1. Introduction
Biological organisms use complex information processing for their survival (Figure 1). This information processing can be interpreted as biological computing (definitions of bold terms can be found in the glossary). Organisms sense their environment as well as internal state to shape their cells and bodies, to adapt their growth and behaviour and to communicate with others. To study the mechanisms and adaptive relevance of these processes, one can study the molecular underpinnings, costs and occurrence of these processes. As a complementary approach, one can also focus on the nature of the computations biological systems perform, drawing on concepts and results from fields such as computer science and information theory.
When we view a cell, tissue, organism, colony or ecosystem as an information processing or computing system, it can fall anywhere on a continuous sliding scale between centralised and distributed computing. Some biological processes can be viewed as controlled by fully centralised computing units, such as the behaviour of higher animals with a specialised computing organ: the brain (Figure 2, left). Others may be better suited to a distributed computing representation, such as a flock of birds or the individual roots in a complex branched root system (Figure 2, right). Yet others may occupy the intermediate niche between centralised and distributed computing, such as a group of animals with dominant individuals, or the arms and brain(s) of an octopus (Figure 2, middle). Centralised computing implies all information is gathered and integrated at a single location, simplifying optimisation and decision-making processes, yet coming at the cost of vulnerability to failure of this single computing location and non-trivial upscaling to more complex problems. In contrast, when information processing is distributed, vulnerability is reduced as risks are spread out over multiple units while upscaling can simply be achieved through adding individual computing units. Here, the complexity lies in how system wide information is generated and how individual entities integrate their local with this system wide information to achieve optimal decision-making in the face of noisy communication and environmental variability.

Fig. 1. Biological computing. All biological organisms, be it primates, snails, plants or single-celled amoebas, sense their environment and internal state, and process, integrate and prioritise this information to compute which action to take next. Figure inspired on earlier work in Scheres and van der Putten (Reference Scheres and van der Putten2017).

Fig. 2. Different spatial organisations of information processing in biological systems. Left: centralised information processing based on a central dominant brain in primates (Image source chimpanzees: https://sciencenorway.no, photographer Etsuko Nogami). Middle: hybrid information processing in an octopus that combines a central brain with semi-autonomous information processing in its arms. (Image source octopus: Octolab TV https://octolab.tv) Right: fully distributed information processing with no central processing organ in plants (Image source plant: New Jersey Agricultural Society Learning Through Gardening program) (Image computer: https://computerkiezen.nl/computer-soorten/desktop-computer/).
A set of computations or information processing steps, be it in a computer or a biological system, can be considered as an algorithm. If we want to understand how a cell, organism or colony makes a decision, we need to decipher the underlying algorithm. At first glance, knowing this algorithm may seem sufficient to fully understand the information processing. However, in reality the physical context in which the algorithm is deployed is essential too (Feinerman & Korman, Reference Feinerman and Korman2013). The context sets limits in which the algorithm has to operate, such as network topology or available memory, but also ecological and evolutionary constraints (Feinerman & Korman, Reference Feinerman and Korman2013; Gordon, Reference Gordon2016).
2. Glossary

Biological systems show embodiment in a myriad of ways. As a striking example, ants memorise views along a foraging route (Wystrach, Reference Wystrach2021). On their way back to the nest, they rotate their body to match their current view with their memories and go forward whenever the image is familiar. Since they can barely rotate their head, the view is familiar precisely when they have headed this way before. This allows these ants to use an extremely simple algorithm and low-resolution images to find their way, in contrast to man-made robots that usually require complicated calculations on high-resolution visual data.
