Streptococcus pneumoniae is a common human lung pathogen. It adheres to the surface mucosa of the nasopharynx by a direct interaction between the bacterial surface-associated proteins and the host epithelia. Interruption of this process has been suggested as a possible strategy for the prevention of pneumococcal disease [Reference Paton and Giammarinaro1]. Adherence to lung epithelia is mediated by choline-binding protein A (CbpA) and less specifically through pneumococcal surface antigen (PsaA) [Reference Ogunniyi, Giammarinaro and Paton2]. These choline-binding proteins are encoded by genes with similar signature sequence motifs [Reference Gosink3], and recently with the availability of total genome sequence data, other genes containing such motifs have been identified encoding choline-binding proteins D, E and G. Disruption of cbpD and cbpE by mutagenesis resulted in major loss of nasopharyngeal colonization in animal models while mutation of cbpG led to loss of adherence in vitro [Reference Gosink3]. A comparison of the available S. pneumoniae genome sequences revealed wide diversity between different serotypes [Reference Hava, LeMieux and Camilli4] and data on the distribution of the cbp genes in vaccine- and non-vaccine-related serotypes are lacking.
Some relationships between penicillin susceptibility and serotype were observed by Azoulay-Dupuis et al. [Reference Azoulay-Dupuis5] who found that strains of pneumococci from invasive infections were more frequently susceptible to penicillin and fell into serotypes 1, 3 and 4 whereas non-invasive strains were less susceptible to penicillin and were of different serotypes. As attachment to host epithelia is a prerequisite for the initiation of infection, the question arises as to whether invasive strains carry the genes for surface proteins to facilitate host cell adherence and whether these genes are also found in isolates from carriage sites. We report here the distribution of cbp genes in pneumococcal isolates from Kuala Lumpur and their correlation with vaccine serotypes (Pneumovax 23; Merck Sharp & Dohme, NJ, USA), penicillin susceptibility and clinical site of origin.
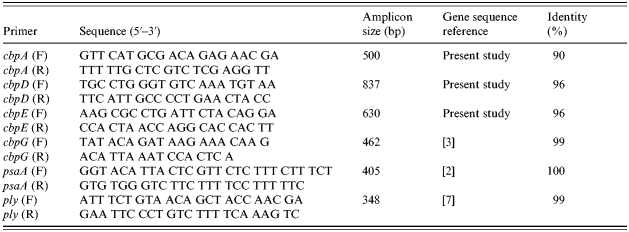
The following genes, cbpA, cbpD, cbpE, cbpG, and psaA, were assayed by PCR using the primers listed in Table 1 to screen 34 well-characterized clinical isolates of pneumococci collected in the year 1999/2000 at the University of Malaya Medical Centre, Kuala Lumpur [Reference Desa6]; ply was also included as it is highly specific for S. pneumoniae and confirmed identification of the species [Reference Salo, Ortqvist and Leinonen7]. Primer sequences were as previously published [Reference Ogunniyi, Giammarinaro and Paton2, Reference Gosink3, Reference Salo, Ortqvist and Leinonen7], except for cbpA, cbpD and cbpE that were designed to amplify a segment within the nucleotide length of the respective cbp genes. Pneumococcal genomic DNA was extracted from a cell suspension by boiling and the PCR cycling condition for all genes comprised one cycle at 94°C for 3 min, followed by 25 cycles of 94°C for 1 min, 52°C for 1 min and 72°C for 1·5 min, and a final elongation at 72°C for 10 min. The PCR products and 100 bp DNA ladder were subsequently run in 1% agarose gel with ethidium bromide incorporated. These products, ranging from 340 bp to 840 bp in size, were sequenced and compared to library sequences in Genbank (www.ncbi.nlm.nih.gov) to confirm their homology to reference sequences. Isolates were grouped into (1) vaccine and non-vaccine types (serotype present or absent from the 23-valent pneumococcal polysaccharide vaccine), (2) susceptibility to penicillin, and (3) invasive and non-invasive sites of isolation.
Table 1. Primer pairs used for amplification of gene fragments by PCR and percentage of identity of the amplified genes in Genbank
Genes cbpD, cbpE, psaA and ply were present in all 34 isolates whereas cbpA and cbpG were found in 47% and 59% of the isolates respectively. The distribution of the latter genes, either singly or in combination, in the three categories of isolates was tested for statistical significance (P<0·05) by χ2 test (Table 2). Isolates belonging to vaccine types were more likely to carry cbpA than non-vaccine types (69% vs. 31%) and this proportion was increased when both genes were present (77% vs. 23%). There was little difference in distribution of these genes in isolates of variable penicillin susceptibility. However, most non-invasive isolates harboured cbpA but less frequently cbpG. None of the differences reached statistical significance.
Table 2. Distribution of cbpA and cbpG according to vaccine and non-vaccine types, penicillin susceptibilities, and invasive and non-invasive sites of the isolates
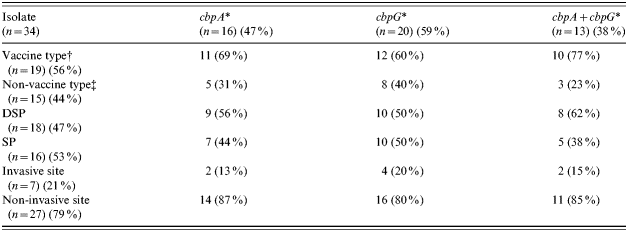
* Detected by PCR; Vaccine/non-vaccine type=serotypes included/not included in the 23-valent Pneumovax vaccines; DSP/SP=decreased susceptibility to penicillin/susceptible to penicillin; Invasive/non-invasive site=sample from sterile (e.g. blood)/non-sterile (e.g. sputum) sites.
† Serotype 1 (n=3), 7F (n=2), 14 (n=1), 19F (n=12), 23F (n=1).
‡ Serotype/group 6A (n=4), 7B/C (n=1), 15A (n=2), 15C (n=3), 16/36/37 (n=2), 23A (n=2), 23B (n=1).
In conclusion we observed differences in the distribution of genes encoding two choline-binding proteins of pneumococci particularly in serotypes found in the current 23-valent pneumococcal vaccine. However, the significance of these differences in terms of pathogenesis of the organisms remains unknown. This might suggest that the two genes are inherent in some serotypes and unrelated to virulence or penicillin susceptibility, however, a larger collection of isolates needs to be investigated. CbpA is the most abundant of surface structures found in streptococcal species [Reference Gosink3] and was present in pneumococci exhibiting increased binding to a human pneumocyte II cell line (A549) compared to poorly binding isolates [Reference Robson, Reed and Horvat8]. It was also reported that cbpA was not present in all pneumococcal isolates [Reference Paton and Giammarinaro1] as confirmed in this study. Genes psaA and ply have been long recognized in the majority of pneumococcal isolates but information on recently identified genes such as cbpD, cbpE and cbpG is lacking. The roles of encoded proteins are currently under investigation in a limited number of pneumococcal strains. Some studies have suggested that CbpD and CbpE may be associated with hydrolase activities [Reference Hammerschmidt9] whereas CbpG is a serine protease with adhesive properties [Reference Mann10]. How such functions relate to nasopharyngeal colonization and adherence or whether cbpA and cbpG act in concert to increase the level of virulence, remain unclear. A shortcoming of this study is that it represents a small number of isolates from a restricted geographical location and a wider collection of pneumococcal isolates from other locations is warranted to establish unequivocally any relationship between the presence of these genes and pathogenic potential.
ACKNOWLEDGMENTS
This study was supported by the University of Malaya Fundamental Grant FP032/2005D.
DECLARATION OF INTEREST
None.




