Introduction
All eukaryotic cells rely on the transport of molecules between the nucleus and the cytoplasm, in an efficient and regulated manner. With few exceptions, transport occurs through nuclear pores that pinch the double-layered membrane of the nucleus. Each nucleus possesses hundreds (yeast and trypanosomes) to thousands (human) of such channels. Nuclear pores are immensely complex multiprotein structures built from 500 to 1000 copies of ~30 different proteins, the nucleoporins (NUPs). Each nuclear pore complex (NPC) is an eight-fold symmetric cylindrical structure, consisting of a symmetric core of one inner and two outer concentric rings (the nucleoplasmic and the cytoplasmic rings) that connect the inner and outer nuclear membrane and form the pore (Alber et al., Reference Alber, Dokudovskaya, Veenhoff, Zhang, Kipper, Devos, Suprapto, Karni-Schmidt, Williams, Chait, Sali and Rout2007b). The nucleoplasmic outer ring is connected to the nuclear basket at the nuclear site of the pore and the cytoplasmic outer ring to eight cytoplasmic filaments at the cytoplasmic site (Alber et al., Reference Alber, Dokudovskaya, Veenhoff, Zhang, Kipper, Devos, Suprapto, Karni-Schmidt, Williams, Chait, Sali and Rout2007b). About one third of all NUPs possess highly unstructured regions enriched in phenylalanine−glycine (FG) motifs (FG NUPs). These FG motifs can phase-separate (Zilman, Reference Zilman2018) and in this way create a passive diffusion barrier for all molecules larger than ~40 kDa (Stanley et al., Reference Stanley, Fassati and Hoogenboom2017). Thus, while molecules up to about 30 kDa can freely enter and exit the nucleus by diffusion, larger proteins and almost all RNA molecules require more complex transport systems and can only pass because they bind to nuclear transporters that specifically interact with the FG-repeat sequences of the central channel (Stanley et al., Reference Stanley, Fassati and Hoogenboom2017).
Efficient and regulated mRNA export is of utmost importance to all eukaryotic cells, as all mRNAs must cross the nuclear envelope to reach their final destination in the cytoplasm. Most importantly, mRNA export is no isolated process restricted to the pore, but is tightly coordinated with the entire nuclear mRNA maturation machinery, starting at transcription (Björk and Wieslander, Reference Björk and Wieslander2017; Wende et al., Reference Wende, Friedhoff and Sträßer2019). An elaborate and collaborative control system ensures, that only mature, fully processed mRNAs can exit to the cytoplasm. mRNAs are transcribed by RNA polymerase II (RNAPII), usually as monogenetic transcripts. The C-terminal domain of the polymerase recruits factors to the transcription site that are needed for the downstream events of RNA processing, including capping, polyadenylation and splicing (Wende et al., Reference Wende, Friedhoff and Sträßer2019). Consequently, many mRNA maturation steps occur co-transcriptionally. During maturation, the pre-mRNA interacts with many proteins and protein complexes and some of these complexes mark the completion of certain mRNA processing steps and recruit the mRNA export factor Mex67-Mtr2 (NXF1-NXT1 or TAP-p15 in human) to the mRNP. The most famous among these complexes is the TREX complex (couples transcription and export) (Sträßer et al., Reference Sträßer, Masuda, Mason, Pfannstiel, Oppizzi, Rodríguez-Navarro, Rondón, Aguilera, Struhl, Reed and Hurt2002; Wende et al., Reference Wende, Friedhoff and Sträßer2019; Ashkenazy-Titelman et al., Reference Ashkenazy-Titelman, Shav-Tal and Kehlenbach2020). Mex67 is the major mRNA export factor (a ‘mobile nucleoporin’ according to some newer studies (Derrer et al., Reference Derrer, Mancini, Vallotton, Huet, Weis and Dultz2019)) and interacts with the FG Nups of the inner pore channel, this way transporting the large mRNP out of the nucleus. At the cytoplasmic site of the pores, the mRNP is remodelled to replace export factors by proteins required for the mRNAs cytoplasmic functions. This process requires the ATP-dependent RNA helicase Dbp5. Faulty RNAs are degraded by the nuclear exosome, aided by the TRAMP complex. In Metazoa, several alternative mRNA export factors and routes exist, and this redundancy may explain why phenotypes after depletion of orthologues to essential yeast export factors are often mild. These factors will mostly not be included here, but are covered well in Scott et al. (Reference Scott, Aguilar, Kramar and Oeffinger2019). Figure 1 provides a simplified overview about nuclear mRNA metabolism.
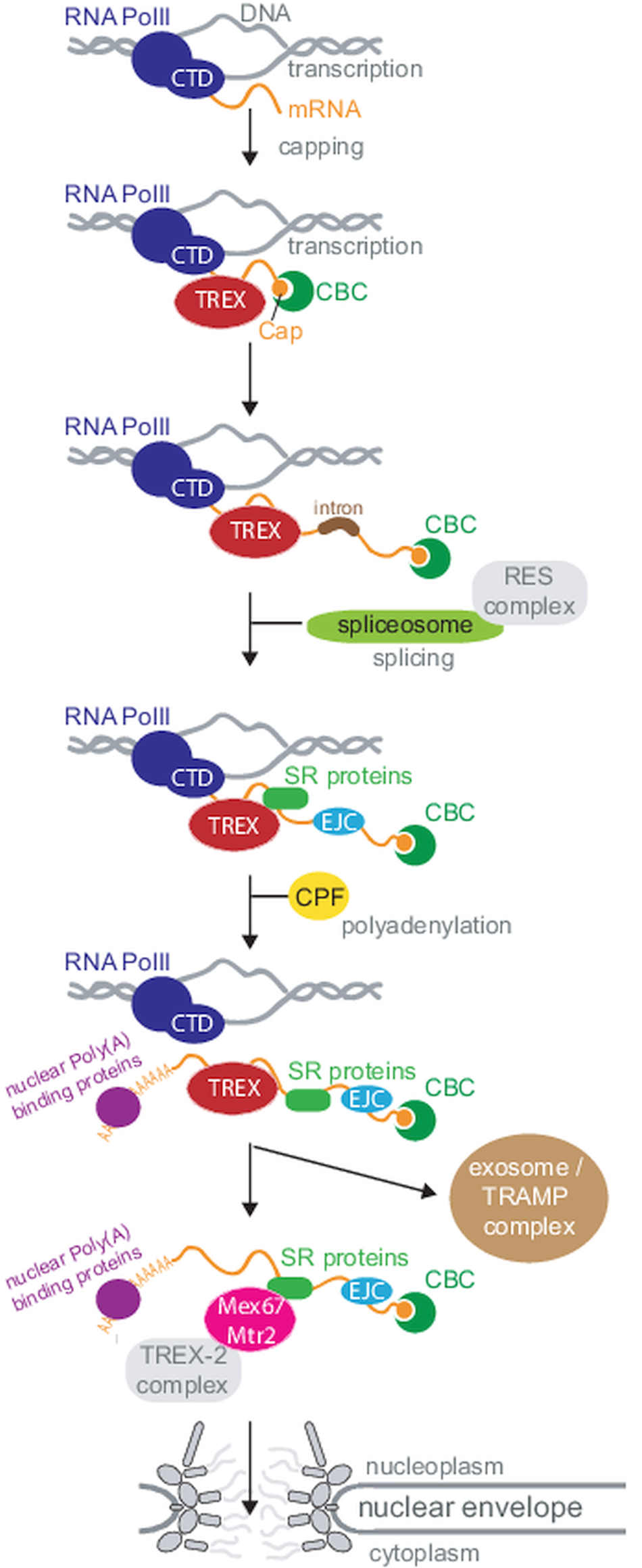
Fig. 1. Overview of nuclear mRNA metabolism. This diagram summarizes the basic steps of nuclear mRNA processing in a simplified version; all steps are discussed in the review in greater detail. Note that not all components are present in all organisms. For clarity, I have sorted processing steps in a pathway, but the order of events is not fully understood and processing steps can also occur in different orders or simultaneously. Abbreviations: CTD: C-terminal domain; CBC: cap binding complex; SR: serine−arginine rich; RNA PolII: RNA polymerase II; CFP: cleavage and polyadenylation factor; TREX: couples transcription and export; TRAMP: Trf4-Air2-Mtr4 polyadenylation.
In this review, I will compare the steps of nuclear mRNA maturation and export and its regulation between opisthokonts (mostly yeast and human), where these processes have been mostly studied, and the African trypanosome Trypanosoma brucei, the causative agent of African sleeping sickness and related cattle diseases. From an evolutionary point of view, these organisms are highly divergent: opisthokonts belong to one of the two major eukaryotic kingdoms, the Amorphea, while trypanosomes belong to the Discoba, which is now considered an extra clade outside of the two major kingdoms (He et al., Reference He, Fiz-Palacios, Fu, Fehling, Tsai and Baldauf2014; Adl et al., Reference Adl, Bass, Lane, Lukes, Schoch, Smirnov, Agatha, Berney, Brown, Burki, Cárdenas, Cepicka, Chistyakova, del Campo, Dunthorn, Edvardsen, Eglit, Guillou, Hampl, Heiss, Hoppenrath, James, Karpov, Kim, Kolisko, Kudryavtsev, Lahr, Lara, Le Gall, Lynn, Mann, Massana i Molera, Mitchell, Morrow, Park, Pawlowski, Powell, Richter, Rueckert, Shadwick, Shimano, Spiegel, Torruella i Cortes, Youssef, Zlatogursky and Zhang2019). A comparison offers the unique opportunity to distinguish features of mRNA export that were present in the last common ancestor of eukaryotes from features that have evolved later.
