite lineages in bats and revealed unexpected phylogenetic relationships (e.g. Duval et al., Reference Duval, Robert, Csorba, Hassanin, Randrianarivelojosia, Walston, Nhim, Goodman and Ariey2007; Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013), which suggests that bats have played an important role in the evolutionary history of malaria parasites (Perkins and Schaer, Reference Perkins and Schaer2016; Galen et al., Reference Galen, Borner, Martinsen, Schaer, Austin, West and Perkins2018). A broader sampling and systematic analysis of bat malaria parasites is essential for better understanding the evolutionary history of haemosporidian parasites, including the human-infecting species (Perkins and Schaer, Reference Perkins and Schaer2016).
Parasites of the genus Hepatocystis infect a wide range of vertebrate hosts including primates, bats, ungulates and rodents, whereas Nycteria parasites exclusively infect insectivorous bats (Garnham, Reference Garnham1966). Historical classification of mammalian haemosporidian parasites was done according to morphological and biological characteristics, resulting in initial misplacement of some chiropteran haemosporidian parasites within the genus Plasmodium. However, the observation that certain bat parasites lack schizogony in erythrocytes, and therefore cannot represent the genus Plasmodium, led to a reclassification of several species, which was later confirmed by molecular phylogenetic studies (e.g. Garnham, Reference Garnham1950; Perkins, Reference Perkins2014). The mammalian Plasmodium clade is paraphyletic as it contains the parasites of the genus Hepatocystis (Galen et al., Reference Galen, Borner, Martinsen, Schaer, Austin, West and Perkins2018). The dipteran vectors for most chiropteran haemosporidian parasites, including Nycteria, remain unknown (Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013, Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015).
Bats are the only flying mammals and exhibit a very high species diversity (Arthur and Lemaire, Reference Arthur and Lemaire1999; Simmons and Cirranello, Reference Simmons and Cirranello2020). They have unique characteristics among mammals and have an ecological diversity of niches that makes them key organisms in maintaining the ecosystem balance (Reis and Guillaumet, Reference Reis and Guillaumet1983; Rodriguez et al., Reference Rodriguez, Hoffmann, Porter and Baker2006; Aziz et al., Reference Aziz, McConkey, Tanalgo, Sritongchuay, Low, Yong, Mildenstein, Nuevo-Diego, Lim and Racey2021). Indeed, numerous studies have demonstrated the important role of the Chiroptera in insect population regulation, pollination and seed dispersal of many ecologically and economically important plants (Tchatat, Reference Tchatat1999; Hutcheon, Reference Hutcheon, Goodman and Benstead2003; Bakwo Fils, Reference Bakwo Fils2009; Kunz et al., Reference Kunz, de Torrez, Bauer, Lobova and Fleming2011; Bakwo Fils et al., Reference Bakwo Fils, Anong and Fohouo2012; Ingala et al., Reference Ingala, Simmons, Wultsch, Krampis, Provost and Perkins2021). This is especially the case in tropical areas, where these animals seem to be among the main agents of seed dissemination due to their abundances and their activities (Reis and Guillaumet, Reference Reis and Guillaumet1983).
This study presents a survey of haemosporidian parasite infections in different bat species in the central region of Cameroon and determines the prevalence, parasitaemia and phylogenetic relationships of haemosporidian infections in bats.
Materials and methods
Bats were sampled in the central region of Cameroon between February 2016 and December 2019 representing both dry and wet seasons. Sampling sites covered the different habitat types: forest, savanna and cultured farmland (Fig. 1). The prevailing climate is equatorial of Guinea type with dry seasons in July to August and December to March and rainy seasons from April to June and September to November (Maurice et al., Reference Maurice, Maurice and Cornelin2011). Temperatures are moderate to high, and constant (mean temperature = 28°C). Precipitation varies between 1500 mm and 3000 mm per year. Vegetation in the region is dominated by a dense and humid tropical forest. Soils are clay and red. The river Nyong (750 km long) is the main water stream of the region.
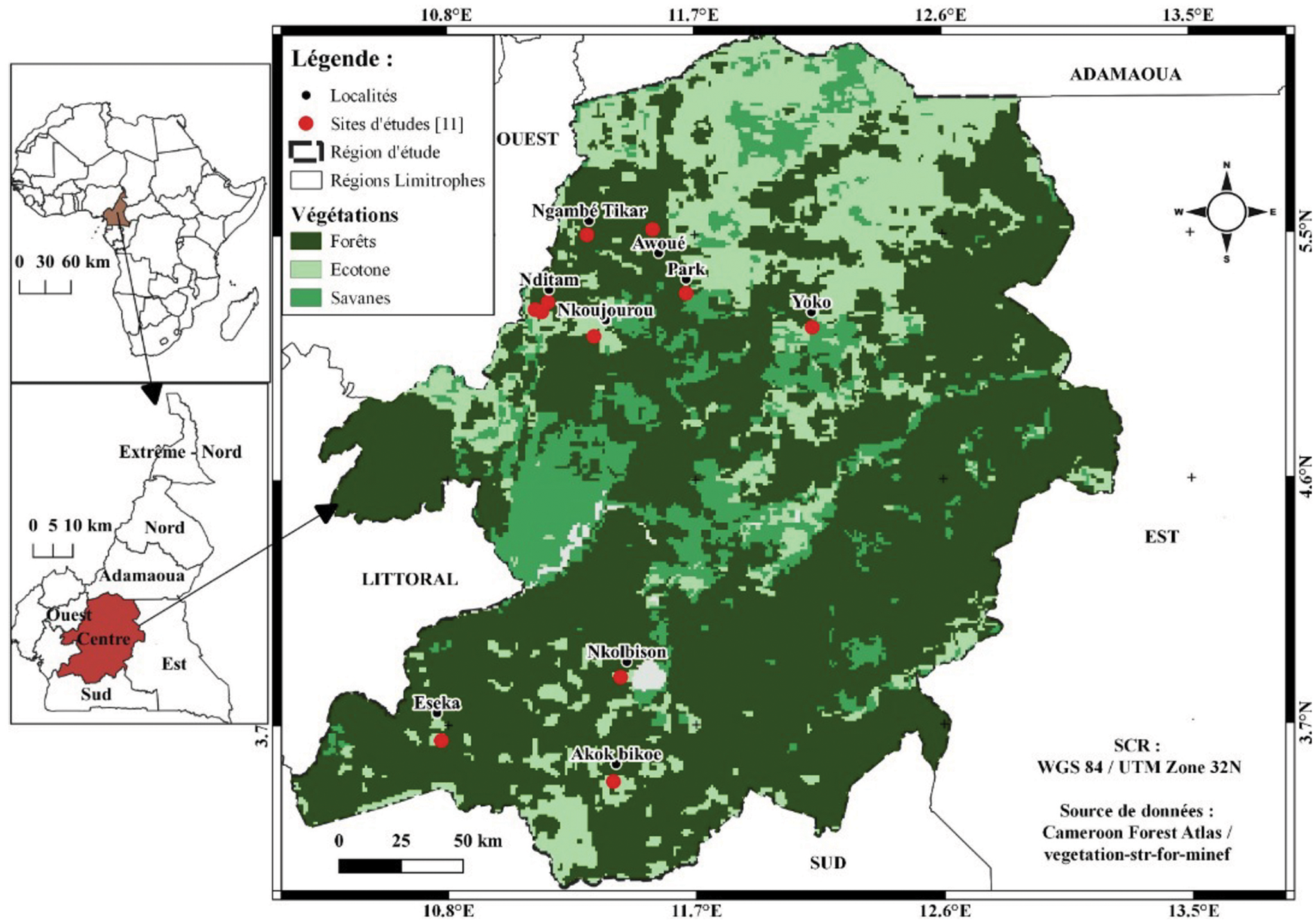
Fig. 1. (A) Map of central region of Cameroon depicting the sampling sites (sampling sites are marked with red dots).
Field sampling and microscopy
Bats were captured using ground-level mist nets. Mist nets were set every capture night from 6 pm to 5 am across potential flight trajectories of bats: above water points, in clearings of farmland, in widespread savannah, in the forest. Sampling was also carried out in diurnal roosts like caves, tunnels, abandoned houses. Standard measurements like sex, age, forearm lengths were recorded, and species were identified using different identification keys (Rosevear, Reference Rosevear1965; Hayman and Hill, Reference Hayman and Hill1971; Patterson and Webala, Reference Patterson and Webala2012; Happold and Happold, Reference Happold and Happold2013). The bats were released, but one adult female and one adult male per species were chosen as voucher specimens and preserved in 70% (vol/vol) ethanol and accessioned in the Biological Laboratory Museum of the University of Maroua.
