INTRODUCTION
Non-typhoidal salmonellosis is an important foodborne problem throughout the world. Even though more than 2500 serovars of Salmonella enterica have thus far been recognized [Reference Farmer1], few serovars appear to cause most foodborne illnesses. One non-host-adapted serovar common to food animals and humans and with wide distribution is Typhimurium. Strains of this serovar in pigs have also been shown to manifest multidrug resistance (MDR) [Reference Gebreyes, Davies, Morrow, Funk and Altier2–Reference Gebreyes, Davies, Funk, Altier and Thakur4].
Diversity among salmonellae has been studied using various phenotypic methods, including antimicrobial resistance profiling and phage typing, and by genotypic methods including pulsed-field gel electrophoresis (PFGE) [Reference Schwartz and Cantor5], amplified fragment length polymorphism (AFLP) [Reference Vos, Hogers and Bleeker6], plasmid profile analysis, ribotyping, and other related methods. One important use of phage typing and antimicrobial resistance profiling was the recognition of the emergence of S. Typhimurium phage type DT104 in the early 1990s [Reference Glynn, Bopp, Dewitt, Dabney, Mokhtar and Angulo7, Reference Threlfall, Frost, Ward and Rowe8]. These methods remain an important part of epidemiological investigations for Salmonella.
Genotyping of Salmonella is becoming an increasingly important epidemiological tool that aids in the identification of sources of infection during outbreak investigations, detection of cross transmission, recognition of particular strains, and monitoring intervention strategies [Reference Gebreyes9]. Among genotyping methods, PFGE is considered the standard method for DNA fingerprinting in Salmonella and other foodborne pathogens [Reference Olive and Bean10], and a system for differentiating epidemic strains from endemic ones has been proposed [Reference Tenover, Arbeit and Goering11]. S. Typhimurium has often been considered very clonal and most genotypic methods including PFGE do not have the discriminatory power to differentiate within phage types [Reference Beltran, Musser and Helmuth12, Reference Antunes, Machado, Sousa and Peixe13]. In the last decade, other methods such as AFLP and repetitive extragenic palindromic–polymerase chain reaction (Rep–PCR) have been developed. The discriminatory power of these methods is evaluated and compared to PFGE in the present study. For ultimate Salmonella strain discrimination, an integrated approach using a highly discriminatory genotyping method together with phenotypic approaches is indispensable as shown previously [Reference Gebreyes9, Reference Pontello, Sodano and Nastasi14–Reference Weigel, Qiao, Barber, Teferedegne, Kocherginskaya and Van der Wolf16].
Modern pig production systems maintain closed herds in a pyramid form with an all-in/all-out management system in multisite production units. Such production systems are expected to limit exchange of infectious agents. Even though cross contamination of pigs and the similarity of Salmonella strains cultured during transport and in holding pens at slaughter has been described in recent years [Reference Hurd, McKean, Wesley and Karriker17, Reference Gebreyes, Davies, Turkson, Morrow, Funk and Altier18], there is very limited information about the clonality or diversity of Salmonella strains within and among different production systems on farms.
The aim of this study is to evaluate three genotyping methods (AFLP, PFGE and Rep–PCR) to determine their discriminatory power, reproducibility, resolution and throughput efficiency when applied to Salmonella serovar Typhimurium strains collected from 18 farms that belonged to two different commercial multiple site swine production systems located in eastern North Carolina.
Methods
Origin of isolates
All the isolates in this study originated from a longitudinal study described previously [Reference Gebreyes, Davies, Funk, Altier and Thakur4, Reference Funk, Davies and Gebreyes19]. Briefly, six nursery and 18 finishing farms of two commercial pig production systems located within two counties in eastern North Carolina were chosen. From each of the cohorts of pigs, 96 faecal specimens were collected once at nursery and once at finishing. Sampling was done in three replicates between 1997 and 2000. Eighteen farms (8 nursery and 10 finishing) that were positive for Salmonella were included in this study.
Isolation and phenotypic characterization
Salmonella isolation was performed using conventional methods as described previously [Reference Bager and Petersen20, Reference Davies, Bovee, Funk, Jones, Morrow and Deen21]. S. Typhimurium (n=196) isolates from 18 farms from all the three replicates collected in a 3-year time period were further investigated as described below.
