INTRODUCTION
Salmonella enterica causes an estimated 1·4 million illnesses and 400 deaths annually in the United States [Reference Voetsch1]. Of 32 219 laboratory-confirmed Salmonella enterica cases with known serotypes reported to the Centers for Disease Control and Prevention (CDC) in 2004, 3325 (10·3%) were serotype Newport [2], making it the third most common serotype causing human illness. Taking into account underreporting, more than an estimated 100 000 people are infected with S. Newport each year. Outbreaks of S. Newport infections have been caused by foods of animal origin, particularly beef [Reference Espie3–Reference Fontaine11], and produce [Reference Kirk12–Reference Van Beneden16]. Outbreaks of multidrug-resistant S. Newport have been due to contaminated beef and horsemeat, while outbreaks due to contaminated produce have tended to be pan-susceptible to antimicrobial agents.
Salmonella isolates may be further differentiated into strains by pulsed-field gel electrophoresis (PFGE). PFGE patterns are tracked by PulseNet, the National Molecular Subtyping Network for Foodborne Disease Surveillance [Reference Swaminathan17]. State public health laboratory officials routinely submit PFGE patterns from Salmonella isolates to PulseNet at CDC. The PulseNet staff compares these patterns within and across states and groups isolates with indistinguishable patterns into potential clusters of illness.
We describe a prolonged multistate outbreak of S. Newport infections due to contaminated tomatoes grown on the eastern shore of Virginia. The 2005 outbreak shared strong commonalities with a 2002 outbreak that resulted in 510 patients with confirmed and probable illness in 26 states [Reference Kretsinger13]. Both outbreaks shared a common serotype, PFGE pattern, geographical distribution of case-patients, epidemiological association with tomatoes, source of tomatoes identified by traceback, and isolation of the outbreak strain of S. Newport from pond water in the tomato-growing region. This report describes the 2005 investigation and highlights the recurrence of tomato-associated outbreaks of salmonellosis linked to on-farm contamination.
METHODS
Case finding, laboratory investigation, and hypothesis generation
On 20 September 2005, New Hampshire State Public Health Laboratory officials informed PulseNet staff of the recent identification of five Salmonella Newport isolates with an indistinguishable PFGE pattern. PulseNet identified 24 additional isolates indistinguishable from the outbreak strain from eight other states. PulseNet notified all state public health laboratories of the outbreak strain pattern, in order to identify additional S. Newport cases with this pattern. Six outbreak isolates were tested for susceptibility to 15 antimicrobial agents using broth microdilution (Sensititre; TREK Diagnostic Systems, Cleveland, OH, USA) by the National Antimicrobial Resistance Monitoring System (NARMS) laboratory at CDC.
On 28 September 2005, the New Hampshire Department of Health and Human Services invited CDC to assist in a field investigation to determine the source of infections and develop recommendations to prevent similar illnesses in the future.
For case ascertainment purposes, a case was defined as culture-confirmed S. Newport infection in the United States with illness onset during 2005 and XbaI restriction enzyme PulseNet PFGE pattern JJPX01.0061. A subset of isolates was also digested with the restriction enzyme BlnI and showed PulseNet pattern JJPA26.0021 (Fig. 1).

Fig. 1. Pulsed-field gel electrophoresis (PFGE) of Salmonella Newport isolates from patients. Lanes 1, 2, XbaI restriction enzyme PulseNet PFGE pattern JJPX01.0061; lanes 3, 4, BlnI restriction enzyme PulseNet PFGE pattern JJPA26.0021; lane S, the PulseNet universal size standard Salmonella Braenderup H9812 digested with XbaI [Reference Hunter34].
To develop hypotheses of possible sources of S. Newport infections, we conducted two telephone and four in-person interviews in the homes of cases in a southern New Hampshire county and a neighbouring Massachusetts county. We also inventoried the refrigerators and pantries of interviewees to search for foods common among them yet absent from our questionnaire. To further assess exposures, we requested case report forms routinely completed by state health departments.
Case-control study
Study questionnaires collected information on demographics, travel history, food sources (grocery stores and restaurants), 35 specific food items deemed important based on hypothesis-generating interviews, and whether foods were prepared inside or outside the home.
Study cases were defined as cases between the ages of 18 and 70 years who had illness onset between 8 August and 12 September 2005. Cases and matched controls were asked about exposures in the 7 days before illness onset of the case.