Genome organization and mRNA metabolism of T. brucei has several highly unusual and often unique features. The parasite possess a very gene-dense genome with only two introns (Mair et al., Reference Mair, Shi, Li, Djikeng, Aviles, Bishop, Falcone, Gavrilescu, Montgomery, Santori, Stern, Wang, Ullu and Tschudi2000; Kolev et al., Reference Kolev, Franklin, Carmi, Shi, Michaeli and Tschudi2010; Siegel et al., Reference Siegel, Hekstra, Wang, Dewell and Cross2010) and almost no regulatory regions (Berriman et al., Reference Berriman, Ghedin, Hertz-Fowler, Blandin, Renauld, Bartholomeu, Lennard, Caler, Hamlin, Haas, Böhme, Hannick, Aslett, Shallom, Marcello, Hou, Wickstead, Alsmark, Arrowsmith, Atkin, Barron, Bringaud, Brooks, Carrington, Cherevach, Chillingworth, Churcher, Clark, Corton, Cronin, Davies, Doggett, Djikeng, Feldblyum, Field, Fraser, Goodhead, Hance, Harper, Harris, Hauser, Hostetler, Ivens, Jagels, Johnson, Johnson, Jones, Kerhornou, Koo, Larke, Landfear, Larkin, Leech, Line, Lord, Macleod, Mooney, Moule, Martin, Morgan, Mungall, Norbertczak, Ormond, Pai, Peacock, Peterson, Quail, Rabbinowitsch, Rajandream, Reitter, Salzberg, Sanders, Schobel, Sharp, Simmonds, Simpson, Tallon, Turner, Tait, Tivey, Van Aken, Walker, Wanless, Wang, White, White, Whitehead, Woodward, Wortman, Adams, Embley, Gull, Ullu, Barry, Fairlamb, Opperdoes, Barrell, Donelson, Hall, Fraser, Melville and El-Sayed2005). Uniquely, genes are arranged head to tail to form about 167 polycistronic transcription units, each consisting out of tens to hundred of transcripts (Berriman et al., Reference Berriman, Ghedin, Hertz-Fowler, Blandin, Renauld, Bartholomeu, Lennard, Caler, Hamlin, Haas, Böhme, Hannick, Aslett, Shallom, Marcello, Hou, Wickstead, Alsmark, Arrowsmith, Atkin, Barron, Bringaud, Brooks, Carrington, Cherevach, Chillingworth, Churcher, Clark, Corton, Cronin, Davies, Doggett, Djikeng, Feldblyum, Field, Fraser, Goodhead, Hance, Harper, Harris, Hauser, Hostetler, Ivens, Jagels, Johnson, Johnson, Jones, Kerhornou, Koo, Larke, Landfear, Larkin, Leech, Line, Lord, Macleod, Mooney, Moule, Martin, Morgan, Mungall, Norbertczak, Ormond, Pai, Peacock, Peterson, Quail, Rabbinowitsch, Rajandream, Reitter, Salzberg, Sanders, Schobel, Sharp, Simmonds, Simpson, Tallon, Turner, Tait, Tivey, Van Aken, Walker, Wanless, Wang, White, White, Whitehead, Woodward, Wortman, Adams, Embley, Gull, Ullu, Barry, Fairlamb, Opperdoes, Barrell, Donelson, Hall, Fraser, Melville and El-Sayed2005). These polycistrons are transcribed by RNA polymerase II from transcription start sites that are epigenetically marked by histone modifications (Siegel et al., Reference Siegel, Hekstra, Kemp, Figueiredo, Lowell, Fenyo, Wang, Dewell and Cross2009). A small number of mRNAs, mostly encoding highly abundant cell surface proteins, are transcribed by RNA polymerase I (Zomerdijk et al., Reference Zomerdijk, Kieft and Borst1991; Chung et al., Reference Chung, Lee and Van der Ploeg1992; Lee and Van der Ploeg, Reference Lee and Van der Ploeg1997; Gunzl et al., Reference Gunzl, Bruderer, Laufer, Schimanski, Tu, Chung, Lee and Lee2003). Polycistronic transcription means that neither 5′ nor 3′ end of the mRNA are accessible for direct capping and polyadenylation. Instead, the 5′ m7G cap is added by trans-splicing the capped 39-nucleotide exon from the spliced leader RNA to each mRNA's 5′ end, coupled with polyadenylation of the downstream transcript (Campbell et al., Reference Campbell, Thornton and Boothroyd1984; LeBowitz et al., Reference LeBowitz, Smith, Rusche and Beverley1993; Ullu et al., Reference Ullu, Matthews and Tschudi1993; Matthews et al., Reference Matthews, Tschudi and Ullu1994). Note that after processing, trypanosome mRNAs do not significantly differ from mRNAs of any other organism: they have a 5′ cap and a poly(A) tail of about 100 nucleotides and the open reading frame is flanked by 5′ and 3′ UTR regions. The only trypanosome-unique mRNA features are specific methylations at the cap structure (Perry et al., Reference Perry, Watkins and Agabian1987; Freistadt et al., Reference Freistadt, Cross and Robertson1988; Bangs et al., Reference Bangs, Crain, Hashizume, McCloskey and Boothroyd1992) and the fact that, as a result of the trans-splicing reaction, every mRNA has exactly the same 39 nucleotides (the miniexon sequence) at its 5′ end. While the trypanosome NPC core structure is conserved, it has features indicating fundamental differences in the mRNA export pathway between trypanosomes and opisthokonts (Obado et al., Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016, Reference Obado, Field and Rout2017; Rout et al., Reference Rout, Obado, Schenkman and Field2017) (Fig. 2). First, trypanosome nuclear pores lack the asymmetric distribution of some NUPs that is required for unidirectionality of transport. Second, apparent orthologues to many NUPs with specific functions in mRNA export and export control are missing and this includes the entire ATP-dependent remodelling complex at the cytoplasmic site. Instead, mRNA export in trypanosomes is likely RanGTP dependent (Obado et al., Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016). Third, trypanosomes can initiate mRNA export co-transcriptionally, indicating that the completion of major processing steps such as polyadenylation and splicing is not required (Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019). These fundamental differences raise the question, how and whether trypanosomes do control mRNA export.
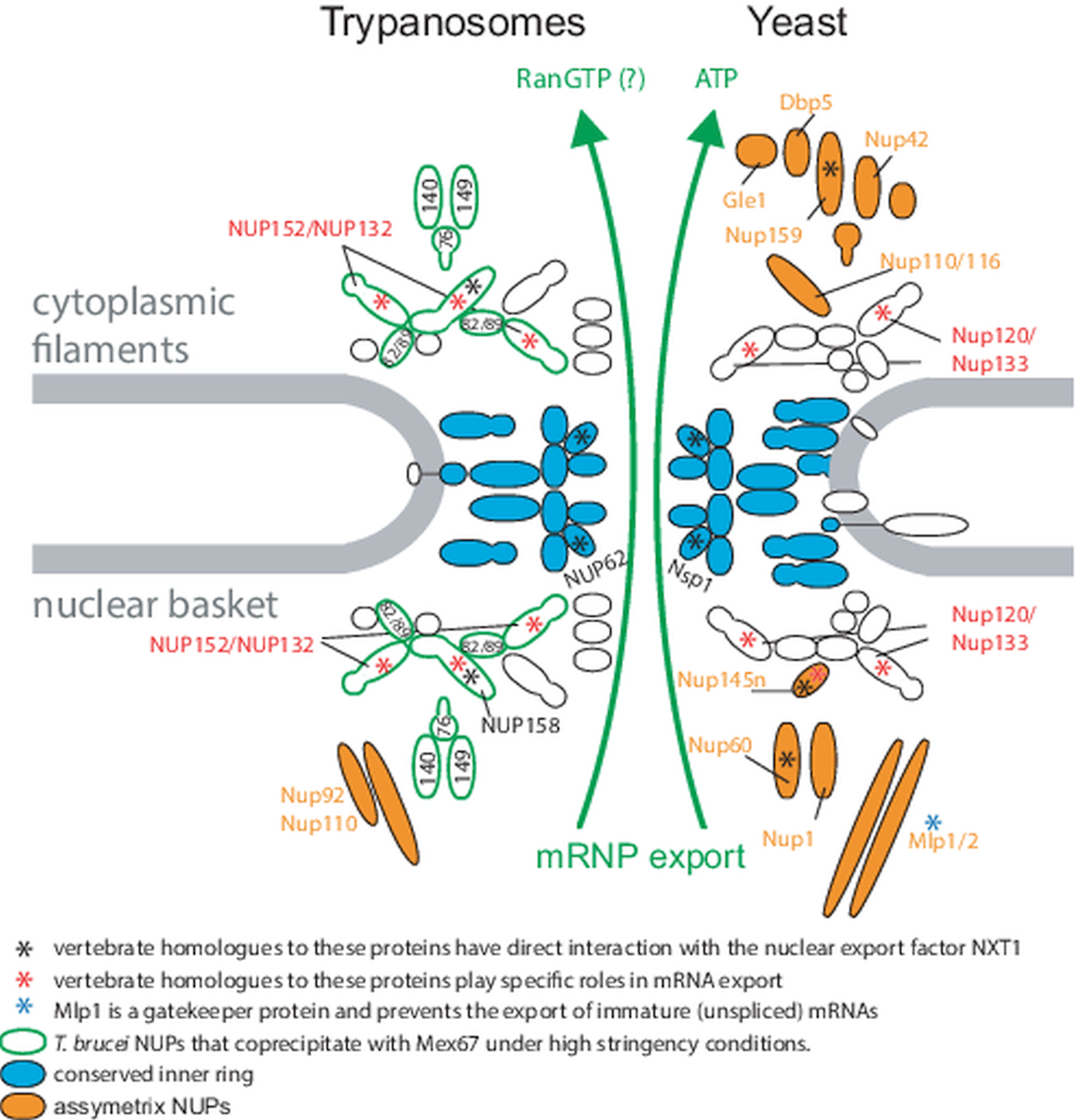
Fig. 2. Nuclear pores in trypanosomes and yeast. Schematics of trypanosome (left) and yeast (right) nuclear pores. The conserved inner ring is shown in blue, and all asymmetric nuclear pore proteins are indicated in orange. Homologues of vertebrate NUPs that were shown to engage in direct interaction with the C-terminal domain of the nuclear export factor NXT1 (Bachi et al., Reference Bachi, Braun, Rodrigues, Panté, Ribbeck, von Kobbe, Kutay, Wilm, Görlich, Carmo-Fonseca and Izaurralde2000; Forler et al., Reference Forler, Rabut, Ciccarelli, Herold, Köcher, Niggeweg, Bork, Ellenberg and Izaurralde2004) are marked with a black asterisks. Homologues to vertebrate NUPs with suspected roles in intermediate mRNA export are marked with red asterisks (Powers et al., Reference Powers, Forbes, Dahlberg and Lund1997; Vasu et al., Reference Vasu, Shah, Orjalo, Park, Fischer and Forbes2001; Blevins et al., Reference Blevins, Smith, Phillips and Powers2003). T. brucei NUPs that coprecipitate with the mRNA export factor Mex67 under high stringency conditions are encircled in green (Obado et al., Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016). The figure is modified from Obado et al. (Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016).
The major part of this review will systematically describe and compare nuclear mRNA processing steps of opisthokonts and trypanosomes, with a particular focus on factors and mechanisms that are relevant for mRNA export (see section ‘Nuclear mRNA processing steps in opisthokonts and trypanosomes’). The review will not discuss the spatial organization of part of the RNA processing machinery into nuclear bodies, as this is well covered by another review in this issue (Faria, Reference Faria2021). The review closes with a discussion on mRNA export control in trypanosomes (see section ‘Co-transcriptional initiation of RNA export indicates the lack of major mRNA export checkpoints in trypanosomes’) and a small outlook.
Nuclear mRNA processing steps in opisthokonts and trypanosomes
The C-terminal domain of RNAPII
mRNAs are transcribed by RNA polymerase II (RNAPII). The C-terminal domain of RNAPII (CTD) contains heptapeptide repeats with the consensus sequence YSPTSPS that get differentially phosphorylated during the different steps of transcription, this way successively recruiting mRNA processing and export factors to the transcription site (Jeronimo et al., Reference Jeronimo, Bataille and Robert2013). At the promoter region, the CTD is hypophosphorylated, during initiation it gets phosphorylated at S5 and S7 and elongation causes dephosphorylation at S5 and increases phosphorylation at Y1, S2 and T4. At the end of the transcription unit, Y1 gets dephosphorylated (upstream of the polyadenylation site) and finally S2 and T4 are dephosphorylated once the polyadenylation site has been passed (Heidemann et al., Reference Heidemann, Hintermair, Voß and Eick2013). RNA processing proteins and their complexes that are recruited specifically via CTD phosphorylation patterns include for example the capping enzyme (Cho et al., Reference Cho, Takagi, Moore and Buratowski1997; McCracken et al., Reference McCracken, Fong, Rosonina, Yankulov, Brothers, Siderovski, Hessel, Foster, Shuman and Bentley1997; Ghosh et al., Reference Ghosh, Shuman and Lima2011), the TREX complex (Meinel et al., Reference Meinel, Burkert-Kautzsch, Kieser, O'Duibhir, Siebert, Mayer, Cramer, Söding, Holstege and Sträßer2013) or, in yeast, the SR protein Npl3 (Dermody et al., Reference Dermody, Dreyfuss, Villen, Ogundipe, Gygi, Park, Ponticelli, Moore, Buratowski and Bucheli2008; Meinel et al., Reference Meinel, Burkert-Kautzsch, Kieser, O'Duibhir, Siebert, Mayer, Cramer, Söding, Holstege and Sträßer2013).
Like many protozoans, trypanosomes have an RNA polymerase II with a non-canonical CTD lacking the repetitive motifs. Still, the CTD is essential for parasite survival and serine-rich with at least 17 phosphorylation sites (Das and Bellofatto, Reference Das and Bellofatto2009; Urbaniak et al., Reference Urbaniak, Martin and Ferguson2013; Das et al., Reference Das, Banday, Fisher, Chang, Rosenfeld and Bellofatto2017). These 17 phosphorylation sites are all in the only stretch of the CTD that is indispensable for RNAPII function, evidence for their importance (Das and Bellofatto, Reference Das and Bellofatto2009). The function of CTD phosphorylation in trypanosomes remains unclear. Trypanosome mRNAs can be processed in the absence of CTD phosphorylation, for example when mRNA is transcribed by a different RNA polymerase, such as RNA polymerase I or by phage RNA polymerase T7 (albeit processing efficiency has never been studied). Moreover, CTD phosphorylation appears not required for transcription per se and also not for co-transcriptional m7G capping (Badjatia et al., Reference Badjatia, Ambrosio, Lee and Günzl2013; Gosavi et al., Reference Gosavi, Srivastava, Badjatia and Gunzl2020). The trypanosome kinase CRK9 was suspected to act as a CTD kinase, because its depletion causes CTD hypophosphorylation (Badjatia et al., Reference Badjatia, Ambrosio, Lee and Günzl2013), but a recent study using analogue-sensitive CRK9 resulted in inhibition of splicing within 5 min, while the loss in CTD phosphorylation took 12–24 h (Gosavi et al., Reference Gosavi, Srivastava, Badjatia and Gunzl2020). These data indicate that there is cross-talk between the mRNA processing machinery and RNAPII, but argue against CRK9 being directly involved in CTD phosphorylation.