Antimicrobial susceptibility testing was performed initially using the Vitek Jr. semi-automated system (Biomerieux. Hazelwood, MO, USA) using break-point panels. Each isolate was first tested against a panel of 10 antimicrobial agents. The antimicrobials and respective resistance MIC break-points were amikacin (Ak) (64 mg/l), co-amoxiclav (Ax) (32/16 mg/l), ampicillin (Am) (32 mg/l), cefotaxime (Cf) (64 mg/l), cephalothin (Ce) (32 mg/l), chloramphenicol (Cm) (32 mg/l), ciprofloxacin (Cip) (4 mg/l), gentamicin (Gm) (16 mg/l), tetracycline (Te) (16 mg/l) and trimethoprim/sulphamethoxazole (T/S) (4/76 mg/l) [22, 23]. In this study, isolates with intermediate MIC break-points were grouped with susceptible organisms in order to not over-estimate occurrence of resistance.
Additional susceptibilities to sulphisoxazole (Su) (0·25 μg), streptomycin (St) (10 μg), kanamycin (Km) (30 μg), and ceftriaxone (Cro) (30 μg) were determined by the Kirby–Bauer disc diffusion method on Mueller–Hinton agar plates using conventional techniques [22, 23]. Results were interpreted according to the NCCLS criteria following performance standards for both human and animal isolates [22, 23]. Escherichia coli strain ATCC 25922 and 35218, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 29213 and Pseudomonas aeruginosa ATCC 27853 were used as quality-control organisms in antimicrobial susceptibility testing according to NCCLS recommendations. Serotyping and phage typing was performed by the National Veterinary Services Laboratories (NVSL) in Ames, Iowa.
AFLP genotyping
The AFLP fingerprinting method was used as described previously with some modifications [Reference Vos, Hogers and Bleeker6]. Briefly, cells were grown overnight on Luria–Bertani broth (BD Diagnostic System, Sparks, MD, USA); genomic DNA was purified using the Qiagen DNAeasy tissue kit (Qiagen, Valencia, CA, USA) and adjusted to a concentration of 50 ng/μl in a volume of 10 μl of water. The DNA was digested with the restriction enzymes EcoRI and MseI at 37°C for 1 h. Adapter oligosequences unique to each end were ligated to the restriction fragments using T4 DNA ligase (New England Biolabs, Beverly, MA, USA) at 16°C overnight. Fragments were then initially amplified using primers complementary to the EcoRI (F, 5′-GACTGCGTACCAAATC) and the MseI (R, 5′-GATGAGTCCTGAGTAA) end. The conditions for amplification were 94°C for 15 s; 60°C for 30 s and increasing at 1 s/cycle for 28 cycles, then incubation at 72°C for 2 min. The amplified products were diluted in deionized water at 1:9 (v/v) ratio and underwent a selective amplification using an infrared-labelled EcoRI primer with an additional adenine base at the 3′-end [EcoRI+A] (Li-Cor, Lincoln, NE, USA). The thermocycling conditions for selective amplification was 13 cycles at 94°C for 10 s, 65°C for 30 s, and 72°C for 1 min, then 25 cycles of 94°C for 10 s, 56°C for 30 s, and 72°C for 1 min, ramping at 1 s/cycle for the final step. The reaction was finally incubated for 2 min at 72°C. Fragments were then separated on a long-ranger polyacrylamide gel using a Li-Cor 4200 DNA sequencer. Amplified bands located between 200-bp and 500-bp fragment lengths were scored and analysed. A detailed AFLP protocol can be found at the following URL (http://www4.ncsu.edu/∼wagebrey/AFLP-Salm.pdf).
PFGE genotyping
PFGE analysis was performed as recommended by the CDC [24]. Briefly, 200 μl overnight culture cells were lysed and intact genomic DNA digested in agarose-embedded plugs with XbaI restriction enzyme. The digested DNA was then separated using a contour-clamped homogeneous electric field (CHEF DRIII, Bio-Rad Laboratories, Hercules, CA, USA) with the following conditions: 0·5× TBE (Tris–borate EDTA), 1% Seakem Gold agarose (FMC BioProducts, Rockland, ME, USA), 14°C, 6 V/cm for 17 h with switch times ranging from 2·2 to 63 s. S. enterica serovar Braenderup ‘Universal Marker’ (kindly provided by Dr Leslie Wolf, NCSLPH, NC, USA) was used as the reference marker. Gels were stained with ethidium bromide for 30 min, destained three times for 20 min each with distilled water and photographed using Alpha imager (Alpha Innotech Corporation, San Leandro, CA, USA). Analysis of PFGE data was performed using Bionumerics software (Applied Maths, Kortrijik, Belgium) using ‘different bands’ algorithm for clustering and the unweighted pair group for arithmetic means (UPGMA) tree-building approach with optimization of 1 and 0·75% position tolerance. Visual inspection of the patterns was performed as a final step for analysis.