Control interviews were conducted by the University of New Hampshire Survey Center in all participating states, using computer-assisted telephone interviewing (CATI) [Reference Choi18]. For each case with a known home telephone number or address, a reverse telephone directory was used to create a list of the case's six neighbours with the closest street addresses. This list was randomized, and neighbours were called until at least four control interviews were completed for each enrolled case. When a household was contacted, the survey centre selected the member of the household between the ages of 18 and 70 years with the most recent birthday. If the selected individual was not available, the interviewer telephoned the next household on the list. If needed, more potential controls were identified progressively further from the case's home, randomized, and then contacted. A control was not enrolled if he or she had a diarrhoeal illness between 1 July 2005 and the date of interview.
Data from the case questionnaires were entered into an Access 2000 (Microsoft Corporation, Redmond, WA, USA) database and merged with data entered via the CATI system from the control questionnaires. Matched statistical analyses were conducted using SAS 9.1 (SAS Institute, Cary, NC, USA). Bivariate and multivariate analyses quantified relationships between each food exposure and disease. All matched odds ratios (mOR) were derived from conditional logistic models. Ninety-five percent confidence intervals (CI) were based on the application of exact methods.
Traceback and environmental investigation
The U.S. Food and Drug Administration (FDA) Center for Food Safety and Applied Nutrition, in conjunction with state and local authorities, conducted a traceback investigation to determine the source of tomatoes eaten by cases, inspected tomato farms, and tested irrigation ponds for Salmonella in October 2005 and July 2006. These inspections were in the eastern shore of Virginia.
Historical incidence of outbreak strain
The incidence of this outbreak strain in PulseNet was compared with the incidence of all S. Newport ascertained in the National Salmonella Surveillance System [2].
RESULTS
Case finding, laboratory investigation, and hypothesis generation
From July to November 2005, 72 laboratory-confirmed S. Newport isolates indistinguishable by PFGE from the outbreak strain were identified in 16 states (Fig. 2). Case-patients ranged in age from 5 months to 75 years, with a median age of 29 years. Of 72 case-patients, 42 (58%) were female. Eight (11%) of the cases resulted in hospitalization. There were no deaths.
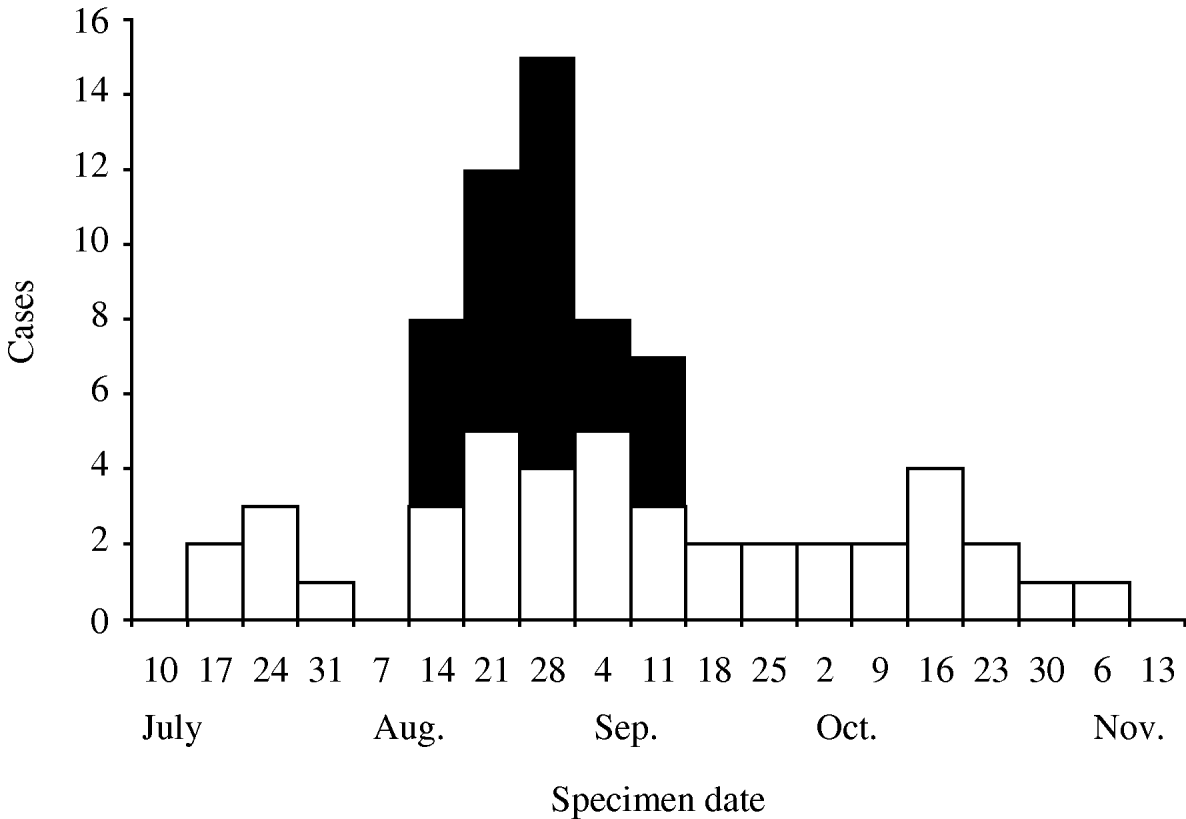
Fig. 2. Case-patients with Salmonella Newport infections due to outbreak strain who were enrolled (■, n=30) and not enrolled (□, n=42) into the case-control study, by week specimen received in state laboratory, 2005.