Despite of not being essential for transcription, the trypanosome CTD mediates correct positioning of RNAPII at transcriptional start sites within the chromatin (Das et al., Reference Das, Banday, Fisher, Chang, Rosenfeld and Bellofatto2017). In trypanosomes, these transcription start sites can stretch over several kilobases (Siegel et al., Reference Siegel, Hekstra, Kemp, Figueiredo, Lowell, Fenyo, Wang, Dewell and Cross2009; Thomas et al., Reference Thomas, Green, Sturm, Campbell and Myler2009; Aslett et al., Reference Aslett, Aurrecoechea, Berriman, Brestelli, Brunk, Carrington, Depledge, Fischer, Gajria, Gao, Gardner, Gingle, Grant, Harb, Heiges, Hertz-Fowler, Houston, Innamorato, Iodice, Kissinger, Kraemer, Li, Logan, Miller, Mitra, Myler, Nayak, Pennington, Phan, Pinney, Ramasamy, Rogers, Roos, Ross, Sivam, Smith, Srinivasamoorthy, Stoeckert, Subramanian, Thibodeau, Tivey, Treatman, Velarde and Wang2010; Kolev et al., Reference Kolev, Franklin, Carmi, Shi, Michaeli and Tschudi2010), as the parasites lack conventional RNAPII promoters, with the exception of the atypical promoter of the spliced leader RNAs (Gilinger and Bellofatto, Reference Gilinger and Bellofatto2001). Interestingly, transcription starts predominantly in the correct direction (Wedel et al., Reference Wedel, Förstner, Derr and Siegel2017); and it remains to be investigated whether the CTD is also involved in defining transcription directionality. Another potential function of trypanosome CTD phosphorylations could be to mediate the pausing of RNAPII that was observed downstream of SL addition sites of each gene and may facilitate trans-splicing (Wedel et al., Reference Wedel, Förstner, Derr and Siegel2017). Such a function (currently purely speculative) would be analogous to the CTD function in higher eukaryotes in connecting mRNA transcription with downstream processing steps.
The TREX complex
The TREX complex, so named because it couples transcription to mRNA export, is one of the first complexes that associates with the newly transcribed transcripts (Sträßer et al., Reference Sträßer, Masuda, Mason, Pfannstiel, Oppizzi, Rodríguez-Navarro, Rondón, Aguilera, Struhl, Reed and Hurt2002; Wende et al., Reference Wende, Friedhoff and Sträßer2019; Ashkenazy-Titelman et al., Reference Ashkenazy-Titelman, Shav-Tal and Kehlenbach2020). It consists of the THO complex, and, in its minimal version, of the RNA helicase Sub2 (UAP56/DDX39B in Metazoa) and the adaptor protein Yra1 (ALYREF/THOC4 in Metazoa) (Table 1). Several further subunits specific to either yeast or human have been described (reviewed in Wende et al., Reference Wende, Friedhoff and Sträßer2019).
Table 1. Proteins of the TREX core complex

Note that only S. cerevisiae TREX complex components are listed with the respective human homologues; TREX subunits unique to human are not listed. The SR proteins Gbp2 and Hrb1 are specific components of the yeast TREX complex and are also not included in this table but discussed in section ‘SR proteins’.
The THO complex is the core of the TREX complex and creates a binding platform for the other TREX proteins on the chromatin during transcription (Aguilera and Klein, Reference Aguilera and Klein1990; Piruat and Aguilera, Reference Piruat and Aguilera1998; Gewartowski et al., Reference Gewartowski, Cuéllar, Dziembowski and Valpuesta2012). In yeast, the THO complex is heteropentameric and consists of Tho2, Hpr1, Mft1, Thp2 and Tex1.
The DEAD box RNA helicase Sub2 (UAP56/DDX39B) plays multiple roles in the TREX complex and has conserved functions in mRNA export (Luo et al., Reference Luo, Zhou, Magni, Christoforides, Rappsilber, Mann and Reed2001; Reed and Hurt, Reference Reed and Hurt2002; Taniguchi and Ohno, Reference Taniguchi and Ohno2008; Dufu et al., Reference Dufu, Livingstone, Seebacher, Gygi, Wilson and Reed2010; Kammel et al., Reference Kammel, Thomaier, Sørensen, Schubert, Längst, Grasser and Grasser2013; Serpeloni et al., Reference Serpeloni, Jiménez-Ruiz, Vidal, Kroeber, Andenmatten, Lemgruber, Mörking, Pall, Meissner and Avila2016). It binds progressively to the newly transcribed mRNA (Kiesler et al., Reference Kiesler, Miralles and Visa2002), promotes spliceosome assembly together with U2AF65 and is essential in the pre-mRNA splicing process (Fleckner et al., Reference Fleckner, Zhang, Valcárcel and Green1997; Shen et al., Reference Shen, Zhang and Zhao2007). One important function of Sub2 is the recruitment of the adaptor protein Yra1 (discussed below) to the mRNP (Strässer and Hurt, Reference Strässer and Hurt2001). Because Sub2 and Mex67 bind to the same domain of Yra1, Sub2 is released from the transcript as soon as Mex67 binds: Sub2 hands over the transcript to the downstream key-players of export (Strässer and Hurt, Reference Strässer and Hurt2001) and this mechanism appears conserved in metazoans (Hautbergue et al., Reference Hautbergue, Hung, Golovanov, Lian and Wilson2008). The function of the helicase in mRNA export is ATP-dependent (Kota et al., Reference Kota, Wagner, Huerta, Underwood and Nickerson2008; Taniguchi and Ohno, Reference Taniguchi and Ohno2008) and thus one of only two energy-dependent steps in mRNA export (the other is the RNP remodelling by RNA helicase Dbp5 at the cytoplasmic site of the pore).
The export adaptor Yra1 (ALYREF/THOC4) is recruited to the mRNP in multiple ways. With the exception of Sub2 (discussed above), these ways differ between yeast and higher eukaryotes. In yeast, a protein of the cleavage and polyadenylation complex (Pcf11 (Johnson et al., Reference Johnson, Cubberley and Bentley2009)), the DEAD-box RNA helicase Dbp2 (Ma et al., Reference Ma, Cloutier and Tran2013), RNA itself (Meinel et al., Reference Meinel, Burkert-Kautzsch, Kieser, O'Duibhir, Siebert, Mayer, Cramer, Söding, Holstege and Sträßer2013) and ubiquitylation of several proteins, e.g. Histone 2B (Vitaliano-Prunier et al., Reference Vitaliano-Prunier, Babour, Hérissant, Apponi, Margaritis, Holstege, Corbett, Gwizdek and Dargemont2012) can recruit Yra1 or contribute to the recruitment. In higher eukaryotes, ALYREF can be recruited to the RNP by the spliceosome via interaction with the exon junction complex (Masuda et al., Reference Masuda, Das, Cheng, Hurt, Dorman and Reed2005; Gromadzka et al., Reference Gromadzka, Steckelberg, Singh, Hofmann and Gehring2016) or by the cap-binding complex (CBC) (Cheng et al., Reference Cheng, Dufu, Lee, Hsu, Dias and Reed2006; Nojima et al., Reference Nojima, Hirose, Kimura and Hagiwara2007; Sen et al., Reference Sen, Barman, Kaja, Ferdoush, Lahudkar, Roy and Bhaumik2019) (see section ‘Cap-binding complex’). ALYREF is also present at the mRNAs 3′ ends, dependent on the cap binding protein CBP80 and the nuclear poly(A) binding protein PABPN1 (Shi et al., Reference Shi, Zhang, Wu, He, Wang, Yin, Tian, Li and Cheng2017). These differences in the loading mechanism of the TREX complex between yeast (mostly transcription dependent) and metazoans (mostly splicing dependent) likely reflect the higher number of introns in metazoans. Independent on how Yra1/ALYREF is recruited to the mRNP, once bound it interacts directly with the export receptor Mex67-Mtr2 (NXF1-NXT1, TAP-p15) and recruits it to the mRNP and thus plays an essential role in mRNA export (Strässer and Hurt, Reference Strässer and Hurt2000; Zenklusen et al., Reference Zenklusen, Vinciguerra, Strahm and Stutz2001; Hautbergue et al., Reference Hautbergue, Hung, Golovanov, Lian and Wilson2008). The binding of NXF1 to ALYREF causes a conformational change in ALYREF that decreases its affinity for the RNP: ALYREF hands over the RNP to Mex67 at the nuclear basket and does not accompany the RNP through the pore (Kim et al., Reference Kim, Yong, Kataoka, Abel, Diem and Dreyfuss2001; Kiesler et al., Reference Kiesler, Miralles and Visa2002; Lund and Guthrie, Reference Lund and Guthrie2005). While Yra1 is the major Mex67 adaptor protein in yeast and essential for mRNA export (Portman and Gull, Reference Portman and Gull2012; Segref et al., Reference Segref, Sharma, Doye, Hellwig, Huber, Lührmann and Hurt1997; Santos-Rosa et al., Reference Santos-Rosa, Moreno, Simos, Segref, Fahrenkrog, Panté and Hurt1998; Zenklusen et al., Reference Zenklusen, Vinciguerra, Wyss and Stutz2002), depletion of the metazoan orthologue has only a mild effect on mRNA export (Gatfield and Izaurralde, Reference Gatfield and Izaurralde2002; Longman et al., Reference Longman, Johnstone and Cáceres2003; Katahira et al., Reference Katahira, Inoue, Hurt and Yoneda2009). The likely reason is that Metazoa have many alternative NXF1 adaptors, including for example organism-specific TREX components or splicing factors of the SR (serine−arginine-rich) protein family (Scott et al., Reference Scott, Aguilar, Kramar and Oeffinger2019; Wende et al., Reference Wende, Friedhoff and Sträßer2019; Ashkenazy-Titelman et al., Reference Ashkenazy-Titelman, Shav-Tal and Kehlenbach2020).
Trypanosomes have an orthologue to the TREX-complex protein Sub2 (Table 1). This helicase has nuclear localization in Trypanosoma cruzi and T. brucei (Serpeloni et al., Reference Serpeloni, Moraes, Muniz, Motta, Ramos, Kessler, Inoue, daRocha, Yamada-Ogatta, Fragoso, Goldenberg, Freitas-Junior and Avila2011a; Dean et al., Reference Dean, Sunter and Wheeler2017; Goos et al., Reference Goos, Dejung, Janzen, Butter and Kramer2017), binds mRNA (Lueong et al., Reference Lueong, Merce, Fischer, Hoheisel and Erben2016) and, importantly, its depletion by RNAi in T. brucei caused growth arrest and accumulation of polyadenylated mRNA in the nucleus, all indicative of an essential function in mRNA export (Serpeloni et al., Reference Serpeloni, Moraes, Muniz, Motta, Ramos, Kessler, Inoue, daRocha, Yamada-Ogatta, Fragoso, Goldenberg, Freitas-Junior and Avila2011a). Still, trypanosomes are unlikely to possess a conventional TREX complex, as obvious orthologues to all other TREX subunits (Yra1, Tho2, Hpr1, Mft1, Thp2 and Tex1) are absent, TbSub2 fails to complement yeast Sub2 (Serpeloni et al., Reference Serpeloni, Moraes, Muniz, Motta, Ramos, Kessler, Inoue, daRocha, Yamada-Ogatta, Fragoso, Goldenberg, Freitas-Junior and Avila2011a) and TbSub2 was not detected in a Mex67 affinity purification approach (Obado et al., Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016). Moreover, TbSub2 differs from its yeast orthologue by a faster ATP hydrolysis rate and an activity less dependent on RNA binding (de Bittencourt et al., Reference de Bittencourt, Serpeloni, Hiraiwa, de Arruda Campos Brasil de Souza and Avila2017). The exact function of TbSub2 in mRNA export remains to be discovered. Interestingly, the Sub2 orthologue of Toxoplasma gondii, a parasite with distant phylogenetic relationship to trypanosomes, also is the only TREX component that can be identified by homology, and its involvement in mRNA export was shown (Serpeloni et al., Reference Serpeloni, Jiménez-Ruiz, Vidal, Kroeber, Andenmatten, Lemgruber, Mörking, Pall, Meissner and Avila2016); one protein with no similarity to Yra1 apart from the presence of an RRM domain could be a functional T. gondii Yra1 orthologue based on its mRNA export phenotype (Serpeloni et al., Reference Serpeloni, Jiménez-Ruiz, Vidal, Kroeber, Andenmatten, Lemgruber, Mörking, Pall, Meissner and Avila2016).