Blood samples of 10–30 μL were collected from each individual by venipuncture of the uropatagial vein. After the blood collection, cotton was placed on the haemostatic vein until the bleeding had stopped; then the individual was released. The blood sample was used to prepare a thin and a thick blood smear shortly after collection. The blood smears were dried and fixed in 99–100% (vol/vol) methanol solution for 3 s. All blood smears were stained with 10% Giemsa solution for 15 to 20 min and dried. The remaining blood sample was dotted on Whatman filter papers. Blood smears and blood dots were immediately air-dried in humid environment in the field with a battery-operated fan and then the samples were dried for at least 3 h in an environment with low humidity. After drying, the filter cards were stored in Ziploc bags containing a sachet with desiccant (silicate gel) and subsequently stored in a freezer at −20°C.
Giemsa-stained blood smears were examined for the presence and parasitemia of haemosporidian parasites and for the analysis of the morphology of observed parasite stages for 20–40 min, using a light microscope (Leica DMLB 1000) at a magnification of ×1000 with immersion oil.
Parasitaemia values (% of infected erythrocytes) were calculated for all bats with confirmed haemosporidian infections. The mean number of erythrocytes per field was determined by counting them in 1–3 fields, and the number of parasites was recorded in 20–100 fields. For this purpose, fields with comparable erythrocyte density were chosen (Schaer et al., Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015). Parasitaemia (in percent) was determined following the calculation: total number of parasites/mean number of erythrocytes per field × number of counted fields.
Molecular methods
DNA was extracted from the dried blood dots on Whatman filter paper (GE Healthcare) using the QIAGEN DNeasy extraction kit (Hilden, Germany) (e.g. Schaer et al., Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015). The protocol for animal tissues was performed with the minor modification of elution of the samples in 50–100 μL AE buffer depending on the density of the blood dot. PCRs were performed using the AllTaq Master Mix Kit (QIAGEN) with 4–5 μL of genomic DNA as the template, and 1 μL of each primer (10 mm). Five genes from the three parasite genomes were targeted for detection and subsequent phylogenetic analysis of haemosporidian parasites: the mitochondrial genes cytochrome b (cytb) and cytochrome oxidase 1 (cox1); the apicoplast caseinolytic protease gene (clpC); and the nuclear genes elongation factor 2 (ef2) and adenylosuccinate lyase (asl). The fruit bats (Pteropodidae spp.) were screened with Hepatocystis-specific cytb-primers (HepF3/HepR3) (Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013). All primers are listed in Supplementary Table S1. All positive PCR products were sequenced with the amplification primers and run on an ABI-373 sequencer. Bat genetic markers comprising the mitochondrial cytochrome b (cytb) and the nuclear introns Acyl-CoA oxidase 2, intron 3 (acox2), Rogdi-like protein gene, intron 7 (rogdi) and Beta-fibrinogen gene, intron 7 (fgb) were amplified and sequenced to verify the morphological identifications of the bat hosts (Supplementary Table S1). All DNA sequences were manually edited using the software Geneious Prime 2021.1 (https://www.geneious.com). Ambiguous base calls or missing data were coded with N´s or the corresponding ambiguity code. Sequences were aligned using the Muscle algorithm (Edgar, Reference Edgar2004). Parasite sequences for the analysis of the phylogenetic relationships of Nycteria included a total of 3243 nucleotides (nt) (1119 nt of cytb, 861 nt of cox1, 528 nt of clpC, 513 nt of the ef2-gene and 222 nt of the asl-gene) and for the phylogenetic relationships of Hepatocystis included a total of 1938 nucleotides (nt) (531 nt of cytb, 993 nt of cox1 and 513 nt of the ef2-gene). Reference sequences were retrieved from GenBank and added to the alignments (all accession numbers are listed in Supplementary Table S2). All individual gene alignments were concatenated, and phylogenetic relationships were evaluated with Bayesian analysis using the taxon Leucocytozoon as outgroup. Data were partitioned according to the number of genes and the software PartitionFinder vs 2 was used to test different DNA substitution models (Lanfear et al., Reference Lanfear, Frandsen, Wright, Senfeld and Calcott2017). MrBayes v3.2.7a (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001) was performed via the CIPRES Science Gateway Web Portal V3.3 (Miller et al., Reference Miller, Pfeiffer and Schwartz2010) with two runs of four chains (three heated, one cold, temperature = 0.03) each for 20 million generations. The first 25% of trees were discarded as burn-in. Mixing and convergence of runs and effective sample size (average ESS > 4000) were assessed with Tracer v1.6 (Rambaut et al., Reference Rambaut, Suchard, Xie and Drummond2014). Figures were created with Microsoft PowerPoint and with BioRender.com.