Table 1 Comparative overview of embodiment aspects for plants and ant colonies discussed in this work

A prime facet of embodiment in distributed systems is whether the involved units in this network occupy a constant position or can move relative to one another, properties which we refer to as solid versus liquid brains (Solé et al., Reference Solé, Moses and Forrest2019). Since ants can move, but plant cells cannot, this affects which cells or ants can have direct connections with each other. Solid brains consist of sessile computing units. Hence, the set of potential connections between units is constant and defined by topology. Still, the solid brain network can tune which connections are actually established, and at which strength. Key mechanisms of tuning connection strengths in plants are, for example, the number and diameter of vascular connections between plant organs and of plasmodesmata between cells, as well as the modulation of plant hormone transport. Examples of solid brains are human brains, artificial neural networks and plants (Piñero & Solé, Reference Piñero and Solé2019; Solé et al., Reference Solé, Moses and Forrest2019). Liquid brains are computing networks with moving units, such as colonies of ants, bees, wasps or termites Piñero & Solé, Reference Piñero and Solé2019; Solé et al., Reference Solé, Moses and Forrest2019; Theraulaz & Bonabeau, Reference Theraulaz and Bonabeau1995). Because their units can move, the liquid brain network of potential connections can change over time and is instead constrained by the space the ants or termites can move in. This type of network is not limited to the social insects: other examples are swarm robots (Vining et al., Reference Vining, Esponda, Moses and Forrest2019) and the immune system (Piñero & Solé, Reference Piñero and Solé2019).
Here, we are interested in the effect of embodiment on distributed computing in plants. We may expect the primarily sessile nature of plants to play a key role in their embodiment and thus the nature of their computations: the cell wall of plant cells is rigid and prevents movement of individual cells, leaves and roots do not slide around on the stem, and once a plant has germinated, it will remain at the same position in the soil until it dies. Still, as an individual plant grows, the orientation of existing organs such as leaves and more importantly the formation of new organs, attached to existing body parts, enables it to explore new territory for light, nutrients or water. Similarly, at a slightly larger scale in clonally reproducing plants, the production of new ramets (individual plant bodies) enables exploration of a wider area, with resources subsequently shared between ramets (Oborny, Reference Oborny2019). Distribution disconnected from, and well beyond, the original plant body occurs mostly through plant sexual reproduction, that is, pollen and seed dispersal.
To discover the effects of embodiment on plant algorithms, here we will compare plant computational processes with corresponding processes in ant colonies. The comparison with ant colonies is chosen for three reasons. Firstly, in both plants and ant colonies computing is spatially distributed. Thus, in both systems the problem of generating and storing system wide information, and integration of local and system wide information in the light of noise and environmental changes need to be solved. Secondly, computational processes in ant colonies are well studied. Finally, ant colonies represent the other extreme on the solid-to-liquid brain computation axis. As a consequence, while the information processing challenges are similar, the possibilities for and constraints on how to solve these are different. We will consider different aspects of embodiment and how these interact with the distributed information processing in these organisms (summarised in Table 1). Our comparison of solid and liquid brain types of embodiment demonstrates differences in whether it is the messenger or rather the message that travels, and whether spatial information can be stored within the system or requires storage in the environment. In addition to differences, similarities are shown to exist in the trade-off between construction costs and signal speed that constrains the formation of substrates for long-distance computations.
3. Facets of embodiment
3.1. How does system wide information arise from local communication?
Plants and ant colonies can only perform distributed computations if their cells or ants can communicate. This communication is essential to transport information as well as to combine each individual outcome into an overall conclusion. Both cells within a plant and ants within a colony communicate with others close to them.