Recently, the retrotransposon hot spot proteins (RHS proteins) were suggested to be the trypanosomes alternative to the TREX complex. This multigene family has 118 members (classified into six groups) that are characterized by a retrotransposon insertion site in the 5′ region of the coding sequence, resulting in ~60% pseudogenes (Bringaud et al., Reference Bringaud, Biteau, Melville, Hez, El-Sayed, Leech, Berriman, Hall, Donelson and Baltz2002). Proteins of five of the six subfamilies (RHS1,3,4,5,6) show nuclear localization, while RHS2 is in the cytoplasm (Bringaud et al., Reference Bringaud, Biteau, Melville, Hez, El-Sayed, Leech, Berriman, Hall, Donelson and Baltz2002; Florini et al., Reference Florini, Naguleswaran, Gharib, Bringaud and Roditi2019). An extensive analysis of retrotransposon hot spot proteins RHS2, RHS4 and RHS6 showed association with active PolII transcription sites (Chip Seq), even for the cytoplasmic RHS2, which can shuttle to the nucleus (Florini et al., Reference Florini, Naguleswaran, Gharib, Bringaud and Roditi2019). RNAi depletion of all proteins caused growth arrest and a global reduction in transcription as well as a block in mRNA export. RHS4 is part of the PolII complex (Das et al., Reference Das, Li, Liu and Bellofatto2006; Devaux et al., Reference Devaux, Lecordier, Uzureau, Walgraffe, Dierick, Poelvoorde, Pays and Vanhamme2006), while RHS2 mostly co-precipitated with ribosomal proteins and translation factors and RHS6 mostly with nuclear proteins involved in transcription and mRNA processing (Florini et al., Reference Florini, Naguleswaran, Gharib, Bringaud and Roditi2019). In conclusion, retrotransposon hot spot proteins have essential functions in transcription and further nuclear and cytoplasmic mRNA processing steps. Whether they act in connecting transcription with export and thus can be considered functional orthologues to the TREX complex remains to be investigated. In particular, direct binding to RNA has not been shown (albeit chromatin interaction appears partially mediated by RNA for RHS2 and RHS4 (Florini et al., Reference Florini, Naguleswaran, Gharib, Bringaud and Roditi2019) and RHS4 co-precipitates with oligo(dT) beads (Lueong et al., Reference Lueong, Merce, Fischer, Hoheisel and Erben2016)). Moreover, it remains unclear, whether the multiple phenotypes observed after RNAi depletion of an RHS protein represent functions in different RNA processing steps (e.g. transcription and export), or, whether block of one RNA processing step (e.g. transcription) subsequently affects multiple downstream pathways without the direct involvement of RHS proteins.
Adding the cap
The first modification added to every newly transcribed mRNA is the 5′ m7G cap (Table 2). mRNA capping is done co-transcriptionally as soon as the first 25−30 nucleotides are transcribed, by capping enzymes that are recruited to the transcription start site; serine 5 phosphorylation of the CTD serves as one recruitment signal (Ramanathan et al., Reference Ramanathan, Robb and Chan2016). mRNA capping involves three enzymatic activities. First, RNA triphosphatase (TPase) removes the γ-phosphate from the triphosphorylated mRNA's 5′ end, creating a 5′ diphosphate mRNA. Second, RNA guanylyltransferase (GTase) transfers a GMP group from GTP to the 5′ diphosphate mRNA, creating the 5′-5′ triphosphate linkage between the cap and the first base of the mRNA. Third, the guanine-N7-methyltransferase (MTase) methylates the N7 amine of the guanine cap using S-adenosylmethionine (SAM) as methyl-donor, to form the cap 0 structure. In Saccharomyces cerevisiae, the three capping activities are on three separate proteins (Cet1, Ceg1 and Abd1) (Martinez-Rucobo et al., Reference Martinez-Rucobo, Kohler, van de Waterbeemd, Heck, Hemann, Herzog, Stark and Cramer2015). In Metazoa, TPase and GTase activities reside on the same protein (Mce1 in mouse) (Yue et al., Reference Yue, Maldonado, Pillutla, Cho, Reinberg and Shatkin1997; Chu et al., Reference Chu, Das, Tyminski, Bauman, Guan, Qiu, Montelione, Arnold and Shatkin2011), while the guanine-N7-methyltransferase activity (Hcm1) is on a separate protein (Saha et al., Reference Saha, Schwer and Shuman1999). In higher eukaryotes, the 2′O of the ribose of the first base or first and second base is methylated, creating the predominant cap 1 and cap 2 structures, respectively (Furuichi et al., Reference Furuichi, Morgan, Shatkin, Jelinek, Salditt-Georgieff and Darnell1975; Bélanger et al., Reference Bélanger, Stepinski, Darzynkiewicz and Pelletier2010; Werner et al., Reference Werner, Purta, Kaminska, Cymerman, Campbell, Mittra, Zamudio, Sturm, Jaworski and Bujnicki2011; Furuichi, Reference Furuichi2015); the responsible human methylases are hMtr1 and hMtr2 (Bélanger et al., Reference Bélanger, Stepinski, Darzynkiewicz and Pelletier2010; Werner et al., Reference Werner, Purta, Kaminska, Cymerman, Campbell, Mittra, Zamudio, Sturm, Jaworski and Bujnicki2011). Ribose methylations are absent in yeast (Mager et al., Reference Mager, Klootwijk and Klein1976; Sripati et al., Reference Sripati, Groner and Warner1976). In metazoans, the 5′ m7G cap is important for the export of spliced mRNAs but not of intron-less mRNAs, probably because it is involved in recruiting the TREX complex upstream to the first exon−exon junction (Cheng et al., Reference Cheng, Dufu, Lee, Hsu, Dias and Reed2006).
Table 2. Enzymes involved in nuclear mRNA capping and cap methylation

a Misleading nomenclature: note that MTr2 is not the MEX67 interacting protein Mtr2.
b The trypanosome enzyme is homologous to the vaccinia virus (VP39) methyltransferase and not to the metazoan Mtr2.
c It is likely but not proven that MTr3 methylates ribose at both the third and fourth nucleotide, as MTr3 -/- cells lack 2′-O ribose methylations on both positions (Arhin et al., Reference Arhin, Ullu and Tschudi2006b).
The process of mRNA capping is unusual in trypanosomes (Table 2). Because of the polycistronic transcription, the 5′ ends of mRNAs are not directly accessible for capping enzymes. Therefore, the cap is added by trans-splicing the capped 39 nucleotide long miniexon of the spliced leader RNA to the 5′ end of each transcript (reviewed in Michaeli, Reference Michaeli2011; Preusser et al., Reference Preusser, Jaé and Bindereif2012). The spliced leader RNA itself is separately transcribed from a tandem array of about 100 SL RNA genes, each copy from its own promoter (these are the only PolII promoters present in trypanosomes) (Günzl et al., Reference Günzl, Ullu, Dörner, Fragoso, Hoffmann, Milner, Morita, Nguu, Vanacova, Wünsch, Dare, Kwon and Tschudi1997; Gilinger and Bellofatto, Reference Gilinger and Bellofatto2001; Srivastava et al., Reference Srivastava, Badjatia, Lee, Hao and Gunzl2017). Capping of the spliced leader RNA is different from both the yeast and metazoan system, in that the RNA triphosphatase activity is on a separate protein (TbCet1, (Ho and Shuman, Reference Ho and Shuman2001) while the RNA guanylyltransferase and the guanine-N7-methyltransferase activity reside on the bifunctional enzyme TbCgm1 (Hall and Ho, Reference Hall and Ho2006; Ruan et al., Reference Ruan, Shen, Ullu and Tschudi2007; Takagi et al., Reference Takagi, Sindkar, Ekonomidis, Hall and Ho2007). Note that trypanosomes also have the RNA guanylyltransferase and guanine-N7-methyltransferase activity on two individual enzymes (Ce1 and Cmt1, respectively) (Silva et al., Reference Silva, Ullu, Kobayashi and Tschudi1998; Hall and Ho, Reference Hall and Ho2006) but these enzymes are cytoplasmic (Dean et al., Reference Dean, Sunter and Wheeler2017) and not involved in SL RNA capping (Ruan et al., Reference Ruan, Shen, Ullu and Tschudi2007; Takagi et al., Reference Takagi, Sindkar, Ekonomidis, Hall and Ho2007; Ignatochkina et al., Reference Ignatochkina, Takagi, Liu, Nagata and Ho2015; Silva et al., Reference Silva, Ullu, Kobayashi and Tschudi1998). One further unique feature of trypanosomes is that the mRNA cap is of the heavily methylated type 4: the first four transcribed nucleotides (AACU) have ribose 2′-O methylations and there are additional base methylations on the first (m62A) and fourth (m3U) position (Perry et al., Reference Perry, Watkins and Agabian1987; Freistadt et al., Reference Freistadt, Cross and Robertson1988; Bangs et al., Reference Bangs, Crain, Hashizume, McCloskey and Boothroyd1992). Cap methylation is essential for trans-splicing (Ullu and Tschudi, Reference Ullu and Tschudi1991) and ribose methylation is required for efficient translation (Zeiner et al., Reference Zeiner, Sturm and Campbell2003b; Zamudio et al., Reference Zamudio, Mittra, Campbell and Sturm2009). Three 2′-O-ribose methyltransferases have been described in trypanosomes, MTr1 (Zamudio et al., Reference Zamudio, Mittra, Foldynova-Trantirkova, Zeiner, Lukes, Bujnicki, Sturm and Campbell2007; Mittra et al., Reference Mittra, Zamudio, Bujnicki, Stepinski, Darzynkiewicz, Campbell and Sturm2008), MTr2 (Hall and Ho, Reference Hall and Ho2006; Zamudio et al., Reference Zamudio, Mittra, Zeiner, Feder, Bujnicki, Sturm and Campbell2006; Arhin et al., Reference Arhin, Ullu and Tschudi2006b) and MTr3 (Zamudio et al., Reference Zamudio, Mittra, Zeiner, Feder, Bujnicki, Sturm and Campbell2006; Arhin et al., Reference Arhin, Li, Ullu and Tschudi2006a); MTr2 and MTr3 are related to the vaccinia virus VP39 methyltransferase. The deletion of some but not all three 2′-O-ribose methyltransferases is viable, with some effects on growth and translation (Zamudio et al., Reference Zamudio, Mittra, Zeiner, Feder, Bujnicki, Sturm and Campbell2006, Reference Zamudio, Mittra, Campbell and Sturm2009; Arhin et al., Reference Arhin, Ullu and Tschudi2006b). The function of the unusual base methylations is unknown, and the responsible enzymes have not yet been identified. The SL RNA is also pseudouridinylated at position −12 relative to the 5′ splice site, but this modification is not essential for growth in culture (Hury et al., Reference Hury, Goldshmidt, Tkacz and Michaeli2009). It is not known yet, whether the trypanosome cap (and its methylations) is required for mRNA export.