Results
Prevalence of haemosporidian infections
A total of 155 bats belonging to five bat families, ten genera and 14 species were investigated. Based on thorough microscopic examinations of thin blood smears (155/155) and PCR screening (153/155), haemosporidian parasites were detected in 28 individuals, corresponding to an overall prevalence of 18.1% (Table 1).
Table 1. Investigated bat species and their corresponding haemosporidian parasites
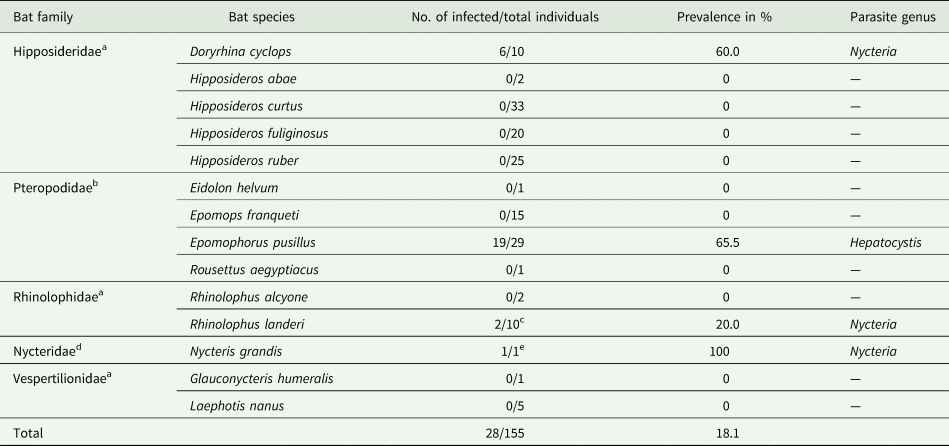
a Insectivorous bats.
b Frugivorous bats.
c Infection in 1/2 samples was only confirmed by microscopy, no DNA available.
d Mainly insectivorous.
e Infection only confirmed by microscopy, no DNA available.
Haemosporidian parasites were detected in four of the five bat families: Pteropodidae, Hipposideridae, Nycteridae and Rhinolophidae. Morphology of the parasitic blood stages (gametocytes), and molecular analysis identified Hepatocystis parasites in Epomophorus pusillus (formerly Micropteropus pusillus) (Pteropodidae) with a high prevalence of 65.5% (19/29). Nycteria parasites were identified in Doryrhina cyclops (formerly Hipposideros cyclops) (Hipposideridae) with a high prevalence of 60.0% (6/10), in Rhinolophus cf. landeri (Rhinolophidae) with a lower prevalence of 20.0% (2/10) and a single Nycteris grandis (1/1). The bat species that comprised Nycteria-infected individuals were exclusively sampled in the forest habitat during the wet season (Supplementary Table S3). Interestingly, none of the E. pusillus individuals that were sampled in the forest during the wet season did feature infections with Hepatocystis parasites (only one individual that was captured in the dry season was infected). However, all 18 E. pusillus that were sampled in the farmland habitat during the wet season were infected with Hepatocystis (Supplementary Table S3). Notably, no haemosporidian infection was observed in the savanna zones during our sampling.
For bats with confirmed haemosporidian infection, parasitaemia values (% of infected erythrocytes) were calculated (Fig. 2C, Table 2, Supplementary Table S4). Mean parasitaemia values in Hepatocystis-infected bats (n = 19) were 0.11 ± 0.10, with maximum values of 0.37% and minimum values of 0.02% (Table 2). Parasite loads in Nycteria-infected D. cyclops individuals (n = 6) ranged between 0.05 and 1.02% (mean = 0.23% ± 0.39). One Rhinolophus sp. individual exhibited 0.71% and the other 0.07% parasitaemia. The single N. grandis individual featured a high parasitaemia of 0.21%.