All but one of the cases resided in states east of the Mississippi River. Of these 16 states, 14 (88%) were also affected by the outbreak of the same strain of S. Newport in 2002 (Fig. 3). Within states during the 2005 outbreak, there was geographical clustering: Ohio's three affected counties were adjacent, and all of Wisconsin's cases resided in one county. All 11 of the New Hampshire and Massachusetts cases resided in two adjacent counties. Despite this geographical clustering, most patients within each state did not report eating at the same restaurants or having other common venue exposures.
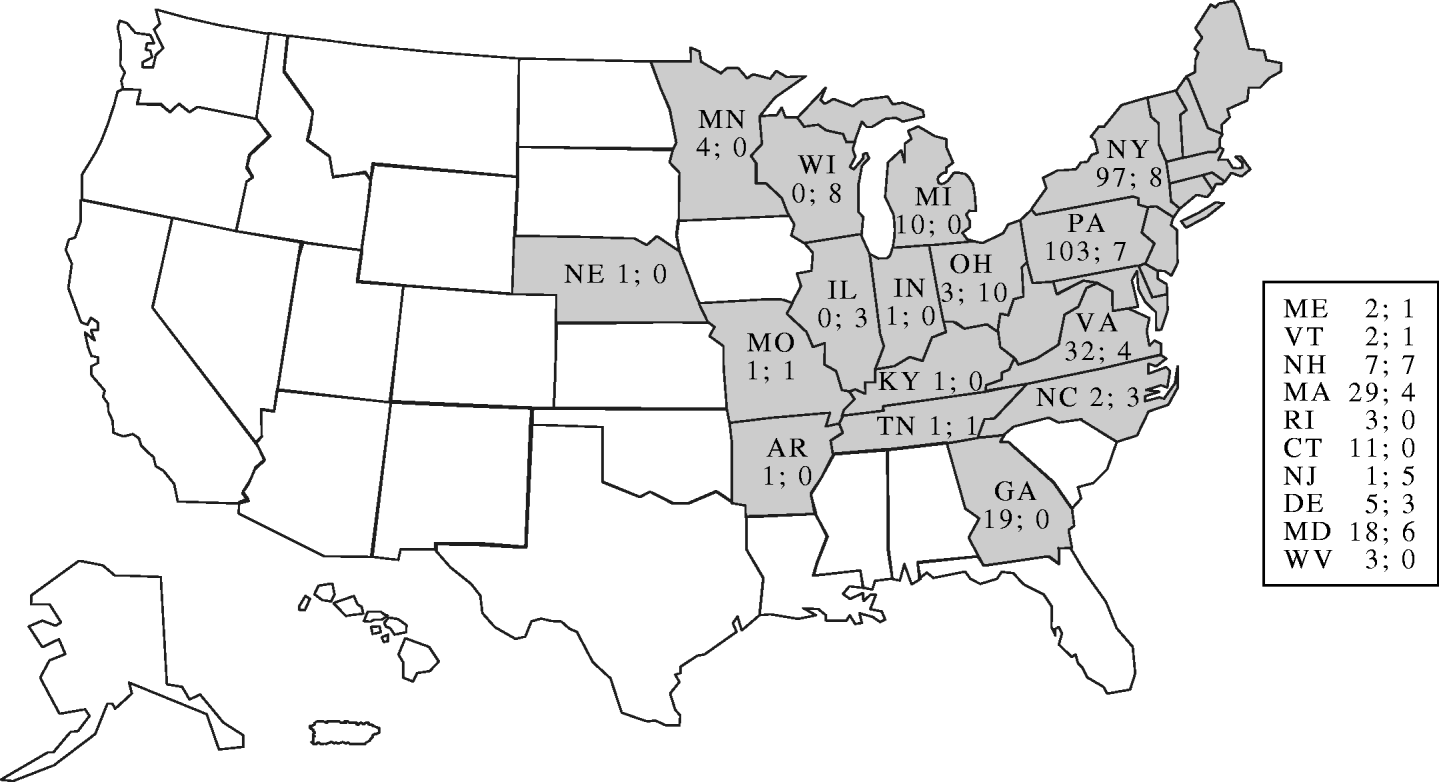
Fig. 3. Distribution of culture-confirmed cases in 2002 and 2005 (separated by semi-colon) due to outbreak strain of Salmonella Newport, by state.