Cap-binding complex
As soon as the m7 G cap is synthesized, it is bound by the nuclear cap-binding complex (CBC), a heterodimer out of Cbp20 and Cbp80 (CBP20/CBP80 or NCBP1/NCBP2 in metazoans) that protects the new transcript from degradation (Table 3). Only Cbp20 binds the m7G cap directly; the Cbp80/CBP80 subunit serves as a binding platform for many regulatory factors with key function in multiple diverse pathways, including transcription, splicing, export and translation (Gonatopoulos-Pournatzis and Cowling, Reference Gonatopoulos-Pournatzis and Cowling2013; Müller-McNicoll and Neugebauer, Reference Müller-McNicoll and Neugebauer2014; Rambout and Maquat, Reference Rambout and Maquat2020). For example, the CBC binds to the Yra1/ALYREF subunit of the TREX complex and recruits it co-transcriptionally to the mRNA's 5′ end, likely facilitating nuclear export (Cheng et al., Reference Cheng, Dufu, Lee, Hsu, Dias and Reed2006; Nojima et al., Reference Nojima, Hirose, Kimura and Hagiwara2007; Sen et al., Reference Sen, Barman, Kaja, Ferdoush, Lahudkar, Roy and Bhaumik2019); in yeast, Npl3, an SR protein which contributes to the formation of an export-competent RNP, is also recruited by the CBC (Sen et al., Reference Sen, Barman, Kaja, Ferdoush, Lahudkar, Roy and Bhaumik2019). The CBC indirectly promotes nuclear export in many ways by promoting various steps in mRNP maturation and it accompanies its RNP target through the pore. However, there is no genetic evidence for a direct involvement of the CBC complex in mRNA export and whether it is strictly required for export is still debated.
Table 3. The cap binding complex in S. cerevisiae, metazoans and trypanosomes

The nuclear cap binding complex of trypanosomes has only one conserved subunit (CBP20) and at least three trypanosome-specific subunits (CBP30, CBP66, CBP110) (Li and Tschudi, Reference Li and Tschudi2005) (Table 3). It is essential and required for trans-splicing and has 15-fold higher affinity to the hypermethylated trypanosome type 4 cap than to a type 0 cap (Li and Tschudi, Reference Li and Tschudi2005). It is not known whether and how the CBP is remodelled during trans-splicing, whether the CBP plays a role in mRNA export and when and where it is replaced by the cytoplasmic cap binding complex.
Spliceosome
All pre-mRNAs that contain introns are processed by the spliceosome in a complex series of reactions that serve to remove the introns and join the 3′ end of each exon with the 5′ end of the next exon (Wahl et al., Reference Wahl, Will and Lührmann2009; Fica and Nagai, Reference Fica and Nagai2017). Splicing is guided by sequence elements within the intronic region, such as the branch point sequence (BPS) that attacks the phosphodiester bond of the 5′ splice site to create a free 5′ exon and the intron-lariat-3′ exon intermediate (Wahl et al., Reference Wahl, Will and Lührmann2009). Another highly conserved intronic sequence element is the polypyrimidine tract (a sequence rich in polypyrimidine) that is located between the branch point site and the 3′ splice site and is required for early steps of spliceosome assembly (Wahl et al., Reference Wahl, Will and Lührmann2009). The spliceosome consists of five small nuclear RNAs (snRNAs) U1, U2, U4, U5 and U6 that associate with both snRNA-specific proteins as well as with Sm proteins (LSm proteins in the case of U6) (Wahl et al., Reference Wahl, Will and Lührmann2009). The Sm/LSm proteins form a ring-like structure around a conserved motif of the respective snRNA, the Sm site (Kambach et al., Reference Kambach, Walke, Young, Avis, de la Fortelle, Raker, Lührmann, Li and Nagai1999). About 75% of splicing events of budding yeast and human occur co-transcriptionally (Neugebauer, Reference Neugebauer2019).
The core spliceosome machinery is highly conserved across all eukaryotes (Will and Lührmann, Reference Will and Lührmann2011), albeit trypanosomes have some variations in the Sm core proteins and Sm sites (Preusser et al., Reference Preusser, Jaé and Bindereif2012). Similar to opisthokonts, splicing in trypanosomes occurs mainly co-transcriptionally (Ullu et al., Reference Ullu, Matthews and Tschudi1993), albeit at least one exception has been reported (Jäger et al., Reference Jäger, De Gaudenzi, Cassola, D'Orso and Frasch2007). The trans-splicing reaction is performed by a variant of the spliceosome that has its U1 snRNP replaced by the SL RNP: the SL RNA serves as both snRNA and trans-splicing substrate (Preusser et al., Reference Preusser, Jaé and Bindereif2012)). Like cis-splicing sites in other organisms, trans-splicing sites are preceded by polypyrimidine tracts (Smith et al., Reference Smith, Blanchette and Papadopoulou2008; Kolev et al., Reference Kolev, Franklin, Carmi, Shi, Michaeli and Tschudi2010). The splicing reaction is analogous to cis splicing, except that a branched Y-structure intermediate instead of a lariat structure is formed, when the SL RNA intron is joined 2´-5′ to an A residue upstream of the polypyrimidine tract. The conventional U1 snRNP is still present in trypanosomes and needed for the cis-splicing of the two intronic RNAs, encoding the poly(A) polymerase PAP1 and the DEAD box RNA helicase DBP2B. Interestingly, these two intronic mRNAs are conserved across Trypanosomatidae (Mair et al., Reference Mair, Shi, Li, Djikeng, Aviles, Bishop, Falcone, Gavrilescu, Montgomery, Santori, Stern, Wang, Ullu and Tschudi2000; Camacho et al., Reference Camacho, la Fuente, Rastrojo, Pastor, Solana, Tabera, Gamarro, Carrasco-Ramiro, Requena and Aguado2019). Moreover, both intronic mRNAs encode for proteins with (potential) functions in nuclear RNA processing: PAP1 presumably adenylates pre-snoRNAs (Chikne et al., Reference Chikne, Gupta, Doniger, Shanmugha Rajan, Cohen-Chalamish, Waldman Ben-Asher, Kolet, Yahia, Unger, Ullu, Kolev, Tschudi and Michaeli2017) and DBP2B is one of two trypanosome homologues of Dbp2 that in yeast recruit Yra1 to the mRNP (Ma et al., Reference Ma, Cloutier and Tran2013). One, purely speculative, model is that the trypanosome cis-splicing is not a random left over of a general loss of introns but could have auto-regulatory functions. Furthermore, interaction of the U1 snRNP protein U1A with the polyadenylation factor CPSF73 (Tkacz et al., Reference Tkacz, Gupta, Volkov, Romano, Haham, Tulinski, Lebenthal and Michaeli2010) and the finding that the U1 snRNP proteins U1C and U1-70K also interact with the SL RNA and the U6 snRNA (Preusser et al., Reference Preusser, Rossbach, Hung, Li and Bindereif2014) indicate additional functions of the U1 RNP in connecting trans-splicing with cis-splicing and polyadenylation. This could explain the relatively high abundance of the U1 snRNP despite the presence of only two introns. An alternative explanation for the high abundance of the U1 snRNP could be yet undiscovered functions of this snRNP unrelated to mRNA processing: the mammalian U1 snRNP for example has been recently shown to regulate chromatin retention of non-coding RNAs (Yin et al., Reference Yin, Lu, Zhang, Shao, Xu, Li, Hong, Cui, Shan, Tian, Zhang and Shen2020).
Several further trypanosome proteins with a function in splicing have been described; next to the SR proteins (see section ‘SR proteins’) these are DRBD3 (=PTB1), DRBD4 (=PTB2) and HNRNPH/F (De Gaudenzi et al., Reference De Gaudenzi, Frasch and Clayton2005; Stern et al., Reference Stern, Gupta, Salmon-Divon, Haham, Barda, Levi, Wachtel, Nilsen and Michaeli2009; Gupta et al., Reference Gupta, Kosti, Plaut, Pivko, Tkacz, Cohen-Chalamish, Biswas, Wachtel, Waldman Ben-Asher, Carmi, Glaser, Mandel-Gutfreund and Michaeli2013; Das et al., Reference Das, Bellofatto, Rosenfeld, Carrington, Romero-Zaliz, del Val and Estevez2015; Clayton, Reference Clayton2019). In addition, the cyclin-dependent kinase CRK9 is essential for the first step in splicing, presumably by phosphorylating proteins of the pre-mRNA processing machinery (Badjatia et al., Reference Badjatia, Ambrosio, Lee and Günzl2013; Gosavi et al., Reference Gosavi, Srivastava, Badjatia and Gunzl2020).
Exon Junction Complex
All splice sites are imprinted to the mRNA by the exon junction complex, EJC, that binds 20−24 nucleotides upstream of exon−exon junctions and has many important cytoplasmic functions (e.g. the recognition of premature stop codons during the pioneer round of translation leading to NMD) (Woodward et al., Reference Woodward, Mabin, Gangras and Singh2017). The EJC consists of the subunits eIF4AIII, Y14 and Magoh; animals have one additional subunit (MLN51) (Table 4). In mammals, the spliceosomal protein CWC22 is involved in recruiting the EJC to the splice site via its interaction with eIF4AIII (Alexandrov et al., Reference Alexandrov, Colognori, Shu and Steitz2012; Barbosa et al., Reference Barbosa, Haque, Fiorini, Barrandon, Tomasetto, Blanchette and Le Hir2012; Steckelberg et al., Reference Steckelberg, Boehm, Gromadzka and Gehring2012). At least in metazoans, splicing appears to be a significant activator of mRNA export: microinjected intronic pre-mRNAs are far more efficiently exported than the same transcripts microinjected without the intron (Luo and Reed, Reference Luo and Reed1999). The likely reason is that the EJC plays a major role in recruiting the TREX complex to the mRNA by directly interacting with several of its components, including UAP56 and ALYREF (Le Hir et al., Reference Le Hir, Gatfield, Izaurralde and Moore2001; Masuda et al., Reference Masuda, Das, Cheng, Hurt, Dorman and Reed2005; Gromadzka et al., Reference Gromadzka, Steckelberg, Singh, Hofmann and Gehring2016; Gerbracht and Gehring, Reference Gerbracht and Gehring2018; Viphakone et al., Reference Viphakone, Sudbery, Griffith, Heath, Sims and Wilson2019). Still, in Drosophila, the EJC appears dispensable for bulk mRNA export (Gatfield and Izaurralde, Reference Gatfield and Izaurralde2002). Moreover, in S. cerevisiae, most mRNAs have no introns and TREX recruitment occurs predominantly via transcription.
Table 4. EJC components in yeast, metazoans and trypanosomes

a interaction with Y14 and Magoh could not be shown (Bercovich et al., Reference Bercovich, Levin, Clayton and Vazquez2009b). Whether this protein is the functional orthologue of eIF4AIII is still debated.
