Introduction
Although tRNAs are smaller than the 40 kDa limit for passive diffusion, the export of tRNAs through the nuclear pore complex (NPC) is an active process facilitated by export factors belonging to the karyopherin-β family called exportins. It is now well accepted that tRNA subcellular trafficking is not unidirectional from the site of transcription in the nucleus to the cytoplasmic site of protein synthesis. Interestingly, tRNAs can traffic from the cytoplasm back to the nucleus via the tRNA retrograde pathway and then again be re-exported to the cytoplasm (for a recent review, see Hopper and Nostramo, Reference Hopper and Nostramo2019). The tRNA retrograde pathway was discovered in yeast, where intron-containing tRNAs travel across the nuclear pore complex to the outer mitochondrial surface where the splicing endonuclease complex is localized (Yoshihisa et al., Reference Yoshihisa, Yunoki-Esaki, Ohshima, Tanaka and Endo2003; Shaheen and Hopper, Reference Shaheen and Hopper2005; Takano et al., Reference Takano, Endo and Yoshihisa2005). Once spliced in the cytoplasm, tRNAs travel back to the nucleus to be further modified. They are then re-exported to the cytosol, where they participate in protein synthesis (Yoshihisa et al., Reference Yoshihisa, Ohshima, Yunoki-Esaki and Endo2007). This ‘shuttling’ mechanism has been documented in several model organisms, including humans, but its biological significance is poorly understood. However, in the yeast Saccharomyces cerevisiae, tRNA retrograde import was proposed as a level of tRNA quality control, which monitors both the end processing and modification state of tRNAs (Kramer and Hopper, Reference Kramer and Hopper2013).
A key players in the nuclear tRNA export/import are exportins Los1 and Msn5 and their homologs in vertebrates exportin-t (Xpo-t) and exportin 5 (Xpo-5) (Okamura et al., Reference Okamura, Inose and Masuda2014). As shown in previous work (Calado et al., Reference Calado, Treichel, Müller, Otto and Kutay2002) and by a recent comprehensive study employing a co-immunoprecipitation approach, these two proteins serve overlapping but distinct roles in tRNA nuclear export (Huang and Hopper, Reference Huang and Hopper2015). Los1 interacts with both spliced and unspliced tRNAs, regardless of whether they are aminoacylated or not, implying that Los1 participates in primary nuclear export and re-export of tRNA to the cytosol. Whereas, Msn5 preferentially binds with spliced and aminoacylated tRNAs establishing its role in tRNA nuclear re-export (Huang and Hopper, Reference Huang and Hopper2015). In addition, translation elongation factor 1 α was identified in a complex with Msn5, possibly providing the specificity of Msn5 for aminoacylated tRNAs (Calado et al., Reference Calado, Treichel, Müller, Otto and Kutay2002; Huang and Hopper, Reference Huang and Hopper2015). In contrast, vertebrate Xpo-5 preferentially exports miRNA, and thus its role in tRNA nuclear export is assumed to be minor (Calado et al., Reference Calado, Treichel, Müller, Otto and Kutay2002). Despite the prominent role of Los1 and Msn5 in the translocation of tRNAs across the NPC, they cannot be the only nuclear exporters since double mutants of these two proteins are viable, suggesting additional export pathways exist and remain uncharacterized (Huang and Hopper, Reference Huang and Hopper2015). Recently, a genome-wide screen in yeast revealed new possible players in the tRNA nuclear export (Wu et al., Reference Wu, Bao, Chatterjee, Wan and Hopper2015). This includes proteins described for their function in rRNA, mRNA and protein export. One of these candidates is represented by the heterodimeric complex of Mex67-Mtr2, well characterized for their essential role in mRNA nuclear export. Inactivation of Mex67-Mtr2 leads to a rapid accumulation of end matured unspliced tRNAs in the nucleus supporting their co-function with Los1 in the primary export pathway. Surprisingly, only four out of ten intron-containing tRNAs were retained in the nucleus, which suggests substrate preference (Chatterjee et al., Reference Chatterjee, Majumder, Wan, Shah, Wu, Huang and Hopper2017).