Rep–PCR genotyping
Template DNA was purified from overnight cultures using a Qiagen DNeasy Tissue kit, (Qiagen). The product was amplified using the Uprime-B1 primers in the repPRO–PCR DNA fingerprinting kit (Bacterial Barcodes, Houston, TX, USA). Cycling parameters included an initial denaturation step at 95°C for 2 min. This was followed by the denaturation cycle at 94°C for 3 s and ramping to 92°C for 30 s. The annealing temperature used was 50°C for 1 min leading into the extension step at 65°C for 8 min. The last three steps were repeated 31 times. There was a final extension step at 65°C for 8 min. The PCR products were analysed by electrophoresis for 1 h using 1·5% agarose gel containing 3 μg/ml ethidium bromide in 1× TAE buffer.
Analysis of phenotypes, genotypes and discriminatory power
Pearson product-moment correlation coefficient (r) was used to compute correlation between predominant MDR patterns and phage types. DNA fingerprints were analysed using the Bionumerics software (Applied Maths). We used 1% optimization for further adjustment as recommended once the background noise was subtracted and the gels were normalized. We used Pearson pair-wise comparisons of band patterns calculated by an index of genetic similarity using the simple matching coefficient. Cluster analysis was carried out with similarity estimates using UPGMA, as recommended previously [Reference Hall25] which also proved consistent in our analysis. The reproducibility of all three genotyping methods was determined from triplicate loadings of 32 isolates.
We applied Simpson's index of diversity to compare the discriminatory power of each of the three genotyping systems used in this study. The formula used to calculate the index of discrimination (DI) was as described previously [Reference Hunter and Gaston26] and is shown here:
where DI=index of discrimination, N=total number of strains in the sample population, S=the total number of types described, n j=the number of strains belonging to the jth type.
The DI value shows the probability that two unrelated strains sampled from the test population would be placed into different cluster types. This value ranges between 0·0 and 1·0. The higher the index, the more discriminatory the method is.
Results
Evaluation of the three genotyping methods and the DI
Using 40 randomly chosen isolates from the pool of 196 isolates, three genotyping methods: AFLP, PFGE and Rep–PCR were evaluated (Fig. 1). Simpson's DI of each of the methods was calculated. AFLP was found to have the highest DI at 0·939 followed closely by PFGE with a DI of 0·925. The Rep–PCR had the lowest DI value of 0·421. No noticeable difference in reproducibility or cost was found. AFLP was superior in its throughput (64 samples processed at a time with a potential to process up to 96 samples) and resolution with the detection of single base-pair differences with a range of 200–500 bp fragment size.
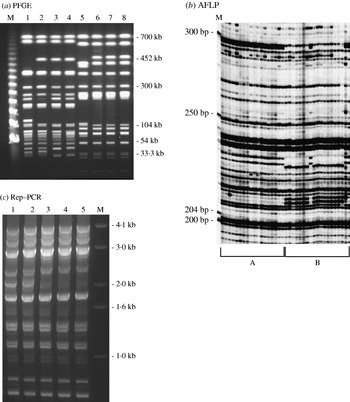
Fig. 1. DNA fingerprint patterns of S. Typhimurium isolates using the three genotyping methods evaluated: (a) PFGE macrorestriction of genomic DNA. Lanes 1–4, S. Typhimurium DT193 isolates with AmKmStSuTe resistance pattern; lanes 5–8, DT104 isolates with AmCmStSuTe resistance pattern are shown. (b) Section of AFLP gel: group A, isolates with AmCmStSuTe pattern and group B, isolates with AmKmStSuTe pattern are shown; (c) Rep–PCR. Lanes 1 and 2 are S. Typhimurium DT104 isolates with AmCmStSuTe resistance pattern; lanes 3–5, DT193 isolates with AmKmStSuTe resistance pattern are shown. Lanes marked as ‘M’ indicate the molecular marker lanes in all the three gels.