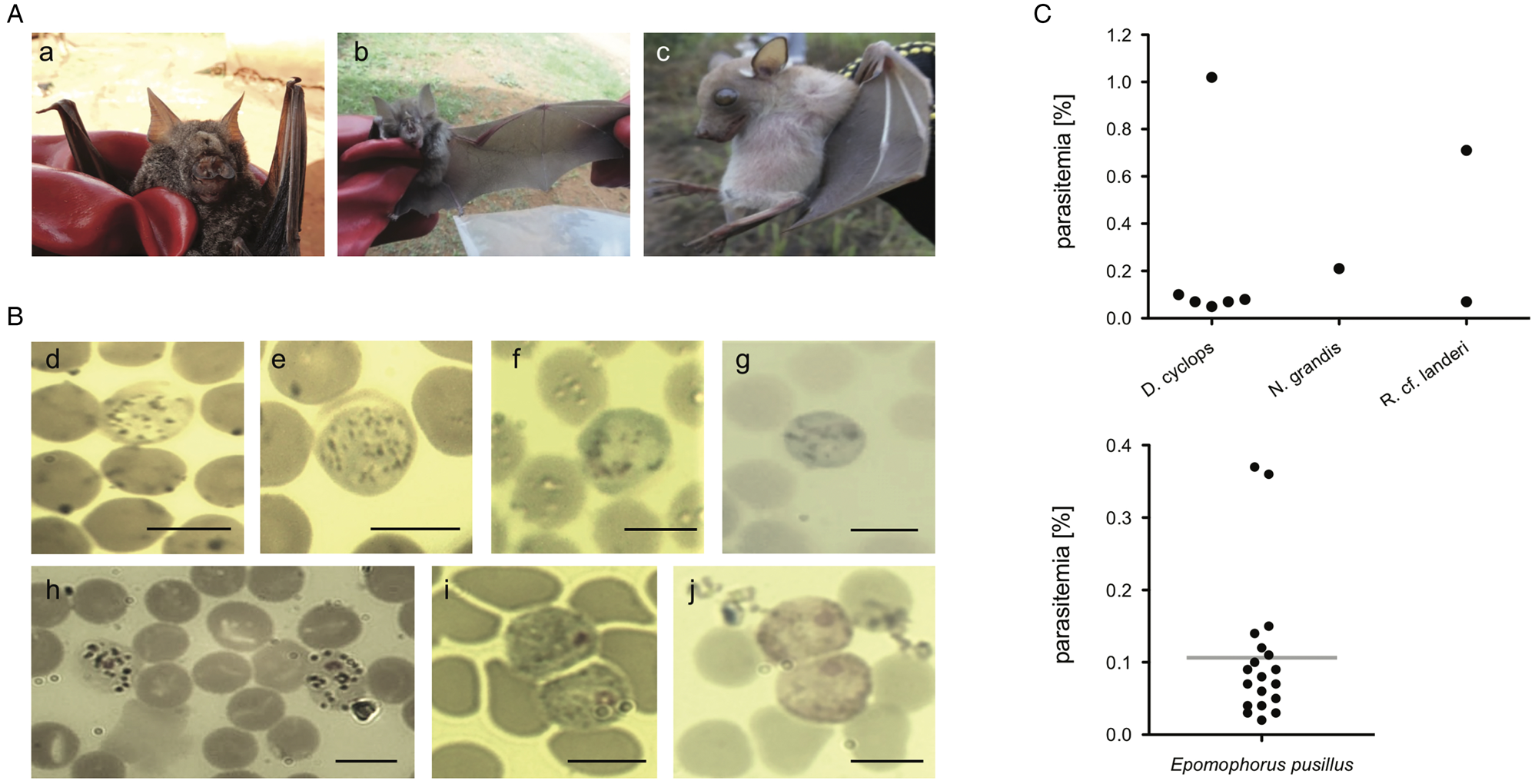
Fig. 2. (A) Pictures of bat host species of the study (a) D. cyclops; (b) R. cf. landeri; (c) E. pusillus; (B) Representative micrographs of Giemsa-stained thin blood films of mature gametocytes of Nycteria parasites: (d, e) gametocytes ex Doryrhina cyclops; (f, g) ex R. landeri; (h) ex Nycteris grandis; (i) representative micrographs of mature macrogametocytes and (j) microgametocytes of Hepatocystis parasites of E. pusillus. Bars = 5 μm. (C) Parasitaemia per bat individual in percent: (k) parasitaemia values of Nycteria and (l) Hepatocystis parasitaemia with a mean of 0.11% (± 0.10), maximum values of 0.37% and minimum values of 0.02% in the wet season.
Table 2. Parasitaemia values of haemosporidian infection for infected bat families.
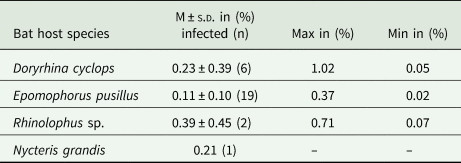
M, mean values of parasitemia; s.d., standard deviation.
Genotyping of infected bat species
The morphological bat species identifications for E. pusillus, D. cyclops and R. landeri were confirmed with molecular barcoding. A part of the mitochondrial cytochrome b (500 bp) was sequenced for four E. pusillus individuals, which featured highest nucleotide identity of 99.4% with an E. pusillus (M. pusillus) reference (KX822887) and 99.4% with an Epomophorus gambianus reference sequence (JF728753). This was followed by sequencing 650 bp of fgb, which resulted in a nucleotide identity of 100% with the E. pusillus reference (JF728439). The nuclear introns acox2 (500 bp) and rogdi (390 bp) were sequenced for three and two individuals of D. cyclops, respectively. The acox2 sequences featured a 100% nucleotide identity with both Doryrhina cf. camerunensis (e.g. MT149618) and D. cyclops (e.g. MT149621). The rogdi sequences featured a 98.5% (e.g. FJ85201) nucleotide identity with both Doryrhina cf. camerunensis (e.g. MT149421) and D. cyclops (e.g. MT149423). However, the morphological identification unambiguously identified the individuals of the study as belonging to the species D. cyclops (mean forearm length = 65 mm; total length = 109 mm) (Happold and Happold, Reference Happold and Happold2013). A part of the mitochondrial cytochrome b (500 bp) was sequenced for one R. landeri individual, which featured highest nucleotide identity of 94.1% with a R. landeri reference sequence (FJ185201), which could mean, according to the identity scores, that the R. landeri samples of the study might belong to another Rhinolophus species.