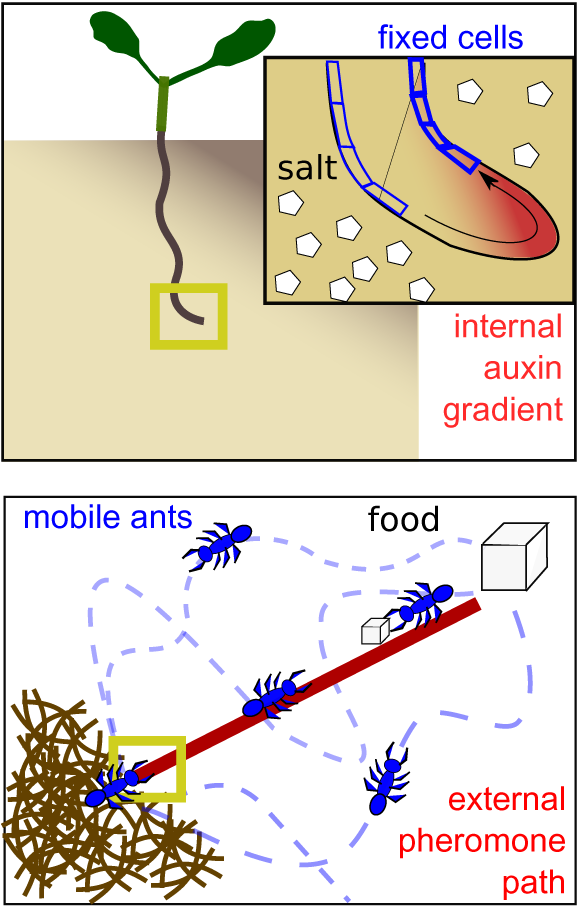
The solid organisation of plant tissue affects the types of signalling needed, specifically the need to coordinate with neighbours’ growth, expansion and differentiation decisions, and the need to transport information about the plant environment from external to internal tissues. In addition to shaping the types of communication needed, it also forms a substrate on which to perform computations and store information. As an example, in halo-tropism, plant roots are able to sense noisy, shallow (5–10% difference) salinity gradients, growing away from the direction with highest salt concentration. Initially, salt induces small cell-level changes in auxin concentration that differ between cells at the left and right side of the root. Through a specific layout of auxin importers and exporters, neighbouring cells exchange and integrate these auxin signals, with a positive feedback of auxin on importers enabling amplification of the initial weak signal into a robust auxin asymmetry dictating asymmetric growth and root bending. Thus, the direction of the salinity gradient is sensed, integrated and computed through modulating auxin transport and stored on the root tip tissue architecture (Figure 3, top) (van den Berg et al., Reference van den Berg, Korver, Testerink and Ten Tusscher2016). Hence, it is the adaptability of connection strengths combined with the stability of connection layout in solid brains that enables complex computations such as pattern recognition (Vining et al., Reference Vining, Esponda, Moses and Forrest2019).

Fig. 3. Storage of spatial environmental information. Top: in the solid brain setting of a plant root, environmental information regarding the direction of a salt gradient can be stored internally in the shape of an auxin asymmetry. Bottom: in the liquid brain setting of an ant colony, environmental information regarding the direction and shortest path towards a food source is stored externally through a pheromone trail.
On a similar note, based on the similarity in dynamics of cellular automata (CA) models performing density computations and plant leaf stomatal aperture behaviour, it has been suggested that stomata compute the optimal fraction of open stomata together (Peak et al., Reference Peak, West, Messinger and Mott2004. The likeness to CA models, in which patterns arise from local interactions only, further suggests that each stoma uses the humidity it senses itself and the stomatal aperture of its nearby neighbours to decide to open or close. The results of these local-information computations are waves of opened stomata travelling across the leaf, akin to the avalanches of activity occurring in self-organised critical systems (Per Bak, 1996: How Nature Works: The Science of Self-Organised Criticality). These waves make relevant information available across the entire surface. For this information processing, the embodiment in the leaf allows the stomata to sense input, do the calculation and implement the result all at once.
Finally, through controlling cell shape and division planes, the precise network of intercellular connections and hence the speed versus reliability of computation can be impacted even within the constraints of a solid plant tissue. Due to their compact size and fixed position, plant cell intercellular contacts are limited to their nearest neighbours, causing plant cellular networks to have a low degree as compared to, for example, the neurons in an animal brain (Bassel, Reference Bassel2018; Duran-Nebreda & Bassel, Reference Duran-Nebreda and Bassel2019). This low degree enhances robustness against errors or death of individual cells, yet limits information transfer speed. The cellular network in the plants apical meristem is even more uniformly connected, and thus robust, than we would expect a priori (Jackson et al., Reference Jackson2019). This organ-level uniformity in connectivity arises from local rules. Firstly, highly connected cells are likelier to divide. Moreover, the new cell wall is placed so that its length is minimised, which corresponds to minimising cell degree. Since these meristems are relatively small organs, signalling speed is likely not limiting, enabling a focus on robustness over connectivity through promoting this uniform connectedness level.