Based on these findings, we adopted AFLP as the primary genotyping method for this epidemiological study to determine the phenotypic and genotypic diversity of 196 S. Typhimurium isolates. To appreciate the discriminatory power of the phenotyping and genotyping methods, we compared the DI of AFLP to that of two phenotyping methods (antimicrobial resistance pattern and phage typing) that were used together with AFLP to characterize all the 196 isolates. AFLP produced 16 clonal clusters with the largest cluster type containing 11·9% of all the strains tested (cluster type 11) (Table). Phage typing had a discriminatory power of 0·628, and produced seven phage types, with 55·4% of all isolates in the largest cluster (DT104). Antimicrobial resistance pattern analysis had the lowest discriminatory index, 0·579, and classified the strains into 12 antimicrobial resistance patterns, with 56·4% of isolates in the largest cluster (AmCmStSuTe).
Table. Clonal cluster types identified from 196 S. Typhimurium isolates from 18 pig farms of two commercial production system located in eastern North Carolina using AFLP genotyping
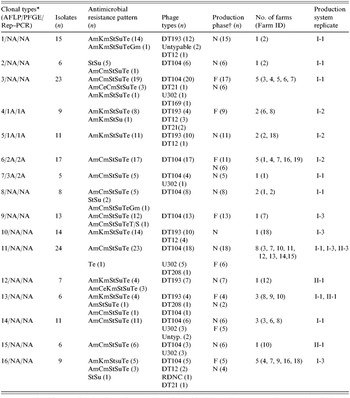
AFLP, Amplified fragment length polymorphism; PFGE, pulsed-field gel electrophoresis; Rep–PCR, repetitive palindromic extragenic–polymerase chain reaction.
* Clonal types for each of the three fingerprinting methods are indicated. AFLP was done for all the 196 isolates. However, only a subset of these (n=40) were analysed with PFGE and Rep–PCR. NA indicates that no isolate from the respective AFLP clonal type was tested by the other two methods.
† N, nursery; F, Finisher.
Genotypic diversity of S. Typhimurium in the two production systems
Overall, 184 of the isolates clustered into 16 distinct AFLP clonal types at an arbitrary threshold break-point of 70% genetic similarity index (Table). The remaining 12 isolates did not cluster with any of the groups and, thus, were considered as sporadic clones and were not included in the Table. Despite the fact that the two major commercial production systems were located in the same geographical area within eastern North Carolina, only two of the 16 clonal types (11 and 13) were shared between the strains of S. Typhimurium identified from the two production systems. On the other hand, within a production system, a widespread similarity of isolates was detected between the two production phases and among the three sampling periods. Clonality of isolates between farms was noted as nine of the 16 clonal clusters were identified from more than one farm each, and six of these groups were identified from more than two farms (Table). Six of the 16 cluster types were identified from both nursery and finishing farms while the remaining 10 were found in one of the production phases but not in both. Clonal type 11 was the most common and widespread both temporally and spatially (Table). This clonal subtype was composed mainly of phage type DT104 isolates (18/24).
Genotypic investigation of isolates collected from nursery farms (n=119) revealed that even though 14 of the 16 cluster types were detected in the nursery phase, 40% of the cluster types were specific to one farm, with little or no detection of similar strains among nursery farms. This may imply that horizontal spread of clonal strains among nursery farms is not common. On the other hand, among isolates from finishing farms (n=77), eight different cluster types were detected (Table). At the finishing stage, in contrast to the nursery farms, only two of the cluster types (types 9 and 16) were unique to a single farm. The remaining six cluster types (75%) were detected on more than one farm, implying a greater likelihood of horizontal spread at the finishing stage.
Antimicrobial resistance patterns, phage types and genotypic clonal types
All 196 S. Typhimurium isolates showed resistance to one or more of the antimicrobials tested. A total of 12 antimicrobial resistance patterns were exhibited (Table). One isolate exhibited resistance to tetracycline alone. Two pentaresistance patterns, AmCmStSuTe (109/196) and AmKmStSuTe (64/196) were found to be the most common. The remaining isolates also showed various resistance patterns at different frequencies including: StSu (n=10), AmCeCmStSuTe (n=3), AmCeKmStSuTe (n=3), StTe (n=1), AmCmStSuTeGm (n=1), AmKmStSuTeGm (n=1), AmKmStSu (n=1), AmStSuTe (n=1) and AmCmStSuTeT/S (n=1).