Morphological and phylogenetic characterization of the haemosporidian parasites
Morphology of gametocyte stages of the Nycteria parasites clearly differed between the different host genera (Fig. 2Bd–h). The mature gametocytes of Nycteria parasites of D. cyclops in the current study resemble the morphology of the Nycteria gametocytes described from some rhinolophid hosts (e.g. Krampitz and de Faveaux, Reference Krampitz and de Faveaux1960; Rosin et al., Reference Rosin, Landau and Hugot1978; Schaer et al., Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015). The gametocytes feature no apparent nuclear area and a distinction between macro- and microgametocytes was not possible in these samples. The cytoplasm stains light blue with fine haemozoin pigment distributed throughout the parasite cell, which does not fill the entire erythrocyte and its round margin is clearly visible (Fig. 2Bd, e). The gametocytes observed in our samples do slightly also resemble the gametocyte morphology that was reported from the same host species, with the exception that we did not detect distinct nuclei (Lutz et al., Reference Lutz, Patterson, Kerbis Peterhans, Stanley, Webala, Gnoske, Hackett and Stanhope2016). The bad quality of the stained blood smears of the Nycteria parasites of R. landeri did not allow a proper morphological investigation (Fig. 2Bf, g). The mature gametocytes of Nycteria parasites of N. grandis featured distinct nuclei with dense chromatin. The dark haemozoin pigment distributed in the cytoplasm had a granular appearance (Fig. 2Bh). Morphology of the gametocytes resembled the reported gametocyte morphology from the same host species in Schaer et al. (Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015), but could not be assigned to known morphospecies.
Phylogenetic analyses were performed to assess the phylogenetic diversity, and possible geographic and/or host-specificity patterns of haemosporidian parasites (Hepatocystis and Nycteria) from Cameroon. The multi-gene phylogeny of Nycteria parasites (two mitochondrial, two nuclear and one apicoplast gene) recovered the Nycteria clade as sister to a clade that contains the lizard and bird Plasmodium species as shown before (Fig. 3) (e.g. Schaer et al., Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015). However, other studies, including the most comprehensive haemosporidian phylogeny, which was based on several nuclear markers, placed Nycteria as basal clade to the mammalian Plasmodium/Hepatocystis parasites (e.g. Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013; Galen et al., Reference Galen, Borner, Martinsen, Schaer, Austin, West and Perkins2018). Representative reference sequences of Nycteria parasites of diverse bat families from Asia and Africa were included in the analysis. The Nycteria sequences of D. cyclops of the current study are grouped closely with Nycteria parasites of D. cyclops from Uganda in one monophyletic clade (Fig. 3). Nycteria parasites of African Rhinolophus host species including the parasite sequence of Rhinolophus sp. from Cameroon also group in their own monophyletic host bat family-specific Nycteria clade that contains sequences from the two species Nycteria cf. gabonensis and Nycteria cf. congolensis (Schaer et al., Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015; Rosskopf et al., Reference Rosskopf, Held, Gmeiner, Mordmüller, Matsiégui, Eckerle, Weber, Matuschewski and Schaer2019). Therefore, the parasites from R. cf. landeri of the current study could represent either N. cf. gabonensis or N. cf. congolensis. The third Nycteria subclade contains parasite sequences from different host bat families in Asia (Pteropodidae, Megadermatidae, Nycteridae and Craseonycteridae). The placement of Nycteria of the Nycteris hosts within this clade is uncertain as no reference sequences are available for cytb, cox1, asl and ef2 for these parasites and thus the concatenated sequences contain a large amount of missing data. Despite using different protocols to amplify and sequence the apicoplast clpC marker, no clpC sequences were successfully generated for the Nycteria parasites of the study. However, the phylogenetic analysis was run with a concatenated dataset that included clpC from reference sequences to analyse the placement of Nycteria parasites from Nycteris hosts (Fig. 3). We also ran a dataset without the clpC data, which resulted in the same tree topology (Supplementary Fig. S1).