Whilst solid brains thus can use spatial structure to represent the outcomes of partial calculations, the mixing of units destroys such helpful spatial distributions in liquid brains (Vining et al., Reference Vining, Esponda, Moses and Forrest2019), necessitating other forms of information processing. As an example, ants convey the task they are performing to ants they meet using smell and physical contact (Gordon, Reference Gordon2016; Piñero & Solé, Reference Piñero and Solé2019). This enables nestmates to estimate the percentage of ants working on a particular task and the implications this has for environmental conditions, and decide between continuing their current task and switching to another task. For example, if a brood-care-worker meets a forager, its estimation of the percentage of foragers increases. Since this may indicate the presence of a rich food source, this brood-worker may then decide to switch to foraging.
Ants can combine this local between-ant information exchange with extensive movement and hence mixing (Vining et al., Reference Vining, Esponda, Moses and Forrest2019). This allows them to communicate directly with many others, and automatically spread and integrate relevant information simply by moving from one neighbourhood to the other. This type of information processing can easily scale up to larger areas and colonies without the need for alternative means of information transfer and processing. Indeed Vining et al. (Reference Vining, Esponda, Moses and Forrest2019) show that walking around enables ant colonies and other liquid brains to solve a more diverse set of consensus problems than plants and other solid brains. They are also able to solve these faster, and their movement patterns can be an essential part of their algorithm.
3.2. How does long-distance communication contribute to information integration?
To enhance signalling speed and reliability, plants and ant colonies use vascular systems and pheromone trails, respectively, to enable communication between not directly neighbouring units. Interestingly, the formation of these information transfer networks is themselves a result of developmental computations, giving rise to a ‘nested distributed computation’. As plant tissues grow, vascular systems develop to support efficient long range transport of water, nutrients and signalling molecules (Conn et al., Reference Conn, Pedmale, Chory and Navlakha2017; Duran-Nebreda et al., Reference Duran-Nebreda, Johnston and Bassel2020). However, since vascular cells do not contribute to primary organ function (e.g., photosynthesis in leaves and water and nutrient uptake in roots), they are also a costly investment (Conn et al., Reference Conn, Pedmale, Chory and Navlakha2017; Duran-Nebreda et al., Reference Duran-Nebreda, Johnston and Bassel2020). Nutrient travel time in tissues depends on both the fraction of vascular tissue and the organ dimensionality (flat or 2D, leaf-like or 2.5D, or fully 3D), and investments in vasculature were demonstrated to pay off only beyond 2D (Duran-Nebreda et al., Reference Duran-Nebreda, Johnston and Bassel2020). In their stems, plants lie along the Pareto front between total vascular branch length and nutrient transport distance, implying optimisation of this trade-off (Conn et al., Reference Conn, Pedmale, Chory and Navlakha2017). Still, different species find different optimal balances (Conn et al., Reference Conn, Pedmale, Chory and Navlakha2017).
To reverse engineer the connectivity rules that operate during vascular network development, Duran-Nebreda et al. (Reference Duran-Nebreda, Johnston and Bassel2020) simulated this in silico. The model works by fusing those nodes (i.e., connecting those cells) in the in silico tissue that reduce across network path length, as an analogy for phloem cells joining together with a sieve plate. Node fusion was performed taking either closeness or betweenness centrality as a measure for minimising average path length. Betweenness centrality was found to perform better than closeness centrality in terms of minimising network construction while maximising transport speed. In other words, the best candidates for vascular cells are not those in the centre of the network, but rather those which transport many molecules. We may therefore expect an internally produced diffusible molecule to drive the selection of vascular cells. Indeed, in leaves, vascular development starts with the main veins (midvein or main parallel veins) growing from the leaf base to its tip, after which more minor veins branch off these major veins (Katifori, Reference Katifori2018). This patterning corresponds to a betweenness centrality-based algorithm, with major vein allowing nearby cells to transport more goods, thereby increasing their chances of becoming a minor vein cell. The plant hormone auxin is a primary candidate for driving this selection of vascular cells (Katifori, Reference Katifori2018). Indeed in models for the prepatterning of leaf venation a phenomenological with-the-flux feedback of auxin on auxin transport rate has been demonstrated to recapitulate experimental observations (see, e.g., Fujita & Mochizuki, Reference Fujita and Mochizuki2006; Stoma et al., Reference Stoma2008).