Phage typing grouped the 196 S. Typhimurium isolates into seven phage types. The two most common phage types detected were DT104 (108/196) and DT193 (49/196). As depicted in the Table, the other less common phage types detected include DT12 (n=13), U302 (n=13), DT21 (n=4), DT208 (n=2) and DT169 (n=1). Five isolates were untypable and one isolate was found to be reactive but did not conform with any known type (RDNC).
Eight of the AFLP clonal types were made up predominantly of isolates that exhibited the AmCmStSuTe resistance pattern. The most common and widespread clonal type in this study (cluster 11) was composed predominantly of phage type DT104. As depicted in Figure 2, 108 isolates of DT104 were analysed using AFLP and nine clonal cluster types were found (Fig. 2, Table).
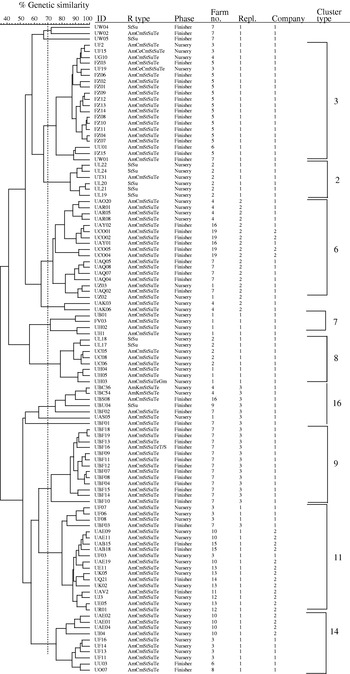
Fig. 2. Dendrogram representing AFLP fingerprints of 108 Salmonella serovar Typhimurium phage type DT104 isolates. The nine predominant AFLP clonal types as categorized in the Table are represented (3, 2, 6, 7, 8, 16, 9, 11 and 14).
The occurrence of this phage type was very highly correlated with this penatresistance pattern (r=0·916, P<0·001). The remaining one cluster type, clonal type 2, was composed mainly of DT104 isolates with StSu resistance pattern. Detailed molecular characterization of these non-pentaresistant DT104 strains has been reported previously [Reference Gebreyes and Altier3]. In contrast to previous reports, using other genotyping methods, that described DT104 as clonally disseminated with limited genotypic diversity [Reference Casin, Breuil, Brisabois, Moury, Grimont and Collatz27], AFLP was able to detect distinct clonal types within DT104. On the other hand, isolates of the phage types DT104 and U302 (both commonly exhibiting AmCmStSuTe pattern) were found in the same cluster type, as shown in the Table. These two phage types appeared together in five of the nine cluster types (clusters 3, 7, 11, 14 and 15) suggesting a close genetic relationship in addition to their phenotypic similarity.
The second most common phage type, also of public health importance, identified from the pigs in this study was DT193. Among the 48 isolates further investigated for genotypic diversity using AFLP, six clonal clusters were identified (Fig. 3). Historically, isolates of phage type DT193 are known to have a high degree of genetic diversity [Reference Hampton, Threlfall, Frost, Ward and Rowe28]. As shown in the Table, four of the six cluster types predominantly composed of DT193 also contained phage type DT12. DT12 phage type isolates also commonly exhibited the same MDR pattern, AmKmStSuTe, to that of DT193. This finding suggests the close phenotypic and genotypic relationship between phage types DT193 and DT12. The AmKmStSuTe resistance pattern, although highly correlated with DT193 (r=0·859, P=0·003), was also exhibited by other relatively rare phage types including DT21 and DT208. Based on fingerprinting, DT21 and DT208 were, however, more often clonally related to DT104 than DT193 (clonal types 3, 11, and 16).
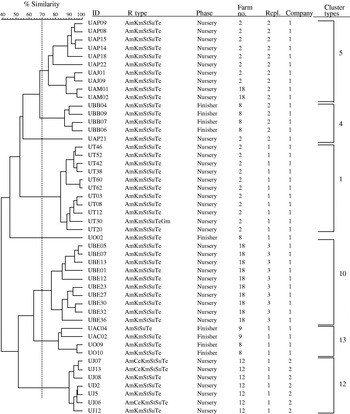
Fig. 3. Dendrogram representing AFLP fingerprints of 48 Salmonella serovar Typhimurium phage type DT193 isolates. The six predominant cluster types as categorized in the Table are represented (5, 4, 1, 10, 13 and 12).