Nuclear export of tRNAs has been extensively studied in several model systems, yet there are still key factors missing and this essential pathway is not fully understood. Compared to other eukaryotes, kinetoplastid parasites show unusual features in nuclear and organellar gene expression. These include processes such as mitochondrial RNA editing, mitochondrial tRNA import and trans-splicing of polycistronic mRNAs. There is also a general lack of transcriptional promoters; consequently, gene expression in trypanosomatids is controlled mostly by posttranscriptional pathways (Daniels et al., Reference Daniels, Gull and Wickstead2010). Nucleo-cytoplasmic tRNA pools may change under certain conditions such as nutrient starvation and oxidative stress (Shaheen and Hopper, Reference Shaheen and Hopper2005; Whitney et al., Reference Whitney, Hurto, Shaheen and Hopper2007; Chafe et al., Reference Chafe, Pierce, Eswara, McGuire and Mangroo2011; Dhakal et al., Reference Dhakal, Tong, Anderson, Kashina, Cooperman and Bau2019; Schwenzer et al., Reference Schwenzer, Jühling, Chu, Pallett, Baumert, Maini and Fassati2019). The trafficking of tRNAs between the nucleus and the cytoplasm might be an additional posttranscriptional event involved in gene regulation with a great importance in the complex life cycle, as these parasites have to face completely different environments with distinct sources of nutrients during the transition between the mammalian and insect stages (Fenn and Matthews, Reference Fenn and Matthews2007).
Whereas mechanisms for rRNA and mRNA transport in these parasites have been described, there is only limited knowledge about tRNA nuclear export (Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013; Bühlmann et al., Reference Bühlmann, Walrad, Rico, Ivens, Capewell, Naguleswaran, Roditi and Matthews2015). Recent findings indicated that similar to other eukaryotes, the canonical nuclear tRNA exporters TbXpo-t and TbXpo-5 are not singularly essential for cell viability in Trypanosoma brucei (Hegedűsová et al., Reference Hegedűsová, Kulkarni, Burgman, Alfonzo and Paris2019). Yet, contrary to yeast, downregulation of both exportins did not result in nuclear accumulation of mature tRNAs, nor did it abolish the export of intron-containing tRNA. With the goal to identify an alternative pathway, the general mRNA exporters TbMex67-TbMtr2 were downregulated, which resulted in a significant increase of nuclearly localized tRNAs. However, contrary to yeast, TbMex67 and TbMtr2 accumulated different subsets of tRNAs in the nucleus. While the elimination of TbMtr2 prevented the export of all tRNAs tested (except for the only intron-containing tRNATyr), the silencing of TbMex67 resulted in nuclear accumulation of tRNAs modified with queuosine (Q). In turn, inhibition of tRNA nuclear export also affected the levels of queuosine tRNA modification (Hegedűsová et al., Reference Hegedűsová, Kulkarni, Burgman, Alfonzo and Paris2019). An overlapping and different role of Mex67 and Mtr2 was also suggested for budding yeast. This is however a different matter, considering that S. cerevisiae lacks the gene for Q-tRNA modification enzyme (Nostramo and Hopper, Reference Nostramo and Hopper2020). These data demonstrate the dynamic nature of tRNA trafficking depending on their modification status and vice versa.