Many species of ants create their own transport and information processing networks using pheromone trails. These trails are created through foragers that leave the nest in search of food and upon returning with food, deposit pheromone trails on their way back to the nest (Beckers et al., Reference Beckers, Deneubourg and Goss1992; Lehue & Detrain, Reference Lehue and Detrain2020; Wendt et al., Reference Wendt, Kleinhoelting and Czaczkes2020). These trails encode quality: the better the food source, the more pheromone the successful foragers deposit. As ants are recruited to these trails, they may also return with food and strengthen the pheromone trail. If enough ants are recruited, this creates a positive feedback loop which strengthens and maintains the path (Lehue et al., Reference Lehue, Collignon and Detrain2020; Wendt et al., Reference Wendt, Kleinhoelting and Czaczkes2020). If food quality is low, not enough ants are recruited, whilst if the food source has been exhausted, returning foragers will no longer deposit pheromone and the trail will disappear (Lehue et al., Reference Lehue, Collignon and Detrain2020).
Additionally, negative feedback mechanisms are at work: whilst naïve foragers prefer occupied food sources, foragers with previous experience of an unoccupied food source will deposit less pheromone at the occupied source, and instead switch to the unoccupied one (Wendt et al., Reference Wendt, Kleinhoelting and Czaczkes2020). Foragers also deposit less pheromone on trails rich in pheromone or other ants. This negative crowding-effect allows the colony to switch to other high-quality sources (Wendt et al., Reference Wendt, Kleinhoelting and Czaczkes2020). Pheromone trails are thus continuously remodelled, with positive feedback promoting efficacy and negative feedback ensuring flexibility.
Just as plant vascular systems need to optimise the trade-off between construction cost and transport efficiency, ant colonies need to balance the amount of unsuccessful foragers against finding the shortest paths to a given resource. When ants lose a pheromone trail, they may discover shortcuts to already found food sources or new possibly more nearby food sources. Because the density of ants on shorter paths is higher, these paths will have more pheromone deposition. Additionally, their total surface area is lower, reducing pheromonal evaporation (Piñero & Solé, Reference Piñero and Solé2019). Together this favours retention of shorter over longer paths, enabling the colony to compute the shortest path. However, this comes at the risk of ants losing the trail entirely and failing to find food. The intensity of pheromone deposition allows the colony to tune between the competing costs of ants getting lost, unsuccessful foragers (construction cost) and excess path length (transport efficiency cost).
Ant pheromone paths and their adaptability depend on the structure of their physical environment. For example, the information sharing between returning foragers to new recruits is an independent process at each nest entrance (Lehue et al., Reference Lehue, Collignon and Detrain2020). Therefore, having multiple nest entrances instead of one interferes with path optimisation, reducing the chances of finding food, establishing stable pheromone trails and effectively choosing the highest quality food source, resulting in a lower overall food gathering (Lehue et al., Reference Lehue, Collignon and Detrain2020; Lehue & Detrain, Reference Lehue and Detrain2020). On the other hand, an increase in nest entrances facilitates the discovery of more food sites, which is useful for species harvesting many small, scattered packets of food, and exploiting more resources simultaneously is also a good bet-hedging strategy against losing resources, for example, to competing colonies or by depletion (Lehue et al., Reference Lehue, Collignon and Detrain2020; Lehue & Detrain, Reference Lehue and Detrain2020).
The branching pattern of plants into branches, leaves and lateral roots is paralleled in vasculature patterns. These branching patterns are shaped by, and store and transmit information on, environmental conditions and internal plant status. As an example, more lateral roots branch off where nutrient levels are higher, yet this response to local soil conditions depends both on how much the plant is in need of that nutrient and the extent to which other parts of the root system have failed to locate this nutrient. This integration of local, systemic and long range information depends on the status storage in the local plant organ as well as on the transport of systemic and long-distance signals through the vasculature (Boer et al., Reference Boer, Teixeira and Ten Tusscher2020; Guan, Reference Guan2017). In contrast, the continuous moving and mixing of ants in an ant colony makes it impossible to store locational or directional information internally in groups of ants themselves. Pheromone paths thus enable storing critical information on where the food is and what the shortest path to it is in the environment instead (Figure 3, bottom).