Of the 72 isolates, 48 (67%) were additionally tested by PFGE with a second enzyme and all showed BlnI pattern JJPA26.0021. Outbreak isolates were susceptible to the standard panel of antimicrobial agents.
Case-control study
Twenty-nine case-patients and 140 matched controls were enrolled in the study. Information was available on a median of five controls (range 4–11) for each case. Excess control enrolment occurred due to a delay in notification when the enrolment target of four controls per case had been achieved. The enrolled cases were residents of Ohio (8), Wisconsin (7), New Hampshire (5), Pennsylvania (4), and one each from Kentucky, Massachusetts, New Jersey, Tennessee, and Vermont. Onset of illness ranged from 9 August to 6 September 2005. The median duration of illness among cases was 8 days (range 4–23 days). The median age of cases was 41 years (range 23–68 years) and of controls was 49 years (range 18–70 years). Seventeen (59%) of 29 cases and 88 (63%) of 140 controls were female. Four (14%) of 28 cases travelled outside their state of residence in the 7 days before illness onset compared with 24 (18%) of 132 matched controls.
Reasons for not enrolling patients included identification of cases after the study period (n=20), failure to meet the study case definition (illness onset before 8 August, n=9; age <18 years, n=10), inability to contact patient (n=3), and travel during the incubation period, making it difficult to identify appropriate controls (n=1).
Illness was associated with eating in a restaurant; 26 (90%) of 29 cases ate at least one of the foods on the questionnaire in a restaurant in the 7 days prior to illness onset compared with 58 (41%) of 140 controls (mOR 15·3, 95% CI 3·6–138·8). No single restaurant or chain was implicated. Patients ate in 26 different restaurant venues.
Three items consumed by at least 40% of patients were significantly associated with illness in bivariate analysis (Table); other items, including some of those identified during hypothesis generation, were not frequently consumed by patients enrolled in the case-control study. More patients (70%) were exposed to uncooked tomatoes in restaurants than any other item. Of 27 patients, 11 (41%) reported eating ‘beefsteak’ tomatoes, and 13 (48%) reported eating ‘other’ types of tomatoes. Twenty-six (90%) of 29 cases and 86 (72%) of 119 controls had any exposure to tomatoes (i.e. any variety of tomato, prepared either in home or in a restaurant).
Table. Risk factors for S. Newport illness among participants of case-control study, restricted to food items consumed by >40% of cases
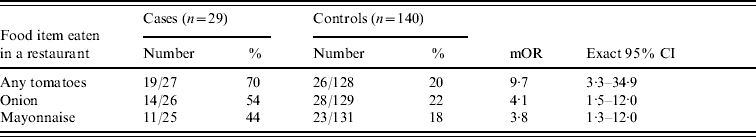
mOR, Matched odds ratio; CI, confidence interval.
For completeness, bivariate matched analyses for tomatoes in restaurants were repeated for the three states with the most enrolled cases. For Ohio, with information for eight cases and 41 controls, the measure of association for illness and tomatoes in restaurants was mOR 6·9 (95% CI 1·0–83·5). For Wisconsin, with six cases and 25 controls the measure of association was mOR 4·9 (95% CI 0·6–57·1) and for New Hampshire, with four cases and 22 controls it was mOR 11·1 (95% CI 0·8–610·0). Risk associated with tomato exposure in restaurants did not vary significantly by state of residence in an overall test for interaction.