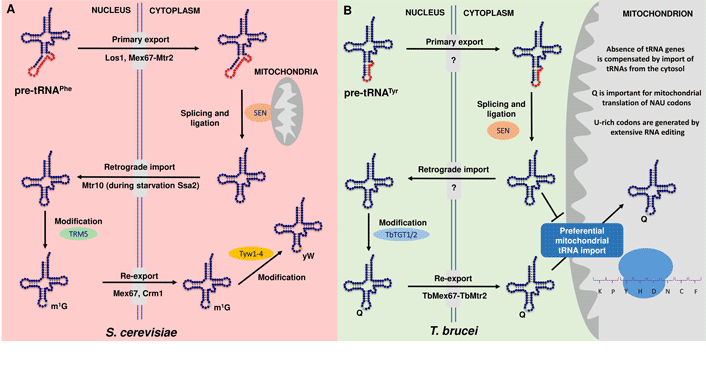
Along these lines, in yeast, the process of the retrograde transport pathway proved necessary for 1-methylguanosine (m1G) formation at position 37 of tRNAPhe, a first step in the synthesis of the hypermodified nucleotide wybutosine (yW). This is because the first step of yW is catalysed by Trm5 methyltransferase that acts only on spliced tRNAs and has a nuclear localization (Ohira and Suzuki, Reference Ohira and Suzuki2011). Consequently, tRNAPhe must be first exported to the cytoplasm by the primary nuclear export pathway to be spliced on the surface of mitochondria, where the tRNA splicing endonuclease is tethered (Fig. 1A). After intron removal, tRNA travels back to the nucleus with the help of the protein Mtr10 to get m1G37 and finally is re-exported to the cytoplasm where the remaining four enzymes (Tyw1-4) for wybutosine biosynthesis reside (Ohira and Suzuki, Reference Ohira and Suzuki2011; Nostramo and Hopper, Reference Nostramo and Hopper2020).

Fig. 1. Conservation and diversity of retrograde nuclear trafficking in S. cerevisiae and T. brucei. (A) Model of the retrograde nuclear transport of tRNAPhe in S. cerevisiae. Transfer RNAs are synthesized as primary tRNAs (pre-tRNAs) in the nucleus and undergo 5′ and 3′processsing, modifications and CCA addition . In this example, pre-tRNAPhe is subsequently exported from the nucleus to the cytoplasm by Los1 and Mex67-Mtr2 in a step called primary nuclear export. tRNAPhe contains an intron, which is removed by splicing endonuclease (SEN) located at the surface of the mitochondria. Spliced tRNA is then modified by several modification enzymes (not shown) and trafficked back to the nucleus with the help of Mtr10 by a process termed tRNA retrograde import. Ssa2 is also involved in this process only under amino acid starvation. In the nucleus, tRNAPhe is the substrate for the methyltransferase Trm5, which methylates G37 (m1G) (in green). In the final step, both Mex67 and Crm1 mediate the constitutive re-export of tRNAPhe to the cytoplasm, where wybutosine (yW) (in yellow) is added to m1G in a sequential series of reactions by Tyw1-4. Notably, the canonical exporters Los1 and Msn5 are dispensable in this transport step (Chatterjee et al., Reference Chatterjee, Nostramo, Wan and Hopper2018; Hopper and Nostramo, Reference Hopper and Nostramo2019; Nostramo and Hopper, Reference Nostramo and Hopper2020). (B) A model for subcellular trafficking and maturation of the tyrosyl-tRNA (tRNATyr) in T. brucei. The tRNA is transcribed in the nucleus containing an 11-nucleotide long intron. In the nucleus, the intron undergoes non-canonical editing prior to the primary export in the cytoplasm (not shown). Only the edited intron-containing tRNATyr is spliced by the SEN complex. After cleavage, tRNATyr undergoes retrograde transport to the nucleus to get modified with queuosine (Q) (in blue) by the nuclear enzyme TbTGT1/2. Finally, Q-containing tRNATyr is re-exported by TbMex67–TbMtr2 to cytoplasm to serve in cytoplasmic translation. Compared to approximately 50% of Q-containing tRNATyr in the cytosol, mitochondria of T. brucei contain nearly fully modified tRNATyr, which could be explained by its preferential import from the cytosol, possibly to play a role in the translation of U-rich tRNAs. The question mark stands for unknown transporter. Note: Except for the only intron-containing (tRNATyr), TbMtr2 serves as a general exporter in the primary tRNA export, while TbMex67 is responsible for the nuclear export of Q-modified tRNAs (Kessler et al., Reference Kessler, Kulkarni, Paulines, Rubio, Limbach, Paris and Alfonzo2017; Hegedűsová et al., Reference Hegedűsová, Kulkarni, Burgman, Alfonzo and Paris2019; Kulkarni et al., 2021, under revision in NAR).