3.3. How do environmental feedbacks enhance information integration?
So far, the information processes we have considered take input and produce output. However, biological computations do not occur in a vacuum but in a physical context consisting of the cells, organisms or colonies, and their direct environment, and output of a previous information processing round often affects this context and thereby the current input state.
Plants, through mechanical feedbacks, can sense, respond to and adapt their own shape. This sensing and responding to their own status is called proprioception (Bastien et al., Reference Bastien, Bohr, Moulia and Douady2013; Meroz, Reference Meroz2021) and enables plants to produce new organs with reproducible shapes (Hamant & Moulia, Reference Hamant and Moulia2016) as well as to control their movement. As an example of the latter, during plant tropisms plant organs grow towards or away from various stimuli, to maximise available light, space, water, nutrients, and so on. It was found that the organ bending observed during tropic growth could not be explained only by, for example, shoots responding to gravity, but must also involve their sensing and responding to their own local curvature (Bastien et al., Reference Bastien, Bohr, Moulia and Douady2013). Interestingly, plant proprioception is not a purely internal process. As an example, plants use the fact that they vibrate in the wind with frequencies dictated by their own weight, stiffness and shape (Hamant & Moulia, Reference Hamant and Moulia2016), allowing them to sense their shape, and modulate branching lengths to avoid breakage. In this case, the sessile plants use externally driven movements to sense their own internal state.
We can also see proprioception at work in ant colonies, when studying their shaping of nest architecture. Nests not only provide shelter but also perform other essential functions, such as ventilation (Ireland & Garnier, Reference Ireland and Garnier2018). Ant colonies do not coordinate their nest construction by direct ant–ant communication. Instead, each ant adds material to or removes it from a piece of the building, and this local structure modification serves as a blueprint for the next construction effort. On the colony level, these local proprioception feedbacks add up to a sophisticated nest architecture and have been described for other social insects (Theraulaz & Bonabeau, Reference Theraulaz and Bonabeau1995). These structure-encoded feedbacks in combination with social feedbacks enable the colony to adapt the nest over time, so that it continues to fit its needs. As an example, volatile pheromones around queen or brood allow ants to enlarge their chambers as they grow (Ireland & Garnier, Reference Ireland and Garnier2018).
Both plant tropisms and ant nest building are the output of information processes, but they also qualitatively change the input of future computations not only for that same process but also for others. For example, only once a seedling has broken through the soil by negative gravitropism, it has light available as an input. The red/far-red ratio in this light can then be an input for a shade avoidance computation, so that the seedling can compete with other plants for this light (Ballaré & Pierik, Reference Ballaré and Pierik2017). Likewise, the construction of nest entrances is a result of colony-level information processing. Yet, as we saw before, the number and position of these entrances also influence the coordination among foragers, and thus the colony’s success in computing the optimal food intake strategy (Lehue et al., Reference Lehue, Collignon and Detrain2020; Lehue & Detrain, Reference Lehue and Detrain2020).
4. Conclusion
We have seen that embodiment is an essential part of distributed information processing for plants as well as for ant colonies. Solid brains, such as plant tissues, use their fixed structure combined with their capacity to adapt and optimise connections between cells and body parts to store and compute information, as we saw in the examples on root halotropism, leaf stomatal aperture regulation and root branching in response to nutrients. In contrast, in ant colonies the extensive movement of individual ants enables substantially faster direct information transfer, yet limits storage of information within the colony itself. Instead information is stored in the environment, in the shape of pheromone trails as well as nest size, shape and entrance numbers. In both cases, embodiment constraints and the trade-off between construction cost and transport speed shape the long-distance signalling architecture, that is, the developing vascular and pheromone trail networks. Whilst local communication between neighbouring plant cells or meeting ants is enough to integrate information across the entire tissue or colony and do complex information processing, these long-distance communication networks enhance signalling reliability and speed. In addition to solid versus liquid brains putting constraints on how information is integrated and where it is stored, it also impacts how information travels. Interestingly, independent of whether it involves direct contacts between cells or ants, or indirect long-distance contacts through vasculature or pheromone trails, plants and ant colonies differ in whether the message, or rather sender and receiver move. In the solid-type plants, cells and organs remain at a fixed position and signalling molecules travel, whereas in the liquid-type ant colonies, pheromones are deposited at a fixed position in space, while the ants wander around.