Trypanosomes and Leishmania have putative orthologues to the three major components of the exon junction complex, Magoh, eIF4AIII and Y14 (Bannerman et al., Reference Bannerman, Kramer, Dorrell and Carrington2018) (Table 4). Whether eIF4AIII (also called FAL1 and HEL54) is a true EJC component is still debated. T. brucei eIF4AIII is a low abundance protein that can be depleted with only minor effect on cell growth and massively overexpressed as wild type or ATPase inactive mutant with no effect on growth (Dhalia et al., Reference Dhalia, Marinsek, Reis, Katz, Muniz, Standart, Carrington and de Melo Neto2006). There are contradicting reports on eIF4EIII localization: eYFP fusions localize to the nucleus and nucleolus (Dhalia et al., Reference Dhalia, Marinsek, Reis, Katz, Muniz, Standart, Carrington and de Melo Neto2006; Dean et al., Reference Dean, Sunter and Wheeler2017) and the same localization was found with immunofluorescence using antiserum raised to the T. brucei protein (Dhalia et al., Reference Dhalia, Marinsek, Reis, Katz, Muniz, Standart, Carrington and de Melo Neto2006). In contrast, antiserum raised to the T. cruzi homologue (called HEL54) indicates cytoplasmic localization of both the T. cruzi and T. brucei proteins, mainly to dots close to the nucleus, in addition to nuclear localization; immuno gold electron microscopy confirmed localization to the outside of the nuclear pore as well as to the nucleus (Inoue et al., Reference Inoue, Serpeloni, Hiraiwa, Yamada-Ogatta, Muniz, Motta, Vidal, Goldenberg and Avila2014). Interestingly, nuclear localization of T. cruzi HEL54 could by enforced by (i) deleting the putative NES, indicating that the protein shuttles between the nucleus and the cytoplasm (ii) inhibition of transcription by Actinomycin D, indicating that export requires the presence of RNA and (iii) RNAi of Mex67, indicating that HEL54 export is Mex67 dependent (RNAi was done in T. brucei for technical reasons) (Inoue et al., Reference Inoue, Serpeloni, Hiraiwa, Yamada-Ogatta, Muniz, Motta, Vidal, Goldenberg and Avila2014). The localization of a shuttling protein to either nucleus or cytoplasm is sensitive to how cells were treated prior to fixation or imaging, a possible explanation for the discrepancy between these datasets. eIF4AIII failed to be co-precipitated by Y14-TAP, possibly because several residues essential for eIF4AIII interaction are mutated in Y14 and Magoh (Bercovich et al., Reference Bercovich, Levin, Clayton and Vazquez2009b). In contrast, the interaction between Y14 and Magoh could be detected both by Y2H and by co-immunoprecipitation (Bercovich et al., Reference Bercovich, Levin, Clayton and Vazquez2009b) and both Y14 and Magoh have nuclear localizations (Bercovich et al., Reference Bercovich, Levin, Clayton and Vazquez2009b; Dean et al., Reference Dean, Sunter and Wheeler2017). RNAi depletion of either Y14 or Magoh only caused minor or no growth effects, respectively, but the reduction in protein levels is not known (Bercovich et al., Reference Bercovich, Levin, Clayton and Vazquez2009b) and only knock-out experiments can answer the question, whether either protein is essential. In addition, the NTF2 domain protein Tb927.10.2240 was co-precipitated with Y14 (Bercovich et al., Reference Bercovich, Levin, Clayton and Vazquez2009b) but the genome-wide localization database TrypTag found cytoplasmic localization with tags at either C- or N- terminus (Dean et al., Reference Dean, Sunter and Wheeler2017), questioning its presence in the EJC. The available data provide evidence for the presence of an EJC in trypanosomes (note that trypanosomes also have a CWC22 homologue) but its composition and function remain unclear. With every mRNA trans-spliced in trypanosomes, the EJC has the potential to mark successful completion of mRNA 5′ end processing and promote nuclear export, however, this model is not supported by any data yet.
RES complex
The RES complex (pre-mRNA REtention and Splicing) was identified in yeast as a trimeric complex consisting of Pml1p, Snu17p and Bud13p, that associate with the spliceosome before the first catalytic step (Dziembowski et al., Reference Dziembowski, Ventura, Rutz, Caspary, Faux, Halgand, Laprévote and Séraphin2004). It has multiple functions in splicing that are only partially understood, but in particular Pml1p deletion caused leakage of pre-mRNAs to the cytoplasm while splicing was hardly affected, indicating that at least this subunit may have a direct role in regulating mRNA export (Dziembowski et al., Reference Dziembowski, Ventura, Rutz, Caspary, Faux, Halgand, Laprévote and Séraphin2004). The RES complex subunits and their spliceosome association are conserved in human (Deckert et al., Reference Deckert, Hartmuth, Boehringer, Behzadnia, Will, Kastner, Stark, Urlaub and Lührmann2006; Bessonov et al., Reference Bessonov, Anokhina, Will, Urlaub and Lührmann2008). Trypanosomes have no readily identifiable orthologues of the RES complex components Pml1p, Snu17p and Bud13p.
SR proteins
SR proteins are multifunctional RNA binding proteins that bind mRNAs throughout their journey from transcription to translation (Änkö, Reference Änkö2014; Wegener and Müller-McNicoll, Reference Wegener and Müller-McNicoll2019) and are conserved across eukaryotes (Busch and Hertel, Reference Busch and Hertel2012). They are best known for their essential roles in splicing and as regulators of alternative splicing but have many functions beyond, including an important role in selective mRNA export and retention (Müller-McNicoll et al., Reference Müller-McNicoll, Botti, de Jesus Domingues, Brandl, Schwich, Steiner, Curk, Poser, Zarnack and Neugebauer2016; Hautbergue et al., Reference Hautbergue, Castelli, Ferraiuolo, Sanchez-Martinez, Cooper-Knock, Higginbottom, Lin, Bauer, Dodd, Myszczynska, Alam, Garneret, Chandran, Karyka, Stopford, Smith, Kirby, Meyer, Kaspar, Isaacs, El-Khamisy, De Vos, Ning, Azzouz, Whitworth and Shaw2017; Zhou et al., Reference Zhou, Bulek, Li, Herjan, Yu, Qian, Wang, Zhou, Chen, Yang, Hong, Zhao, Qin, Fukuda, Flotho, Gao, Dongre, Carman, Kang, Su, Kern, Smith, Hamilton, Melchior, Fox and Li2017). Classical SR proteins consist of an N-terminal RRM domain, a glycine−arginine-rich spacer region of variable length and a C-terminal RS domain with at least 40% RS dipeptide content (Manley and Krainer, Reference Manley and Krainer2010). SR protein activity is tightly regulated by posttranslational modifications, in particular by reversible phosphorylations of the serine residues within the RS domain through a range of kinases and phosphatases (Zhou and Fu, Reference Zhou and Fu2013). Most SR proteins are adaptors for NXF1 and are required for selective nuclear export of specific mRNA isoforms (Müller-McNicoll et al., Reference Müller-McNicoll, Botti, de Jesus Domingues, Brandl, Schwich, Steiner, Curk, Poser, Zarnack and Neugebauer2016). The binding to NXF1 is mediated by two pairs of arginine residues flanking a glycine-rich region in the SR protein linker region (Lai and Tarn, Reference Lai and Tarn2004; Huang and Steitz, Reference Huang and Steitz2005; Hargous et al., Reference Hargous, Hautbergue, Tintaru, Skrisovska, Golovanov, Stévenin, Lian, Wilson and Allain2006; Tintaru et al., Reference Tintaru, Hautbergue, Hounslow, Hung, Lian, Craven and Wilson2007; Botti et al., Reference Botti, McNicoll, Steiner, Richter, Solovyeva, Wegener, Schwich, Poser, Zarnack, Wittig, Neugebauer and Müller-McNicoll2017). Importantly, SR proteins bind NXF1 only in their hypophosphorylated stage (Zhou and Fu, Reference Zhou and Fu2013), and, given that SR protein dephosphorylation is required for the release of the splicing machinery this suggests a possible mechanism for the selective export of spliced mRNAs (Huang and Steitz, Reference Huang and Steitz2005). Just like ALYREF of the TREX complex, the SR proteins SRSF3 and SRSF7 increase the RNA binding ability of NXF1 upon binding, possibly by inducing a structural change (Hautbergue et al., Reference Hautbergue, Hung, Golovanov, Lian and Wilson2008; Viphakone et al., Reference Viphakone, Hautbergue, Walsh, Chang, Holland, Folco, Reed and Wilson2012; Müller-McNicoll et al., Reference Müller-McNicoll, Botti, de Jesus Domingues, Brandl, Schwich, Steiner, Curk, Poser, Zarnack and Neugebauer2016). However, while ALYREF hands the mRNA over to NXF1, SR proteins bind close to NXF1 and remain in the complex during export (Müller-McNicoll et al., Reference Müller-McNicoll, Botti, de Jesus Domingues, Brandl, Schwich, Steiner, Curk, Poser, Zarnack and Neugebauer2016; Botti et al., Reference Botti, McNicoll, Steiner, Richter, Solovyeva, Wegener, Schwich, Poser, Zarnack, Wittig, Neugebauer and Müller-McNicoll2017). SR proteins add another level of complexity to the regulation of mRNA export: in mammalian cells, more than 1000 endogenous mRNAs require specific SR proteins for export (Müller-McNicoll et al., Reference Müller-McNicoll, Botti, de Jesus Domingues, Brandl, Schwich, Steiner, Curk, Poser, Zarnack and Neugebauer2016). The function of SR proteins in nuclear export is not restricted to mammals: even though S. cerevisiae lacks classical SR proteins, it has three SR-like proteins that share the basic SR protein domain structure: Npl3, Gbp2 and Hrb1. All these shuttle between the cytoplasm and the nucleus, when bound to newly transcribed mRNA (Lee et al., Reference Lee, Henry and Silver1996; Windgassen and Krebber, Reference Windgassen and Krebber2003; Häcker and Krebber, Reference Häcker and Krebber2004). Gbp2 and Hrb1 are yeast-specific subunits of the TREX complex and important mRNA surveillance factors: they bind their mRNA targets via the THO complex (Hurt et al., Reference Hurt, Luo, Röther, Reed and Sträßer2004) and recruit either Mex67 or the TRAMP complex (discussed in section ‘The nuclear exosome, the TRAMP complex and the NNS complex’), targeting the mRNA for export or decay, respectively (Hackmann et al., Reference Hackmann, Wu, Schneider, Meyer, Jung and Krebber2014). Npl3 acts as an adaptor protein for Mex67 and mediates mRNA export, regulated by a very similar phosphorylation and dephosphorylation cycle of Npl3 as described for mammalian SR proteins (Gilbert and Guthrie, Reference Gilbert and Guthrie2004).
Not counting the auxiliary splicing factor U2AF65 (=RBSR4), trypanosomes have at least five SR proteins: RBSR1 (Tb927.9.6870), RBSR2 (Tb927.9.6870), RBSR3 (Tb927.3.5460), TRRM1 (=RRM1, Tb927.2.4710) and TSR1 (Tb927.8.900) (Clayton, Reference Clayton2019) and all localize primarily to the nucleus (Manger and Boothroyd, Reference Manger and Boothroyd1998; Ismaïli et al., Reference Ismaïli, Pérez-Morga, Walsh, Mayeda, Pays, Tebabi, Krainer and Pays1999; Dean et al., Reference Dean, Sunter and Wheeler2017; Wippel et al., Reference Wippel, Malgarin, de Martins, Vidal, Marcon, Miot, Marchini, Goldenberg and Alves2019b). Studies indicate essential functions of trypanosome SR proteins in cis- and trans splicing, mRNA stability, processing of snoRNA, rRNA, and snRNAs, and modelling of chromatin structure (Manger and Boothroyd, Reference Manger and Boothroyd1998; Ismaïli et al., Reference Ismaïli, Pérez-Morga, Walsh, Mayeda, Pays, Tebabi, Krainer and Pays1999; Gupta et al., Reference Gupta, Chikne, Eliaz, Tkacz, Naboishchikov, Carmi, Waldman Ben-Asher and Michaeli2014; Levy et al., Reference Levy, Bañuelos, Níttolo, Ortiz, Mendiondo, Moretti, Tekiel and Sanchez2015; Naguleswaran et al., Reference Naguleswaran, Gunasekera, Schimanski, Heller, Hemphill, Ochsenreiter and Roditi2015; Wippel et al., Reference Wippel, Malgarin, de Martins, Vidal, Marcon, Miot, Marchini, Goldenberg and Alves2019b). Even though none of the SR proteins was co-purified with T. brucei MEX76 in cryomill-affinity purification (Obado et al., Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016), at least RRM1 has a potential function in mRNA processing and export: RRM1 co-precipitates with the nuclear non-canonical poly(A) polymerase NPAPL/ncPAP1, a putative subunit of the trypanosome TRAMP-complex ((Cristodero and Clayton, Reference Cristodero and Clayton2007; Etheridge et al., Reference Etheridge, Clemens, Gershon and Aphasizhev2009), see section ‘The nuclear exosome, the TRAMP complex and the NNS complex’), and also with retrotransposon hot spot proteins (Naguleswaran et al., Reference Naguleswaran, Gunasekera, Schimanski, Heller, Hemphill, Ochsenreiter and Roditi2015), which may in trypanosomes connect transcription with mRNA export (Florini et al., Reference Florini, Naguleswaran, Gharib, Bringaud and Roditi2019). T. brucei DRBD2 was suggested to be the orthologue of the yeast SR-like protein Gbp2, but it has cytoplasmic localization and no RS domain (Wippel et al., Reference Wippel, Malgarin, Inoue, Leprevost, Carvalho, Goldenberg and Alves2019a) and is unlikely a functional orthologue.
