The authors apologise for errors that appear in the molecular phylogenetic analyses of the ND1 locus. The ND1 sequences of Brachylaima mesostoma, NCBI Acc. Nos. KT074969 and KT074970, were accidentally replaced with incorrect ones, which resulted in a slightly longer branch of the species in Fig. 2B and related supplementary material Fig. S4, inclusion of incorrect sequences in Table S5, different maximum likelihood fits in Table S9, and incorrect estimates of interspecific variability between B. mesostoma and other tested Brachylaimidae in Table 3. Please find below the corrected materials. The conclusions of the study are not affected by this correction.

Fig. 2. Maximum-likelihood analysis of sequences of mitochondrial DNA loci [CO1 (A) and ND1 (B)] of Brachylaimoidea.
The interspecific variability between B. mesostoma and the other species, reported initially in Table 3, was as follows: L. paradoxum 1.350 ± 0.301, L. vogtianum 1.491 ± 0.327, L. subtilis 1.337 ± 0.287 base differences per site generated by averaging over all sequence pairs between groups (means ± S.E.). There was no intraspecific variability between the two analyzed B. mesostoma sequences.
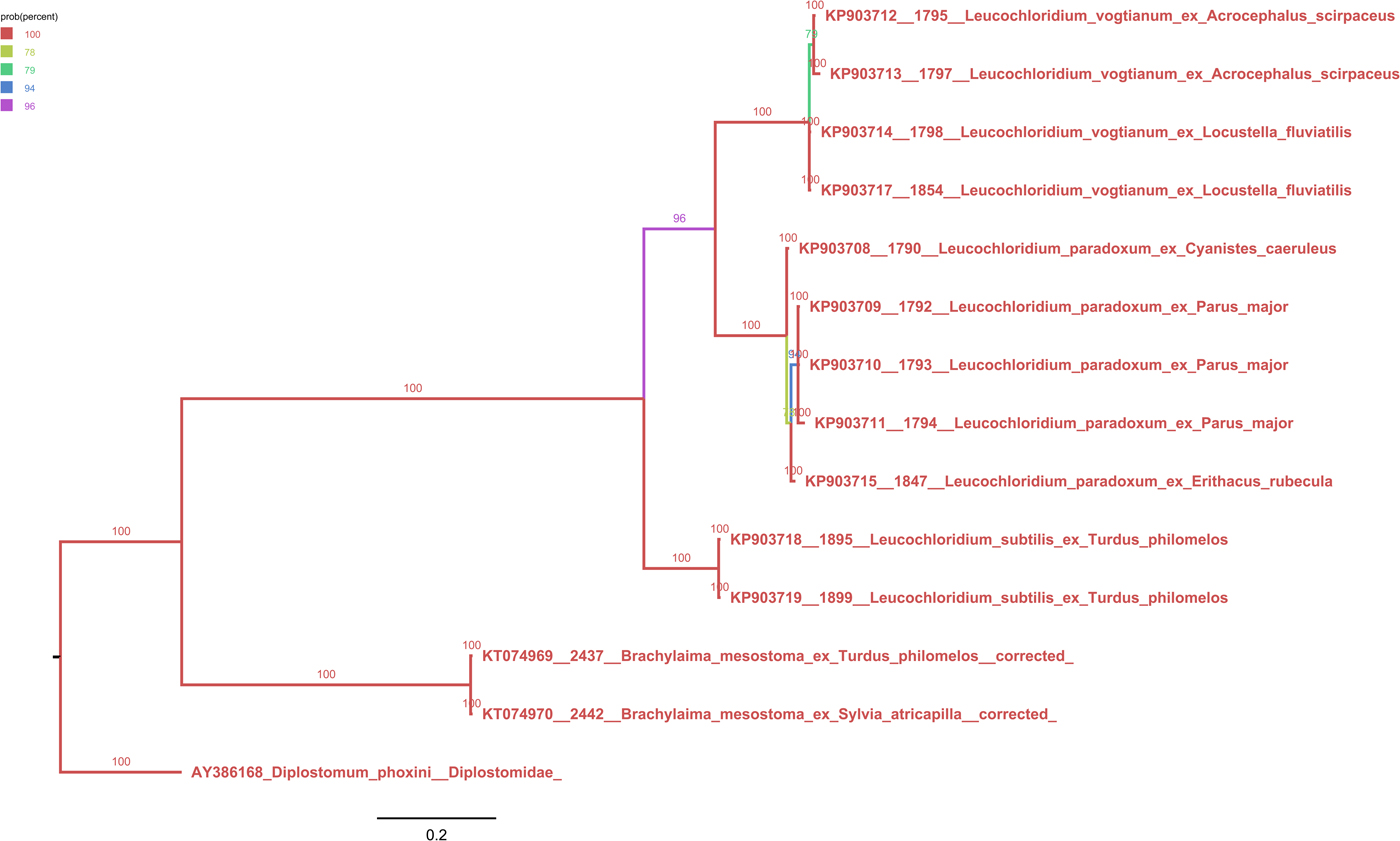
Figure S4. Phylogenetic tree (50% majority-rule consensus) based on Bayesian inference of Brachylaimoidea ND1 locus. Color of branches indicates Bayesian posterior probabilities. We obtained the following summary statistics for the for analysis performed: average standard deviation of split frequencies 0.0028, maximum standard deviation of split frequencies 0.0100, average potential scale reduction factor 1.005, and maximum potential scale reduction factor 1.008.