The frequency of unknown and missing responses was assessed. For tomatoes in a restaurant, two (7%) of 29 cases and 12 (9%) of 140 controls had unknown or missing responses. For a sensitivity analysis, we assumed that the missing values biased the odds ratio estimate towards the null. That is, the two cases with missing values were assumed to have not eaten tomatoes, while the 12 controls with missing values were assumed to have eaten them. Even in this most extreme case, the odds ratio for tomatoes was still large and statistically significant (mOR 4·8, 95% CI 1·9–13·4). The proportion of missing and unknown values for other exposures was checked, and no evidence for bias was detected.
Onion and mayonnaise consumption were also significantly associated with illness in bivariate analysis. In a multivariate logistic model with tomatoes, neither onion nor mayonnaise (all prepared in restaurants) was individually or jointly significant, and the odds ratio for tomatoes remained essentially unchanged from bivariate analysis. Onion and mayonnaise are often consumed along with tomatoes in sandwiches or salads and were significantly correlated with tomato consumption.
Traceback and environmental investigation
Selected restaurants at which cases ate meals containing tomatoes were contacted to determine the source of tomatoes served. All six restaurants contacted reported using large, whole round tomatoes that were sliced at the restaurant. The traceback was based on the sources of tomatoes for two restaurants at which more than one patient ate a meal containing tomatoes in the 7 days prior to illness onset.
Results of the traceback indicated that the tomatoes were of the ‘red round’ variety and originated from two growers/packing houses on the eastern shore of Virginia. Farms in this region supplied only the eastern and central United States at the time of this outbreak, matching the national distribution of cases of the outbreak pattern of S. Newport.
An irrigation pond water sample taken during a visit to a farm on the eastern shore of Virginia in October 2005 yielded S. Newport matching the outbreak strain. This irrigation water entered the soil bed and did not directly contact fruit. In addition, pond water samples taken during the July 2006 environmental inspection of the farms specifically implicated in the 2005 outbreak yielded Salmonella serotype Javiana and S. Newport (which did not match the outbreak strain). In the 2006 inspection, large numbers of geese and turtles were observed in ponds. One of the growers used pond water for application of pesticides to tomato plants. Based on how pipes were laid between ponds and wells, cross-connections were possible, and contaminated pond water may have seeped through the sandy soils from the ponds into nearby wells. Well water is used to recharge pond water throughout the area, and depending on the farm, both water sources may be used for irrigation and mixing with pesticides. Well water samples collected during this inspection from several wells close to ponds were negative for Salmonella.
Historical incidence of outbreak strain
Of all S. Newport isolates from 1998 to 2005 in the PulseNet database, 589 (4·03%) of 14 600 isolates demonstrated XbaI pattern JJPX01.0061. Of these 589 isolates, DNA from 244 (41%) was also digested by the second enzyme, BlnI; 228 (93%) of 244 demonstrated BlnI pattern JJPA26.0021. Of isolates with XbaI pattern JJPX01.0061, four have a known non-human source: an isolate from cantaloupe submitted in September 2002; an isolate from salad submitted in October, 2002; and two isolates from irrigation pond water for tomato farms, one submitted in September 2003 and another submitted in October 2005.
This outbreak strain has caused illness during the summer and autumn each year (Fig. 4). For the months of July–November for each year, 2002–2005, the outbreak strain represented 12·2%, 1·9%, 3·3%, and 4·1%, respectively, of all S. Newport isolates in PulsetNet. PFGE distinguished the annual increase in cases due to this outbreak strain from the background of all salmonellosis caused by S. Newport.