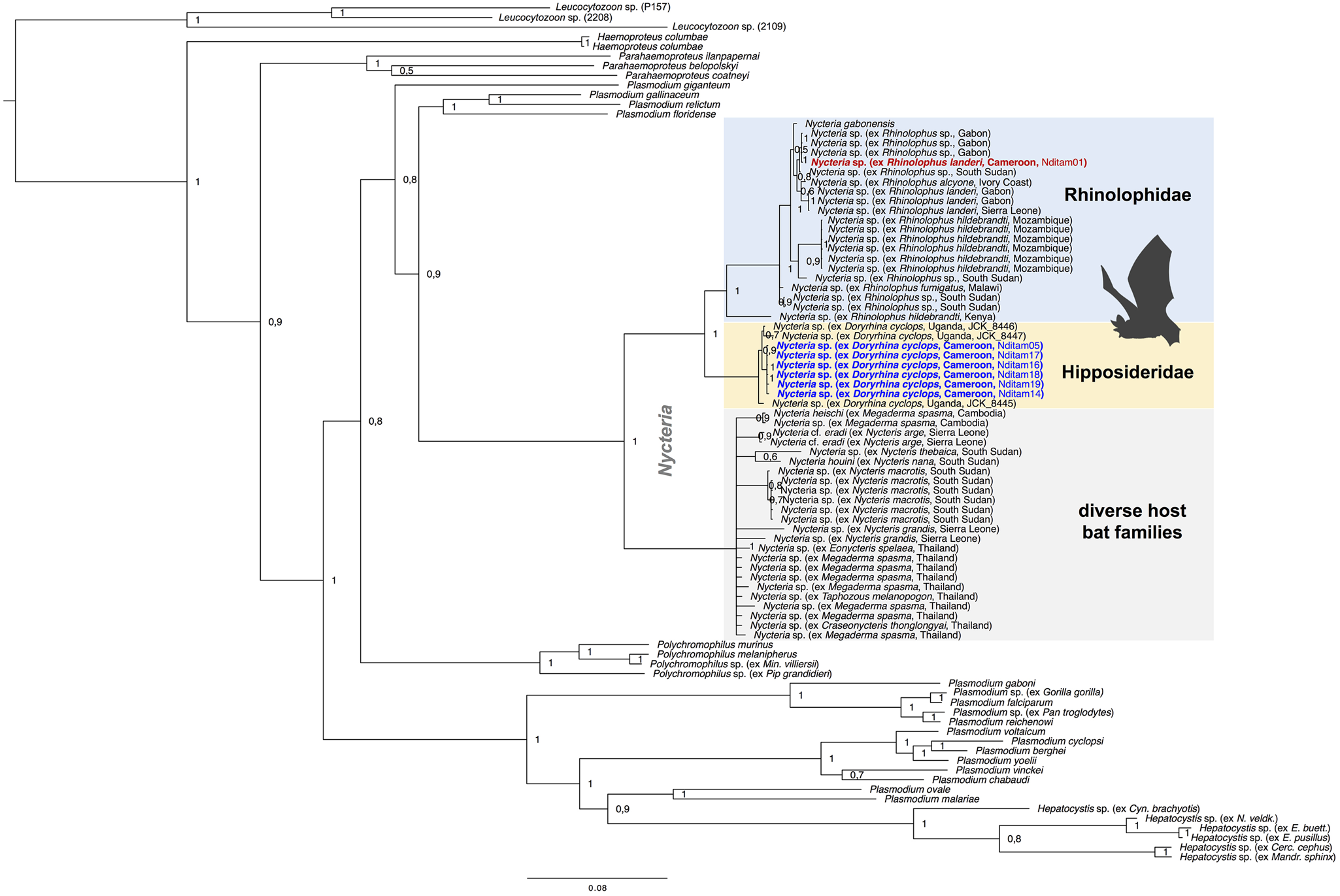
Fig. 3. Multi-gene phylogeny of Nycteria parasites in the context of the major haemosporidian parasite clades recovered by Bayesian analysis. Posterior probabilities values are given. The analysis is based on the concatenated alignment (total of 3243 bp) of the mitochondrial genes cytb (1119 bp) and cox1 (861 bp), the apicoplast marker clpC (528 bp) and the nuclear ef2-gene (513 bp) and asl-gene (222 bp). Placement of Nycteria parasites as sister to a clade that contains the lizard and bird Plasmodium species. The monophyletic Nycteria clade contains representative reference sequences of Nycteria parasites of diverse bat families from Asia and Africa. The samples of the current study are highlighted in bold blue (ex Doryrhina cyclops) and bold red (ex R. landeri). Nycteria parasites of African Rhinolophus host species (blue clade) and the African hipposiderid Doryrhina hosts (yellow clade) group in their own host bat family specific clades. The grey clade contains Nycteria sequences from different host bat families in Asia (Pteropodidae, Megadermatidae, Nycteridae and Craseonycteridae).
The morphology of the gametocyte blood stages of the Hepatocystis parasites of E. pusillus in Cameroon corresponds to the descriptions of Hepatocystis epomophori parasites of epauletted fruit bats from other African countries (Garnham, Reference Garnham1966; Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013, Reference Schaer, Perkins, Ejotre, Vodzak, Matuschewski and Reeder2017) (Fig. 2B). The sexually dimorphic macro- and microgametocytes were distinguished after Giemsa staining, the macrogametocytes stained blue, with a red staining nucleus of smaller size (in comparison with the nucleus of the microgametocyte) (Fig. 2Bi). The microgametocytes featured pale brown cytoplasm with evenly distributed haemozoin except for the nuclear zone (Fig. 2Bj). Bayesian analyses confirmed Hepatocystis as a monophyletic clade with high support (posterior probability of 1) and as sister clade to a clade that comprises the Plasmodium (Plasmodium) and Plasmodium (Vinckeia) parasites (Fig. 4). The analysis also recovered all major clades [Leucocytozoon, Haemoproteus, Parahaemoproteus, Polychromophilus, Plasmodium (Plasmodium), Plasmodium (Vinckeia), Plasmodium (Laverania)] as monophyletic. The Hepatocystis clade is comprised of two main subclades, the first including all parasites of Asian and African primates, the second main group contains African chiropteran Hepatocystis parasites including the Hepatocystis parasites of the study (Fig. 4B). Within the African bat Hepatocystis clade, parasite sequences do not cluster in host genus or species-specific clades but represent several close related taxa or cryptic species as shown before (Schaer et al., Reference Schaer, Perkins, Ejotre, Vodzak, Matuschewski and Reeder2017). Further, sequences from different countries and locations in both West Africa and Central-/East Africa fall in several different places across the tree, showing no pattern of clustering according to countries. The sequences of E. pusillus from Central Cameroon group closely together, confirming high sequence similarities between each other. However, together they group with West and Central-/East African Hepatocystis parasites of E. pusillus and other epauletted fruit bat species on a polytomous branch, with whom they also share very close phylogenetic relationships.