Table S5. Alignment of trimmed ND1 locus corresponding to nt. 25–424 (400 bp) of Leucochloridium paradoxum KP903708, which consisted of partial ND1 coding sequence.
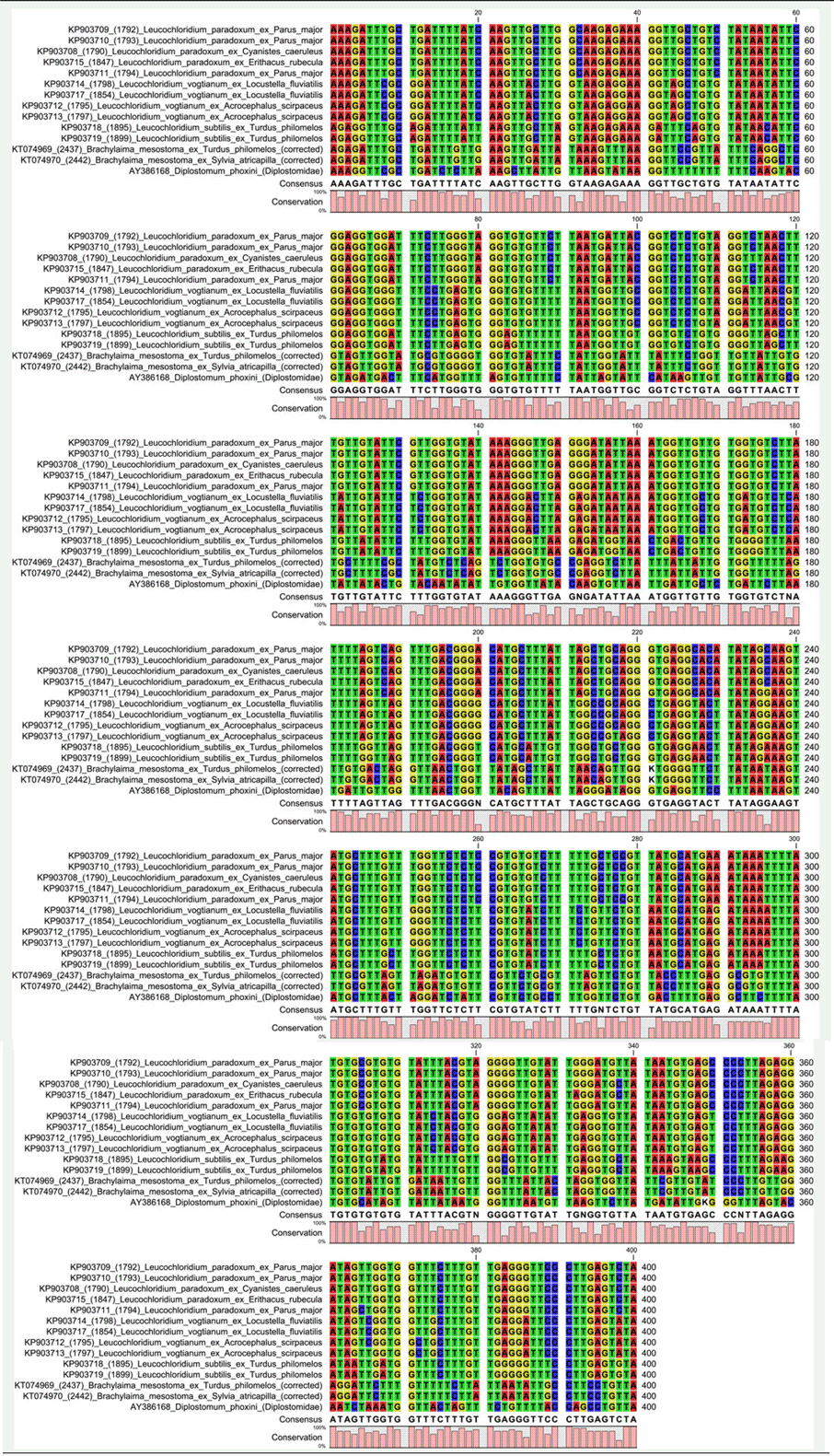
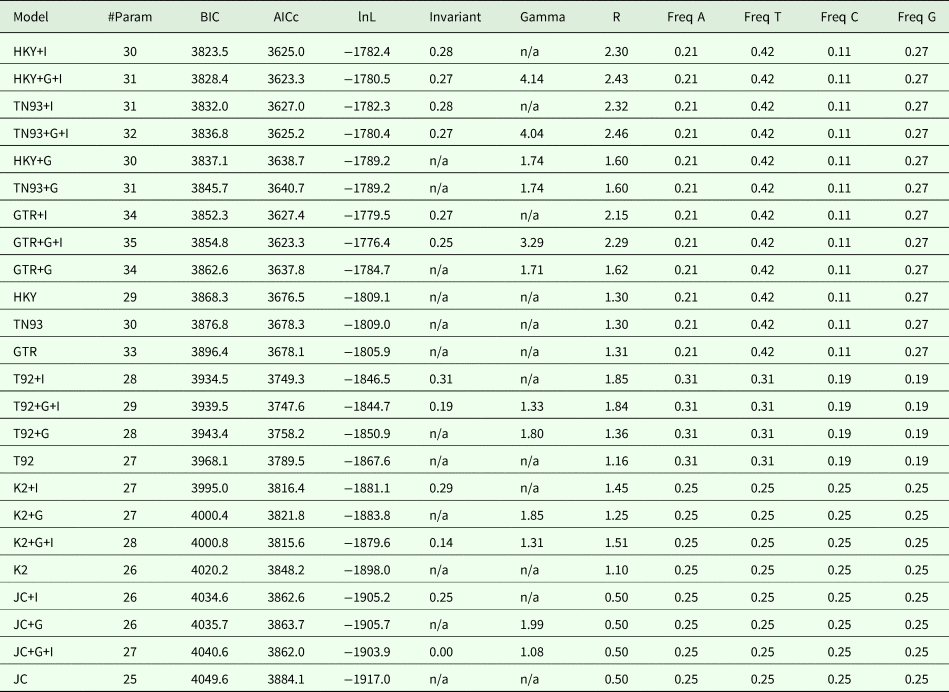
Table S9. Maximum likelihood fits of 24 nucleotide substitution models for the ND1 locus (14 sequences with a total of 398 positions in the final dataset), with all sites used for the analyses, including the gaps. For each model, we calculated the Bayesian information critetion, Akaike information criterion (corrected) and maximum likelihood values.
Acknowledgements
We thank Miroslav Těšínský for expert technical assistance.