An interesting issue that has thus far attracted limited attention is the impact of growth on plant information processing. As plants grow and age, they acquire an increasing number of branches, leaves and roots, resulting in an increasingly large and complex network. This increase in size and network complexity can be expected to lengthen communication times and increases the complexity of signal integration. Whilst plant branching patterns have been extensively studied (see, e.g., Chandrasekhar & Navlakha, Reference Chandrasekhar and Navlakha2019; Conn et al., Reference Conn, Pedmale, Chory and Navlakha2017; Walker & Bennett, Reference Walker and Bennett2018), to our knowledge the precise effects of growth on processing delays and signal integration are still unknown. An intriguing possibility for future research is whether as plants grow during development, or between plant species of different sizes and branching patterns, molecular signalling networks vary to fulfil the different constraints that come with their varying network sizes. On a similar note, ant colonies of varying sizes may tune the balance between following versus losing a pheromone trail differently based on the different constraints arising from their colony sizes.
Finally, when people think of cognition, information processing usually is at its heart. Since plants can perform complex computations, a logical follow-up question is whether plants can be considered cognitive. Much of the plant cognition debate is taken up by two competing perspectives: plant neurobiology and the collection-of-parts perspective (Cazalis & Cottam, Reference Cazalis and Cottam2021). This latter perspective views the plant as a non-cognitive collection of parts, with individual cells, leaves, roots and so on, each processing information and making separate decisions. Proponents of the plant neurobiology perspective view the plant as an individual organism, endowed with consciousness and intelligence. This perspective makes extensive use of comparisons to animal brains, nerves and synapses and focusses on electrical signals as the major source of plant information processing. The neurobiological perspective is controversial and has been criticised among other things for its largely speculative nature and its insistence on fitting plant behaviour to a neural animal mould (Cazalis & Cottam, Reference Cazalis and Cottam2021). Given the importance of embodiment as, for example, discussed in this review and the differences between plant and neural animal bodies, why would one expect to find strictly analogous structures and methods for information processing?
Although plant neurobiology thus seems a poor fit, a plant is also more than the sum of its parts, arguing against the collection-of-parts perspective as a suitable alternative. Instead, the ‘distributed-computing’ perspective taken in this review and by others (e.g., (Bassel, Reference Bassel2018; Duran-Nebreda & Bassel, Reference Duran-Nebreda and Bassel2019; Peak et al., Reference Peak, West, Messinger and Mott2004) provides a viable perspective. It considers plants as more than the sum of their parts but eschews brain-like comparisons, instead taking into account the specifics of plant embodiment. In this review, we hope to have demonstrated this to be a fruitful perspective, with consideration of embodiment serving as an informative means to gain a better understanding of not only how but also why plants use particular types of information processing.
Financial support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Conflict of interest
The authors declare none.
Authorship contributions
L.L.M.v.S. co-designed the literature research and wrote the master literature thesis on which this article was based. B.L.S. provided supervision and wrote the article. K.H.t.T. co-designed the literature research, provided supervision and wrote the article.
Data availability statement
Not applicable given the review character of the manuscript.







Comments
Dear Editor,
Hereby I would like to submit our article titled "" for consideration in Quantitative Plant Biology.
Our article discusses the importance of whether biological information processing occurs in a centralized or distributed manner, and -in case of distributed computing- whether individual computing units are fixed or flexible in position, for the nature of the information processing and storage of information. To highlight the particularities of solid brain, distributed information processing in plants a comparison is made with the liquid brain, distributed information processing in ant colonies, that has been particularly well studied. We end with a dicussion on how taking into account embodiment, the physical context in which information processing occurs, and considering the solid brain, distributed nature of plant computing may help resolve the debate on plant cognition.
We believe our article to be of interest to an audience of plant biologists keen to integrate quantitative, information and computer science perspectives into their research on plant development, adaptation and signalling, and as such believe it is highly suitable for Quantitative Plant Biology.
Thank you for your consideration,
Kirsten ten Tusscher