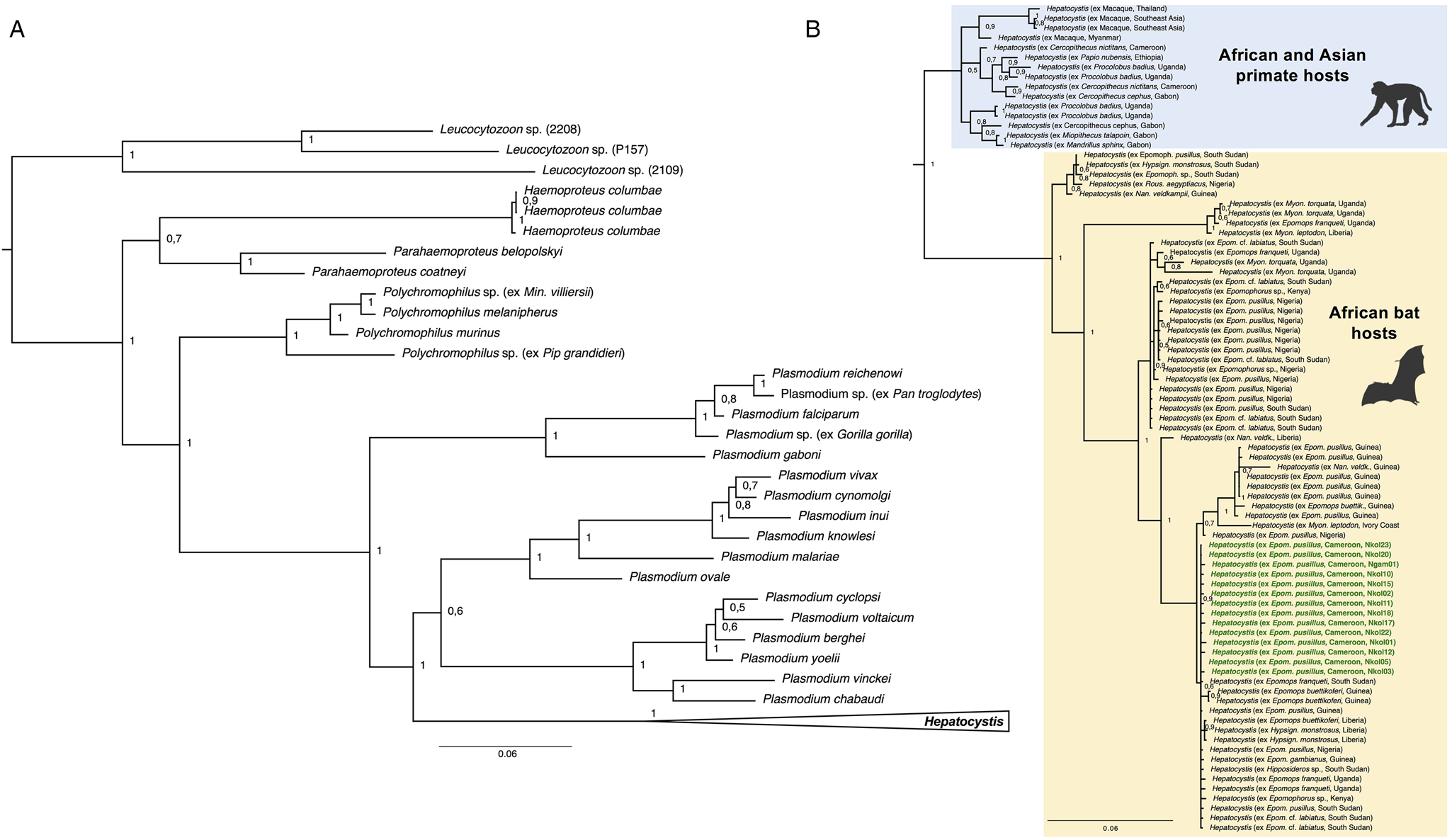
Fig. 4. (A) Phylogenetic analysis of Hepatocystis parasites in the context of some major haemosporidian clades recovered by Bayesian analysis. Posterior probabilities are given. The analysis is based on the concatenated alignment (total of 1983 bp) of the mitochondrial cytochrome b (531 bp) and cytochrome oxidase 1 (993 bp) and the nuclear elongation factor 2 (513 bp). (B) Placement of Hepatocystis as collapsed clade as sister clade to the mammalian Plasmodium (Plasmodium) and Plasmodium (Vinckeia) clade. (B) The uncollapsed clade contains representative reference sequences of Hepatocystis of primate hosts from Asia and Africa as well as African bat hosts. The samples of the current study are highlighted in bold green. Sequences of the Hepatocystis parasites from E. pusillus bats in Cameroon closely group within the Hepatocystis parasite clades of epauletted fruit bat species from different African countries and no host species specificity is apparent.
Discussion
This study provides the first information on haemosporidian parasite infections in bats in Cameroon. Bats are one of the most species-rich groups of mammals in the Central region of Cameroon in the equatorial tropical area, that features different habitat types (forest, savanna and cultivated farms) (Atagana et al., Reference Atagana, Bakwo Fils, Mbeng, Tsague and Sévilor2018; Mbeng et al., Reference Mbeng, Bakwo Fils, Atagana, Tsague and Joseph2019). Haemosporidian parasite infections were identified in 28 out of 155 investigated bats, belonging to four different bat families. Hepatocystis parasites were discovered in E. pusillus with a high prevalence of 65.5%, which is in line with previous reports from Hepatocystis infections in epauletted fruit bats in other African countries (e.g. Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013, 2017; Lutz et al., Reference Lutz, Patterson, Kerbis Peterhans, Stanley, Webala, Gnoske, Hackett and Stanhope2016; Boundenga et al., Reference Boundenga, Ngoubangoye, Mombo, Tsoubmou, Renaud, Rougeron and Prugnolle2018; Atama et al., Reference Atama, Manu, Ivande, Rosskopf, Matuschewski and Schaer2019). No infections were detected in the other sampled epauletted fruit bat species, E. franqueti, which has also been recorded as a host of Hepatocystis in previous studies (e.g. Lutz et al., Reference Lutz, Patterson, Kerbis Peterhans, Stanley, Webala, Gnoske, Hackett and Stanhope2016; Schaer et al., Reference Schaer, Perkins, Ejotre, Vodzak, Matuschewski and Reeder2017). All E. franqueti were captured in the forest habitat in the wet season, along with nine E. pusillus that were also uninfected. Strikingly, all 18 E. pusillus individuals that were investigated in the cultured farmland habitat during the wet season were infected with Hepatocystis, which points to high transmission rates in the cultured habitat due to a potentially high abundance of the invertebrate vector in the farmland in contrast to the forest habitat.