Discussion
North Carolina is the second largest pig producing state in the United States with more than 10 million head marketed each year. Modern swine production units are concentrated in few counties within the eastern part of the state. Higher prevalence of Salmonella among pigs in modern commercial facilities in this region has been reported previously [Reference Funk, Davies and Gebreyes19, Reference Davies, Bovee, Funk, Jones, Morrow and Deen21]. The modern pig production systems maintain closed herds in a pyramid form with all-in/all-out, multi-site production units. One major advantage of such production systems is that they limit exchange of infectious agents among production systems. Even though cross contamination of pigs and similarity of Salmonella strains in holding pens at slaughter has been described in recent years [Reference Hurd, McKean, Wesley and Karriker17, Reference Gebreyes, Davies, Turkson, Morrow, Funk and Altier18], there is a very limited knowledge as to the clonality or diversity, both at phenotypic and genotypic levels, of Salmonella strains within and among production systems on farms. We reported previously that antimicrobial use in the study groups was associated with the frequency of the serovar Typhimurium as well as antimicrobial resistance mainly to tetracycline and β-lactam class antimicrobials. Antimicrobial use as a feed additive was more common in production system 1 [Reference Gebreyes, Davies, Funk, Altier and Thakur4].
Molecular methods with high discriminatory power are essential to differentiate among bacterial isolates of clonal descent such as strains of S. Typhimurium. Therefore, in the present study, we evaluated three genotyping methods: PFGE, AFLP and Rep–PCR. These methods have previously proven useful to characterize foodborne pathogens. PFGE has been used as a standard technique for subtyping foodborne pathogens in the United States [24]. The use of PFGE has been shown to have limited discriminatory power for some Salmonella serovars such as Enteritidis [Reference Liebana, Garcia-Migura, Breslin, Davies and Woodward29]. The other two methods employed in this study have been developed relatively recently. AFLP has been used since 1995 [Reference Vos, Hogers and Bleeker6] in eukaryotic and prokaryotic organisms and has gained acceptance in recent years as a useful tool for the discrimination of Salmonella [Reference Vos, Hogers and Bleeker6, Reference Gebreyes9, Reference Olive and Bean10, Reference Lindstedt, Heir, Vardund and Kapperud30]. These three methods have also recently been evaluated in the epidemiological investigation of E. coli O157:H7 [Reference Hahm, Maldonado, Schreiber, Bhunia and Nakatsu31]. Although that study identified PFGE as the most discriminatory, it recommended the other techniques as well; AFLP for high throughput and Rep–PCR for its consistency. It should also be noted that AFLP and Rep–PCR have a relatively lower reproducibility than PFGE. This is mainly since these two methods employ PCR as one of the steps in the genotyping process. The use of PCR increases the likelihood of non-specific band amplification due to potential contaminants that in turn affects the reproducibility.
Under field conditions, using AFLP, Tamada et al. also reported a considerable degree of genetic diversity within Typhimurium isolates [Reference Tamada, Nakaoka and Nishimori32]. On the other hand, a high degree of clonality among Typhimurium isolates collected from different regions has also been reported [Reference Walker, Lindsay, Woodward, Ward and Threlfall33]. Several investigators have carried out population genetic analyses of Salmonella serovars. Beltran et al. [Reference Beltran, Musser and Helmuth12] determined the genetic structure of natural populations by multi-locus enzyme electrophoresis and the relationships of antigenic variations using 23 enzyme-encoding chromosomal genes. The study revealed that the genetic structure of natural populations of Salmonella to be highly clonal with most isolates existing as clones with worldwide distribution. Several genotypic studies performed on MDR Salmonella, mainly Typhimurium phage type DT104, also report this phage type as being clonally disseminated [Reference Murphy, McNamara and Hill34, Reference Rubino, Muresu and Solinas35]. In the present study, AFLP was found to have the highest discriminatory index of 0·939. With no noticeable difference in reproducibility and with its advantage in high throughput and resolution, we employed AFLP as a method of choice to subtype the S. Typhimurium strains.