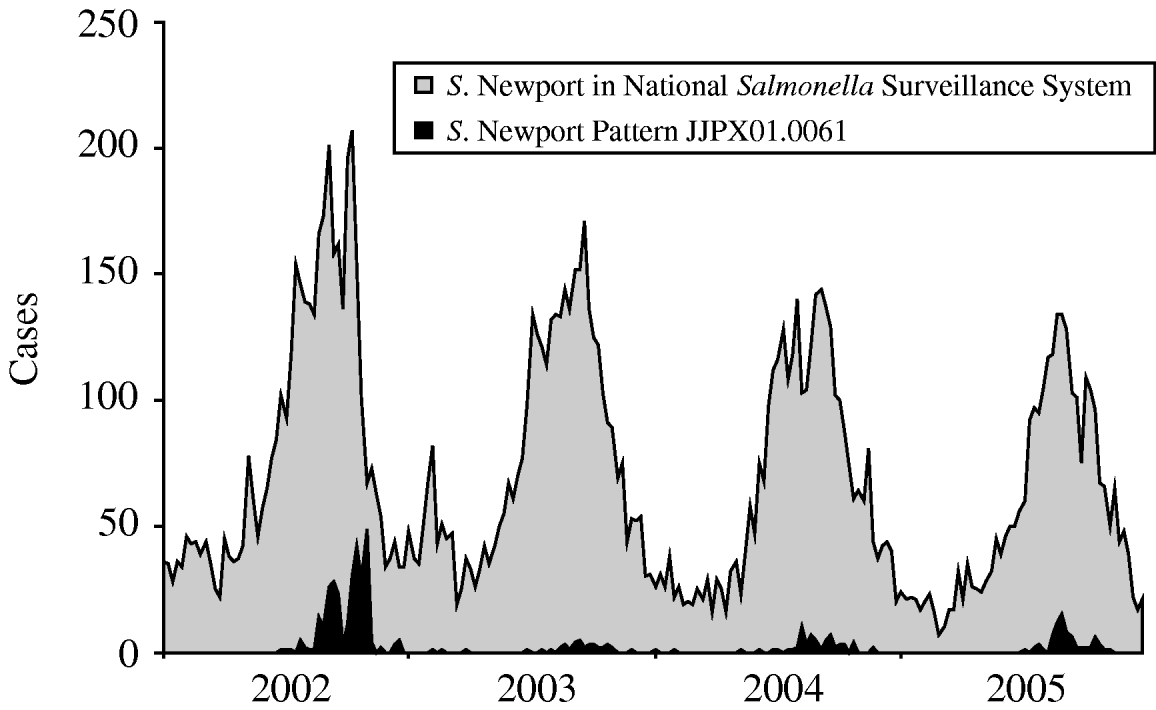
Fig. 4. Number of all human Salmonella Newport cases (gray) with subset that were subtyped as the outbreak PFGE pattern (black), by week, 2002–2005. The large tomato-associated outbreaks in 2002 and 2005 are visible. Small clusters of the outbreak pattern were also detected in the same seasons of 2003 and 2004, but were not investigated.
DISCUSSION
A single strain of S. Newport caused illness among at least 72 laboratory-confirmed patients in 16 states in 2005. Given that about one of every 38 cases of sporadic, laboratory-confirmed Salmonella infection is ascertained by public health surveillance [Reference Voetsch1], we estimate that more than 2500 patients may have been affected by this outbreak. The epidemiological and traceback investigations demonstrated that tomatoes from the eastern shore of Virginia caused this outbreak.
When analysis of the case-control study was completed in October 2005, examination of the epidemic curve revealed little evidence of ongoing transmission. By then, contaminated tomatoes associated with this outbreak were likely to have been consumed or destroyed. This outbreak was probably due to a small fraction of all tomatoes on the market, but until the traceback was completed several months later, the specific tomatoes involved could not be identified. Therefore, no recall was undertaken.
The outbreak strain was susceptible to all antimicrobials tested. Multidrug-resistant and pan-susceptible S. Newport infections frequently appear to have different sources, for both outbreaks and sporadic infections. In a large study of sporadic S. Newport illnesses, multidrug resistant S. Newport illnesses were associated with foods of animal origin including ground beef, while pan-susceptible S. Newport illnesses were linked to contact with reptiles or amphibians [Reference Varma19]. This outbreak contributes to the list of pan-susceptible S. Newport outbreaks that were produce-associated.
Tomatoes have repeatedly been demonstrated as a vehicle for multistate Salmonella outbreaks [Reference Kretsinger13, Reference Srikantiah20–24]. Round, Roma, and grape tomatoes have all been implicated in outbreak investigations, in spite of the challenge that tomatoes are rarely the principal ingredient of a dish, and therefore interviewees may fail to recall eating tomatoes. In addition, eating in restaurants was associated with illness in these outbreaks. It is possible that restaurants may have different sources of tomatoes compared to groceries, and might store and handle them in ways (e.g. commingling) that spread contamination through a larger quantity of food and/or amplify bacterial growth (e.g. cutting and holding).