Adding the poly(A) tail
The 3′ end processing of an mRNA also occurs co-transcriptionally (Kumar et al., Reference Kumar, Clerici, Muckenfuss, Passmore and Jinek2019; Stewart, Reference Stewart2019a). In yeast, the 3′ end processing machinery is composed of the cleavage and polyadenylation factor (CPF) and the two accompanying cleavage factors CF1A and CF1B. The yeast CPF is a large multiprotein complex with three enzymatic activities. The first two activities are required for the addition of the poly(A) tail: the endonuclease (Ysh1/Brr5) cleaves the new transcript and the poly(A) polymerase (Pap1) successively adds AMP to the resulting free hydroxyl group at the 3′ end (Kumar et al., Reference Kumar, Clerici, Muckenfuss, Passmore and Jinek2019). The third activity links mRNA 3′ end processing to regulation of transcription elongation and termination and consists of two phosphatases that dephosphorylate serine 5 and tyrosine 1 of the CTD (Krishnamurthy et al., Reference Krishnamurthy, He, Reyes-Reyes, Moore and Hampsey2004; Schreieck et al., Reference Schreieck, Easter, Etzold, Wiederhold, Lidschreiber, Cramer and Passmore2014) (compare section ‘The C-terminal domain of RNAPII’). CF1A and CF1B contribute to RNA recognition and nuclease activation and bind specific RNA sequences (Yang and Doublié, Reference Yang and Doublié2011; Xiang et al., Reference Xiang, Tong and Manley2014). The human homologue of CPF is the cleavage and polyadenylation specificity factor (CPSF), which shares many orthologues with the yeast machinery (Kumar et al., Reference Kumar, Clerici, Muckenfuss, Passmore and Jinek2019). In human, the highly conserved AAUAAA motif of the polyadenylation signal (PAS) directs the cleavage of the pre-mRNA 10−30 nucleotides downstream (Hu et al., Reference Hu, Lutz, Wilusz and Tian2005; Derti et al., Reference Derti, Garrett-Engele, Macisaac, Stevens, Sriram, Chen, Rohl, Johnson and Babak2012; Chan et al., Reference Chan, Huppertz, Yao, Weng, Moresco, Yates, Ule, Manley and Shi2014; Schönemann et al., Reference Schönemann, Kühn, Martin, Schäfer, Gruber, Keller, Zavolan and Wahle2014; Gruber et al., Reference Gruber, Schmidt, Gruber, Martin, Ghosh, Belmadani, Keller and Zavolan2016) and this motif is conserved in fission yeast (Mata, Reference Mata2013; Schlackow et al., Reference Schlackow, Marguerat, Proudfoot, Bähler, Erban and Gullerova2013), albeit less well in budding yeast (Zhao et al., Reference Zhao, Hyman and Moore1999). Further cis-acting sequences contribute to poly(A) site recognition, but these are less well conserved between species. Whether and how poly(A) addition is connected to nuclear export is still largely unknown; an attractive model is that the release of the CPF from the RNP signals the completion of the export competent RNP (Stewart, Reference Stewart2019a).
The cleavage and polyadenylation complex is mostly conventional in trypanosomes (Hendriks et al., Reference Hendriks, Abdul-Razak and Matthews2003; Bercovich et al., Reference Bercovich, Levin and Vazquez2009a; Tkacz et al., Reference Tkacz, Gupta, Volkov, Romano, Haham, Tulinski, Lebenthal and Michaeli2010; Koch et al., Reference Koch, Raabe, Urlaub, Bindereif and Preusser2016), except that it contains at least two trypanosome-specific subunits (Tb927.11.13860 and Tb927.8.4480) and no potential CTD phosphatase is among the CPF components (Koch et al., Reference Koch, Raabe, Urlaub, Bindereif and Preusser2016). However, the recognition of the poly(A) site is non-conventional: instead of recognizing specific cis-elements on the mRNA, the cleavage takes place at a conserved distance to the polypyrimidine tract used for the trans-splicing of the upstream gene. This distance varies between different species of Trypanosomatida and is about 100 nucleotides in T. brucei (Campos et al., Reference Campos, Bartholomeu, daRocha, Cerqueira and Teixeira2008; Kolev et al., Reference Kolev, Franklin, Carmi, Shi, Michaeli and Tschudi2010; Clayton and Michaeli, Reference Clayton and Michaeli2011; Dillon et al., Reference Dillon, Okrah, Hughitt, Suresh, Li, Fernandes, Belew, Corrada Bravo, Mosser and El-Sayed2015). The likely reason for this unusual poly(A) site recognition is the strict coupling of trans-splicing with polyadenylation of the upstream transcript (LeBowitz et al., Reference LeBowitz, Smith, Rusche and Beverley1993; Ullu et al., Reference Ullu, Matthews and Tschudi1993; Matthews et al., Reference Matthews, Tschudi and Ullu1994), which is also reflected by the finding that RNAi knock-down experiments of most CPF proteins inhibit both polyadenylation and trans-splicing (Hendriks et al., Reference Hendriks, Abdul-Razak and Matthews2003; Koch et al., Reference Koch, Raabe, Urlaub, Bindereif and Preusser2016). Whether the presence of a poly(A) tail supports mRNA export is, like in other systems, not known. However, the fact that trypanosomes can export mRNAs co-transcriptionally ((Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019) discussed below), indicate that polyadenylation is at least not essential for export.
Poly(A) binding proteins
Once the poly(A) tail is synthesized, it is covered by nuclear poly(A) binding proteins, that come in two domain variants for RNA binding, either with zinc-finger domains (Nab2 in S. pombe and S. cerevisiae and ZC3H14 in human) or with a single RRM domain (Pab2 in S. pombe and PABPN1 in human) (Table 5). The S. cerevisiae zinc-finger protein Nab2 (which belongs to the SR protein family) plays a major role in mRNA export, as it shuttles between the nucleus and the cytoplasm, recruits Mex67-Mtr2 to the mRNA and interacts with the nuclear pore-associated protein Mlp1 (Fasken et al., Reference Fasken, Corbett and Stewart2019); less mechanistic information is available for the orthologous human zinc-finger protein ZC3H14 or the single RRM domain protein PABPN. Once the mRNA reaches the cytoplasm, its nuclear poly(A) binding proteins are replaced by cytoplasmic poly(A) binding proteins, that all have four RRM domains (Table 5). Most of these proteins can shuttle to the nucleus and also function in mRNA export; this is best established for Pab1 of S. cerevisiae (Brune et al., Reference Brune, Munchel, Fischer, Podtelejnikov and Weis2005; Dunn et al., Reference Dunn, Hammell, Hodge and Cole2005; Brambilla et al., Reference Brambilla, Martani, Bertacchi, Vitangeli and Branduardi2019).
Table 5. Poly(A) binding proteins in yeast, human and trypanosomes

a Mammals have several further cytoplasmic PABP isoforms: tPABP (testis-specific), ePABP (embryonic) and PABP4 (Gray et al., Reference Gray, Hrabálková, Scanlon and Smith2015).
Trypanosomatidae have no obvious homologues to yeast or human nuclear poly(A) binding proteins with CCCH zinc-finger domains or single RRMs (Table 5). Instead, they have two (T. brucei) or three (T. cruzi and Leishmania) essential poly(A) binding proteins with four RRM domains that have a dominant cytoplasmic localization and co-purify with translating polysomes (Bates et al., Reference Bates, Knuepfer and Smith2000; da Costa Lima et al., Reference da Costa Lima, Moura, Reis, Vasconcelos, Ellis, Carrington, Figueiredo and de Melo Neto2010; Kramer et al., Reference Kramer, Bannerman-Chukualim, Ellis, Boulden, Kelly, Field and Carrington2013). In T. brucei, PABP2 appears to be the major poly(A) binding protein for bulk mRNAs, as it co-purifies a diverse range of RNA binding proteins and localizes across both small and large polysomal fractions; while PABP1 is found in small polysomes only and binds to only few proteins (Zoltner et al., Reference Zoltner, Krienitz, Field and Kramer2018). Interestingly, T. brucei PABP2 and Leishmania PABP2 and PABP3 (but not PABP1 of either organism) can be trapped inside the nucleus under certain conditions (da Costa Lima et al., Reference da Costa Lima, Moura, Reis, Vasconcelos, Ellis, Carrington, Figueiredo and de Melo Neto2010; Kramer et al., Reference Kramer, Bannerman-Chukualim, Ellis, Boulden, Kelly, Field and Carrington2013). This indicates that these isoforms are shuttling, fulfilling both the function of a nuclear and a cytoplasmic PABP, analogous to yeast Pab1. Whether PABP2 is involved in nuclear export is not known, but given that nuclear export can occur co-transcriptionally prior to poly(A) tail synthesis ((Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019), see section ‘Co-transcriptional initiation of RNA export indicates the lack of major mRNA export checkpoints in trypanosomes’), its binding to the poly(A) is at least not essential for export.
The TREX-2 or THSC complex
Just like TREX-1, the TREX-2 or THSC complex has multiple roles in nuclear mRNA metabolism, ranging from regulation of transcription to mRNA export (García-Oliver et al., Reference García-Oliver, García-Molinero and Rodríguez-Navarro2012; Stewart, Reference Stewart2019b). It consists of the Sac3 (GANP) scaffold, bound to Thp1 (PCID2), Cdc31 (centrin-2/CENP), Sem1 (DSS1) and two copies of Sus1 (ENY1) (Stewart, Reference Stewart2019b). In resting cells, TREX-2 is mainly found at the nucleoplasmic site of the NPC, where it interacts (via Sac3) with the export factor Mex67-Mtr2 (NXF1-NXT1) as well as the NUPs Nup1 (NUP153) and, at least in vertebrates, TPR (the metazoan homologue to Mlp1, Fig. 2) (Ullman et al., Reference Ullman, Shah, Powers and Forbes1999; Fischer et al., Reference Fischer, Sträßer, Rácz, Rodríguez-Navarro, Oppizzi, Ihrig, Lechner and Hurt2002; Soop et al., Reference Soop, Ivarsson, Björkroth, Fomproix, Masich, Cordes and Daneholt2005; Wickramasinghe et al., Reference Wickramasinghe, McMurtrie, Mills, Takei, Penrhyn-Lowe, Amagase, Main, Marr, Stewart and Laskey2010; Rajanala and Nandicoori, Reference Rajanala and Nandicoori2012; Umlauf et al., Reference Umlauf, Bonnet, Waharte, Fournier, Stierle, Fischer, Brino, Devys and Tora2013; Jani et al., Reference Jani, Valkov and Stewart2014; Aksenova et al., Reference Aksenova, Smith, Lee, Bhat, Esnault, Chen, Iben, Kaufhold, Yau, Echeverria, Fontoura, Arnaoutov and Dasso2020). The TREX-2 complex is essential for mRNA export (Fischer et al., Reference Fischer, Sträßer, Rácz, Rodríguez-Navarro, Oppizzi, Ihrig, Lechner and Hurt2002; Wickramasinghe et al., Reference Wickramasinghe, McMurtrie, Mills, Takei, Penrhyn-Lowe, Amagase, Main, Marr, Stewart and Laskey2010; Umlauf et al., Reference Umlauf, Bonnet, Waharte, Fournier, Stierle, Fischer, Brino, Devys and Tora2013; Jani et al., Reference Jani, Valkov and Stewart2014). In yeast, TREX-2 also interacts with complexes involved in transcription (Rodríguez-Navarro et al., Reference Rodríguez-Navarro, Fischer, Luo, Antúnez, Brettschneider, Lechner, Pérez-Ortín, Reed and Hurt2004; García-Oliver et al., Reference García-Oliver, García-Molinero and Rodríguez-Navarro2012; Schneider et al., Reference Schneider, Hellerschmied, Schubert, Amlacher, Vinayachandran, Reja, Pugh, Clausen and Köhler2015; García-Molinero et al., Reference García-Molinero, García-Martínez, Reja, Furió-Tarí, Antúnez, Vinayachandran, Conesa, Pugh, Pérez-Ortín and Rodríguez-Navarro2018) and it has been suggested to play a role in repositioning transcribed genes to the NPC (Jani et al., Reference Jani, Lutz, Marshall, Fischer, Köhler, Ellisdon, Hurt and Stewart2009), a phenomenon called ‘gene gating’ (Ben-Yishay et al., Reference Ben-Yishay, Ashkenazy and Shav-Tal2016). TREX-2 may therefore play a major role in connecting mRNA transcription with export, but the detailed function of the complex remains to be explored. Trypanosomes have no obvious orthologues to the core components of the yeast TREX-2/THSC complex, Sac2, Thp1, Sem1 and Sus1.