The parasites of E. pusillus from Central Cameroon might belong to one of the diverse lineages of the Hepatocystis epomophori species complex as the parasites feature a close relationship with Hepatocystis parasites of diverse fruit bat host species from West and Central-/East Africa, which comprise parasites that have tentatively been assigned to the morphospecies H. epomophori (Schaer et al., Reference Schaer, Perkins, Ejotre, Vodzak, Matuschewski and Reeder2017). Diverse assemblages of cryptic species have also been detected in Hepatocystis parasites of primate hosts (Thurber et al., Reference Thurber, Ghai, Hyeroba, Weny, Tumukunde, Chapman, Wiseman, Dinis, Steeil, Greiner, Friedrich, O'Connor and Goldberg2013).
Nycteria parasites were discovered in D. cyclops with a high prevalence of 60.0%. This is only the second time that this parasite genus has been reported from this host species and again, with a high prevalence (Lutz et al., reported three out of three infected D. cyclops in Uganda) (Lutz et al., Reference Lutz, Patterson, Kerbis Peterhans, Stanley, Webala, Gnoske, Hackett and Stanhope2016). Despite the geographical distance, our phylogenetic analysis recovered a very close relationship between the Nycteria parasites of D. cyclops from Uganda and Cameroon, with no nucleotide differences in the partial cytb sequences and less than 1% sequence divergence for the partial cox1 gene sequences. No Nycteria morphospecies has been described from D. cyclops and based on the distinct monophyletic phylogenetic relationship of the parasite sequences of D. cyclops, these parasites might represent a distinct/new Nycteria species, which is host species specific to D. cyclops with a corresponding distribution range in Africa. Interestingly, the bat species D. cyclops has also been identified as host species of another haemosporidian species, Plasmodium cyclopsi (Landau and Chabaud, Reference Landau and Chabaud1978; Schaer et al., Reference Schaer, Perkins, Decher, Leendertz, Fahr, Weber and Matuschewski2013). In both cases, Nycteria and P. cyclopsi, the identity of the dipteran vector species and even family is unknown. Nycteria parasites were also identified in one of two investigated Rhinolophus cf. landeri individuals from Cameroon, a host species that has been repeatedly reported as host of Nycteria parasites in several other African countries, which renders R. cf. landeri as common host of Nycteria (e.g. Krampitz and de Faveaux, Reference Krampitz and de Faveaux1960; Schaer et al., Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015; Rosskopf et al., Reference Rosskopf, Held, Gmeiner, Mordmüller, Matsiégui, Eckerle, Weber, Matuschewski and Schaer2019). The study confirms a specificity of Nycteria parasites in regard to their bat host families for the parasites of rhinolophid and hipposiderid hosts (e.g. Schaer et al., Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015; Lutz et al., Reference Lutz, Patterson, Kerbis Peterhans, Stanley, Webala, Gnoske, Hackett and Stanhope2016). A previous study found Nycteria parasites of the Nycteris hosts also to be monophyletic (Schaer et al., Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015). Our phylogenetic analysis recovered the Nycteria parasites of African Nycteris hosts within a clade that comprises parasite sequences from the different host bat families Pteropodidae, Megadermatidae, Emballonuridae and Craseonycteridae from Asia. However, this placement must be considered with caution, as for most of the Asian parasites the molecular data are limited to short reference sequences of the cytb gene (e.g. Arnuphapprasert et al., Reference Arnuphapprasert, Riana, Ngamprasertwong, Wangthongchaicharoen, Soisook, Thanee, Bhodhibundit and Kaewthamasorn2020). Further, for most parasites of Nycteris hosts, reference sequences for cytb and cox1 (as well as for asl and ef2) are missing and thus the concatenated sequences contain a large amount of missing data (Schaer et al., Reference Schaer, Reeder, Vodzak, Olival, Weber, Mayer, Matuschewski and Perkins2015). Additionally, a previous study has shown that the genes in the mitochondrial genome of Nycteria parasites of Nycteris hosts are rearranged and thus the mitochondrial genome differs from that of other Nycteria parasites (of other host genera) and to that of all other haemosporidian genera (Karadjian et al., Reference Karadjian, Hassanin, Saintpierre, Gembu Tungaluna, Ariey, Ayala, Landau and Duval2016).
In conclusion, our survey of haemosporidian parasites in bats in Central Cameroon shows that Hepatocystis infections in African epauletted fruit bats varies among bat host species and also between populations of the same bat species in different geographic locations. A high prevalence was confirmed in E. pusillus on the one hand, but infections were lacking in E. franqueti on the other hand. Habitat type and seasonality may play a role in transmission of Hepatocystis parasites on a local scale. This study also adds important information on the distribution and host specificity of the neglected haemosporidian genus Nycteria.
Cameroon is known as ‘Africa in miniature’ as it mirrors the continent´s diverse habitats and biodiversity. Future systematic sampling and longitudinal studies across the country with its diverse bat fauna are needed to assess the full diversity of haemosporidian parasites in bats in Cameroon, to study the transmission biology of haemosporidian parasites, their effects on the bat hosts and to identify their insect hosts.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182021001542
Data
All sequences of the study are available at GenBank (NCBI) with the accession numbers MZ451636-MZ451644, MZ460913-MZ460953, MZ460988-MZ460992.
Acknowledgements
We thank François Nguetsop for the coaching in microscopic work, as well as Lara Palm and Olga Wawrzynczak for helping with the molecular work in the laboratory.
Author contributions
KT and TT conceived and designed the study. KT, DM, VN, JA and EF carried out field work and bat sampling. JS performed molecular work and phylogenetic analysis. All authors conducted data gathering and wrote the article.
Financial support
The field work was supported by the special funds from the Cameroon government allocated for the modernization of research in the state Universities (T. Tchuinkam and EM Bakwo-Fils); through the Presidential Decret No: 2009/121 of 08 April 2009. JS is funded by an individual research grant from the German Research Foundation (DFG; project number 437846632).
Conflict of interest
The authors declare there are no conflicts of interest.
Ethical standards
All surveys were reviewed and approved by the authorization from the Ministry of scientific Research and Innovation under permit No 0000039/MINRESI/BOO/COO/C10/C11 from 16 February 2016. All work was performed in accordance with the relevant guidelines and regulations regarding care and use of animals.