An analogous pathway affecting queuosine (Q) modification of the anticodon of tRNATyr in T. brucei was reported (Kessler et al., Reference Kessler, Kulkarni, Paulines, Rubio, Limbach, Paris and Alfonzo2017) (Fig. 1B), where like in yeast, tRNA splicing occurs in the cytoplasm (Yoshihisa et al., Reference Yoshihisa, Yunoki-Esaki, Ohshima, Tanaka and Endo2003). Notably, the tRNA-guanine transglycosylase (TGT), the modification enzyme responsible for Q-tRNA formation, resides in the nucleus and it is not able to add Q to an intron-containing tRNA. Therefore, after transcription, processing of the 5′ and 3′ ends and non-canonical intron editing (Rubio et al., Reference Rubio, Paris, Gaston, Fleming, Sample, Trotta and Alfonzo2013), the intron-containing tRNATyr is exported from the nucleus to be spliced in the cytoplasm by SEN (complex of tRNA splicing endonuclease). After ligation of both exons, tRNATyr is imported back to the nucleus to obtain Q and subsequently re-exported to the cytoplasm to fulfil its function in protein synthesis (Kessler et al., Reference Kessler, Kulkarni, Paulines, Rubio, Limbach, Paris and Alfonzo2017; Kulkarni et al., 2021, under revision in NAR).
However, the situation in T. brucei is even more complicated given the fact that the mitochondrial genome is entirely devoid of tRNA genes and all tRNA molecules in the cell have a nuclear origin (Tan et al., Reference Tan, Pach, Crausaz, Ivens and Schneider2002). Surprisingly, compared to approximately 50% of Q-containing tRNATyr in the cytosol, the level of Q modification in the mitochondria is almost 100%, which could be justified by the preferential import of Q-tRNAs and also their ability to translate the mitochondrial predominantly U-rich mRNAs resulting from U-insertion editing (Kulkarni et al., unpublished manuscript, under revision in NAR) (Fig. 1B). To elucidate the mechanism of mitochondrial tRNA import, a recent study revealed that tRNAs and proteins may use the same import pathway across the mitochondrial outer membrane but it seems that these two import pathways are not linked (Niemann et al., Reference Niemann, Harsman, Mani, Peikert, Oeljeklaus and Warscheid2017). Still the factors involved in preferential import of Q-modified tRNAs remain to be identified. In addition, the role of tRNA fragments in translation modulation and/or a shortening of the bulk of cellular tRNAs as a result of nutritional stress was recently reported in vertebrate cells as well as in trypanosomes (Fricker et al., Reference Fricker, Brogli, Luidalepp, Wyss, Fasnacht, Joss, Zywicki, Helm, Schneider, Cristodero and Polacek2019; Schwenzer et al., Reference Schwenzer, Jühling, Chu, Pallett, Baumert, Maini and Fassati2019; Cristodero et al., Reference Cristodero, Brogli, Joss, Schimanski, Schneider and Polacek2021).
Clearly, the complex tRNA biology of kinetoplastid parasites has the potential to provide additional control steps of regulation of gene expression. In conclusion, T. brucei provides an ideal model to study the crosstalk between the tRNA trafficking and modification. Nevertheless, further studies based on differences in tRNA pools after silencing of the export factors or facing different nutritional and stress environment together with more complex analyses such as ribosome profiling could identify potential regulatory loops important for the complex lifecycle of these parasites.
Acknowledgements
I would like to thank Professor Juan D. Alfonzo and all members of the Paris laboratory for their comments and useful suggestions.
Author contribution
Z.P. wrote the article.
Financial support
This work was partially supported by the Czech Science Foundation (20-11585S to Z.P.) and the ERDF/ESF project Centre for Research of Pathogenicity and Virulence of Parasites (CZ.02.1.01/0.0/0.0/16 019/0000759 to Z.P).
Conflict of interest
None.
Ethical standards
Not applicable.