In this study, two important phenotyping methods, antimicrobial resistance pattern and phage typing, were also used in support of the AFLP genotyping for the epidemiological investigation of S. Typhimurium in pigs. Although these phenotyping methods usually have a lower discriminatory power, they have proven useful in identification of outbreaks as well as in understanding the extent of antimicrobial resistance among Salmonella isolates [Reference Ward, Threlfall and Van der Wolf36]. The use of phenotypic approaches in addition to genotyping has often been recommended as some genetic clones may show distinctly different phenotypes [Reference Gebreyes9, Reference Pontello, Sodano and Nastasi14–Reference Weigel, Qiao, Barber, Teferedegne, Kocherginskaya and Van der Wolf16, Reference Liebana, Garcia-Migura, Breslin, Davies and Woodward29]. This was also demonstrated in the present study; phage types DT104 and U302, in addition to DT193 and DT12 were clustered within the same genotypic clonal types. We acknowledge the fact that microbiological approaches have the inherent limitations that the strains collected from these farms do not represent the overall strain population in the pig groups. However, we also believe that valuable data on the clonality and diversity of these strains can be generated without extensive extrapolation using these phenotypic and genotypic approaches.
One predominant phage type of importance that is known to show MDR is DT104. Since it was first recognized in the United Kingdom in 1984 [Reference Threlfall, Frost, Ward and Rowe8, Reference Threlfall, Ward, Skinner and Rowe37], it has been identified essentially in all continents and various host species [Reference Glynn, Bopp, Dewitt, Dabney, Mokhtar and Angulo7, Reference Threlfall, Frost, Ward and Rowe8, Reference Threlfall, Ward, Skinner and Rowe37, Reference Kariuki, Gilks, Kimari, Muyodi, Waiyaki and Hart38]. This phage type was identified as the most common in pigs in the present study. Most DT104 strains have previously been shown to exhibit pentaresistance with the AmCmStSuTe pattern [Reference Glynn, Bopp, Dewitt, Dabney, Mokhtar and Angulo7, Reference Threlfall, Frost, Ward and Rowe8, Reference Threlfall, Ward, Skinner and Rowe37, Reference Kariuki, Gilks, Kimari, Muyodi, Waiyaki and Hart38]. Similar to these previous reports, we found a very high correlation (r=0·916, P[0·001) between AmCmStSuTe resistance and occurrence of DT104. We also detected DT104 isolates with other resistance patterns. Of particular interest were isolates that showed resistance only to StSu (n=10). Although rare, they represented an important phenomenon in that they were genotypically closely related to the pentaresistant DT104 isolates (clusters 2, 8 and 16). This finding also shows the importance of using genotypic data in addition to phenotypic findings to identify and further characterize clonal strains.
In this study, we were able to identify 16 AFLP clonal types among the total of 196 Typhimurium isolates. One striking finding was that isolates from the two major pig production systems were for the most part dissimilar, despite the fact that all of the 18 farms that belong to two production systems were located within the same geographic area. Only two of the clonal types were common to both production systems. One major factor that this rare clonality of isolates between the two systems resulted in may be attributed to different management conditions. First, as stated earlier, the commercial pig production systems in the area use pyramid systems where pigs from the same breeding unit are moved into farrowing units that supply weaned pigs to specific nursery farms. Weaned pigs are then moved to finishing units at a different site before marketing at approximately 6 months of age. This closed pyramid system may help to control dissemination of infectious agents across production systems.
Conversely, clonality within a production system differed depending on the production phase. At the nursery, 40% of the clonal types detected were specific to one farm. On the other hand, 75% of the clonal types detected at finishing originated from more than one farm. This can be explained by random error or may also be attributed to the different management systems between nursery and finishing unit. Even though we did not have the specific management data for the groups included in this study, it is common that biosecurity measures are more stringent at nursery stage than at finishing. This is because nursery pigs tend to be more susceptible to various infectious agents once the maternal immunity wanes and pigs become stressed due to separation from the sow and the establishment of their dominance order in the nursery units. In addition, co-mingling of the pigs at the nursery level is not common and weaned pigs from specific farrowing units are moved into designated nursery sites. However, it is relatively more common that pigs from different nursery units are co-mingled at a finishing site.
In this study, we identified AFLP to be an invaluable genotyping method with superior discriminatory power that can differentiate among S. Typhimurium strains, particularly DT104. Unique clonal types were detected between the two major production systems based on the 196 samples collected from 18 farms. DT104 was the most common phage type isolated from 17 of the 18 farms. Such a high frequency of occurrence of MDR DT104 among apparently healthy pigs indicates the importance of pigs as reservoirs of Salmonella serovars of public health importance.
ACKNOWLEDGEMENTS
This work was supported by funding from FDA (FD-U-001880-01) and USDA-CSREES (2001-51110-11460). We thank Dr David White (FDA-CVM) and Dr Shelley Rankin (U. Penn.) for critical review of the manuscript.