Salmonella outbreaks due to contaminated tomatoes have been large and widely dispersed, suggesting that contamination is occurring early in the distribution chain, such as at the farm or packing house, rather than at the individual restaurants. The point of contamination for this outbreak probably occurred on the farm, although the precise mechanism of contamination was not determined. Possible sources of environmental Salmonella contamination include faeces from domestic or wild animals (e.g. reptiles, amphibians, or birds). On at least one farm, pond water potentially contaminated with Salmonella was used to dilute pesticides sprayed on tomato plants. Laboratory studies show that the survival of Salmonella is not markedly affected by pesticides at concentrations used for crop protection [Reference Guan, Blank and Holley25, Reference Ng, Fleet and Heard26].
Tomato stems and flowers inoculated with Salmonella can yield fruits contaminated with the bacteria [Reference Guo27]. Once contaminated, cut tomatoes provide an excellent medium for bacterial amplification, and food handlers should refrigerate tomatoes after slicing [Reference Lin and Wei28]. Growers, harvesters, retailers, re-packers, and food-service employees should follow available guidelines for good manufacturing practices and good agricultural practices when handling tomatoes [29–Reference Beckman31]. These guidelines do not stipulate that all water used for agricultural purposes meet potable drinking water standards, but rather, more generally, that water be suitable for its intended use. Surface waters are more vulnerable to contamination than protected wells, and some water applied in growing fields may not be suitable for its intended use if it contacts the edible portion of the crop near harvesting.
S. Newport cases with the outbreak PFGE pattern occurred each year during 2002–2005, and tomatoes were possibly the source of cases in all four of these years. The 2002 and 2005 recurrent tomato-associated outbreaks, coupled with the isolation of the outbreak strain from pond water, suggest a persistent source of environmental contamination in tomato fields or packing houses in the eastern shore of Virginia. Some counties in this region have consistently had an elevated incidence of S. Newport infections from 1968 to 1998 [32]. This suggests that human infections and the source of contamination for tomatoes grown in this region may have a common environmental source.
As in most case-control studies, we were concerned about the possibility of misclassifying exposures. Thirty percent of patients did not recall consuming tomatoes in restaurants, and some of these patients may represent background illness unrelated to the outbreak. However, tomatoes in restaurants remained significantly associated with illness in an analysis that assumed that missing values biased the odds ratio estimate towards the null. Furthermore, tomatoes in restaurants independently had elevated associations with illness within different states, although the power of a combined, multistate analysis was necessary to show a clear significant association. Ultimately, the results of the epidemiological study were corroborated by the isolation of the outbreak strain from pond water near tomato fields identified in the traceback.
This outbreak advances an increasing awareness of produce-associated outbreaks [Reference Sivapalasingam33]. To prevent future tomato-associated outbreaks, further environmental and laboratory research by industry, academia, and government agencies is necessary to determine the source of pathogens, mechanisms by which the pathogens come in contact with the fruit, the stages of development at which plants are most susceptible to contamination persisting on or in the tomato, and procedures by which contamination can be reduced or eliminated. Future research and tracebacks should focus at all levels of tomato production, including the seed nursery, the field, and the packing house. Because tomatoes are used as a raw ingredient in many dishes in which they are not cooked to kill bacteria, prevention of contamination is paramount.
ACKNOWLEDGEMENTS
We gratefully acknowledge the following individuals for their contributions to study design and data collection: E. Barrett (Virginia Department of Health); E. Berl (Massachusetts Department of Public Health); M. Deasy (Pennsylvania Department of Health); M. P. Fage (New York State Department of Health); A. Ingram (Tennessee Department of Health); D. H. Johnson (Wisconsin Department of Health and Family Services); M. Malavet (New Jersey Department of Health and Senior Services); S. Nowicki (Ohio Department of Health); M. Spayne (Vermont Department of Health); and A. E. Smith and M. Belanger (University of New Hampshire Survey Center). We thank the following individuals for serving as scientific advisors: M. A. Smith (Center for Food Safety and Applied Nutrition, FDA); and F. Angulo and C. Braden (Enteric Diseases Epidemiology Branch, CDC). We also acknowledge the important contributions from public health, food regulatory, and laboratory personnel from the affected states and our FDA partners.
DECLARATION OF INTEREST
None.