Mex67-Mtr2 (NXF1-NXT1 or TAP-p15): the major mRNA export factor
The Mex67-Mtr2 heterodimer (NXF1-NXT1 or TAP-p15 in metazoans) is the major mRNA export complex, conserved across most eukaryotes (Segref et al., Reference Segref, Sharma, Doye, Hellwig, Huber, Lührmann and Hurt1997; Katahira et al., Reference Katahira, Strässer, Podtelejnikov, Mann, Jung and Hurt1999). It binds its mRNA targets directly, or more often indirectly (for example via the Yra1/ALYREF/THOC4 subunit of the TREX complex) and then mediates the export of its cargo by interacting with FG NUPs of the NPC. Mex67/NXF1/TAP has five domains: (i) the N-terminal arginine-rich RNA binding domain binds RNA and this activity is essential for RNA export (Zolotukhin et al., Reference Zolotukhin, Tan, Bear, Smulevitch and Felber2002; Hautbergue et al., Reference Hautbergue, Hung, Golovanov, Lian and Wilson2008). This domain becomes accessible to RNA by a conformational change of the protein induced by its binding to the TREX complex (Viphakone et al., Reference Viphakone, Hautbergue, Walsh, Chang, Holland, Folco, Reed and Wilson2012). (ii and iii) the pseudo RRM (RNA recognition motif) domain and the LRR (leucine-rich repeat) domain of NXF1/TAP are both involved in export mostly by binding splicing factors of the SR (serine−arginine rich) protein family (Huang et al., Reference Huang, Gattoni, Stévenin and Steitz2003; Müller-McNicoll et al., Reference Müller-McNicoll, Botti, de Jesus Domingues, Brandl, Schwich, Steiner, Curk, Poser, Zarnack and Neugebauer2016). (iv and v) the NTF2L and UBA (ubiquitin associated) domains mediate the interactions between NSF1/TAP and the FG Nups of the NPC, allowing transport (Fribourg et al., Reference Fribourg, Braun, Izaurralde and Conti2001). The smaller partner of the complex, Mtr2/NXT1, also has an NTF2-like fold, which binds to the NTF2L domain of Mex67/NXF1. Recent data indicate that the interaction of Mex67/NXF1 with the nuclear pores is independent on mRNA (Ben-Yishay et al., Reference Ben-Yishay, Mor, Shraga, Ashkenazy-Titelman, Kinor, Schwed-Gross, Jacob, Kozer, Kumar, Garini and Shav-Tal2019; Derrer et al., Reference Derrer, Mancini, Vallotton, Huet, Weis and Dultz2019) and, at least in yeast, is not even interrupted by the Dbp5 remodelling in the cytoplasm (Derrer et al., Reference Derrer, Mancini, Vallotton, Huet, Weis and Dultz2019). Strikingly, a fusion of Mex67 and Nup116 is sufficient to compensate for Mex67 deletion, indicating that at least the essential function of Mex67 is fully restricted to its nuclear pore localization (Derrer et al., Reference Derrer, Mancini, Vallotton, Huet, Weis and Dultz2019).
The trypanosome Mex67 protein was identified by homology searches in the trypanosome genome (Schwede et al., Reference Schwede, Manful, Jha, Helbig, Bercovich, Stewart and Clayton2009). Affinity purification of Mex67 resulted in the identification of the trypanosome Mtr2 homologue, a 15.2 kDa protein with an NTF2 domain that shares higher similarity to the human p15 than to yeast Mtr2 (Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013). Trypanosome Mex67/Mtr2 fulfils all characteristics expected of a functional mRNA export complex: (i) Mex67 localizes to the nuclear pores (Kramer et al., Reference Kramer, Kimblin and Carrington2010; Dean et al., Reference Dean, Sunter and Wheeler2017) and Mtr2 to the nucleus (Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013; Dean et al., Reference Dean, Sunter and Wheeler2017) and possibly to the nuclear pores (Dean et al., Reference Dean, Sunter and Wheeler2017). (ii) Depletion of either Mex67 or Mtr2 causes a growth effect and accumulation of polyadenylated mRNAs in the nucleus (Schwede et al., Reference Schwede, Manful, Jha, Helbig, Bercovich, Stewart and Clayton2009; Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013). (iii) In affinity capture experiments Mex67 co-isolates the Nup76 complex (Nup76, Nup140, Nup149) and the NUPs Nup152, Nup158 and Nup89; and, under low stringency conditions, many further NUPs, indicating interactions with the nuclear pore (Obado et al., Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016) (Fig. 2). Uniquely, TbMex67 possess a CCCH type zinc finger at its N-terminus (Kramer et al., Reference Kramer, Kimblin and Carrington2010; Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013) that is essential for its function (Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013). It is tempting to speculate, that the CCCH finger mediates mRNA binding of Mex67, perhaps even specific to the miniexon sequence that is present on every mRNA (Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013). Consistent with this hypothesis is the absence of a TREX-1 and TREX-2 complex in trypanosomes (see sections ‘The TREX complex’ and ‘The TREX-2 or THSC complex’) and the fact that no putative Mex67 adaptor protein co-purified with Mex67 (Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013; Obado et al., Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016). Two independent studies have analysed Mex67 interacting proteins (Table 6): The first study was a classical immunoprecipitation using TbMex67 with a C-terminal PTP tag as a bait (Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013). This resulted in two equally strong bands on a Coomassie gel that were identified by mass spectrometry as Mtr2 (as expected) and, surprisingly, importin 1 (IMP1): importins transport proteins from the cytoplasm to the nucleus. TbIMP1 has nuclear pore localization (Dean et al., Reference Dean, Sunter and Wheeler2017) and its depletion by RNAi is lethal and causes poly(A) accumulation in the nucleus, indicating an important role in mRNA export, perhaps as a transporter of Mex67 (Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013). The closest homologue to IMP1 in human is the karyopherin TRN2 (transportin 2). TNR2 has been shown to interact with NXF1 in two independent studies; however, the studies contradict each other: in one study this interaction was RanGTP dependent (Shamsher et al., Reference Shamsher, Ploski and Radu2002), in the other it was RanGTP sensitive (Güttinger et al., Reference Güttinger, Mühlhäusser, Koller-Eichhorn, Brennecke and Kutay2004), indicating a function in protein export or protein import, respectively. To date, a function in protein import is considered more likely (Twyffels et al., Reference Twyffels, Gueydan and Kruys2014). It remains unclear, whether T. brucei IMP1 is an importin or exportin and whether it has other targets than Mex67 (TNR2 has many additional cargoes (Güttinger et al., Reference Güttinger, Mühlhäusser, Koller-Eichhorn, Brennecke and Kutay2004)). Interestingly, upon depletion of IMP1 in trypanosomes, cell fractionation experiments detected a shift of Mex67 from the cytoplasmic to the nuclear fractions (Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013), indicative of IMP1 functioning in Mex67 export. In this context, the results from the second Mex67 interaction study (based on cryomilled trypanosomes and mass spectrometry detection of all Mex67 co-purified proteins) are highly interesting: next to Mtr2 and IMP1 and many nuclear pore proteins (Fig. 2) Mex67 also co-purified stoichiometric amounts of the small GTPase Ran and Ran binding proteins (RanBP1 and GAP TBC-RootA) (Obado et al., Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016). It is unlikely that Mex67 binds Ran directly: binding to Ran would probably be via the NTF2-like domain of Mex67 (as NTF2 binds to and imports Ran-GDP into the nucleus (Nehrbass and Blobel, Reference Nehrbass and Blobel1996; Ribbeck et al., Reference Ribbeck, Lipowsky, Kent, Stewart and Görlich1998; Smith et al., Reference Smith, Brownawell and Macara1998; Stewart et al., Reference Stewart, Kent and McCoy1998) but the structure of the mammalian TAP/p15 complex shows that the NTF2-like domain is not accessible to Ran (Fribourg et al., Reference Fribourg, Braun, Izaurralde and Conti2001) and a high confidence model of the trypanosome complex based on this structure shows the same (Obado et al., Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016). One model consistent with all data would be (i) Inside the nucleus, Mex67-Mtr2 binds its mRNA cargo via the zinc-finger domain of Mex67 and it also binds to IMP1-RanGTP. (ii) This complex passes the nuclear pore. (iii) At the cytoplasmic site, Ran hydrolyses the bound GTP aided by GTPase activating protein (GAP) and RanBP1, resulting in disassembly of the complex and release of the mRNA cargo to the cytoplasm. The last step would be analogous to the ATP-dependent remodelling of the RNP export complex by the DEAD box RNA helicase Dbp5 in opisthokonts, potentially compensating for the absence of Dbp5 in trypanosomes (see section ‘Nuclear pores and NUPs’). Note that this model is purely speculative and more experimental work is required to determine the exact functions of all Mex67 interacting proteins.
Table 6. Summary of Mex67 interactions partners in trypanosomes, identified by two independent studies

T. brucei Mex67/Mtr2 has additional functions in tRNA export ((Hegedűsová et al., Reference Hegedűsová, Kulkarni, Burgman, Alfonzo and Paris2019), review in this issue from Zdenek Paris) and in ribosome biogenesis (Rink and Williams, Reference Rink and Williams2019; Rink et al., Reference Rink, Ciganda and Williams2019).
mRNA export by the rRNA transporters NMD3 and XPO1?
The proteins Crm1/Xpo1 (exportin 1) and Nmd3 mediate transport of the large ribosomal subunit subunits through the pore: the nuclear export signal containing protein Nmd3 acts as an adaptor to recruit the Crm1/Xpo1 export receptor to the pre-60S subunit, facilitating its export (Johnson et al., Reference Johnson, Lund and Dahlberg2002; Baßler and Hurt, Reference Baßler and Hurt2019). Trypanosomes have orthologues for both Nmd3 and Xpo1 (Zeiner et al., Reference Zeiner, Sturm and Campbell2003a; Prohaska and Williams, Reference Prohaska and Williams2009) and the function in nuclear export of the large ribosomal subunit appears conserved: TbXPO1 depletion causes nuclear accumulation of ribosomal RNAs (Biton et al., Reference Biton, Mandelboim, Arvatz and Michaeli2006), TbNMD3 depletion inhibited processing of the large ribosomal subunit (Droll et al., Reference Droll, Archer, Fenn, Delhi, Matthews and Clayton2010; Rink et al., Reference Rink, Ciganda and Williams2019) and both XPO1 and NMD3 associate with T. brucei 60S ribosomal subunits (Prohaska and Williams, Reference Prohaska and Williams2009).
Surprisingly, unlike in other systems, trypanosome NMD3 and XPO1 appear also involved in mRNA export (Bühlmann et al., Reference Bühlmann, Walrad, Rico, Ivens, Capewell, Naguleswaran, Roditi and Matthews2015): RNAi depletion of NMD3 caused the poly(A) FISH (fluorescence in situ hybridization) signal to shift from being mainly cytoplasmic to being almost entirely nuclear (Bühlmann et al., Reference Bühlmann, Walrad, Rico, Ivens, Capewell, Naguleswaran, Roditi and Matthews2015), exactly like RNAi depletion of MEX67 (Schwede et al., Reference Schwede, Manful, Jha, Helbig, Bercovich, Stewart and Clayton2009; Dostalova et al., Reference Dostalova, Käser, Cristodero and Schimanski2013). Moreover, RNAi depletion of either NMD3, XPO1 or MEX67 have identical effects on mRNA levels: there is a minor stabilization of most mRNAs, and a pronounced stabilization of mRNAs encoded by the so-called PAG genes (procyclin-associated genes), short-lived transcripts that are co-transcribed with the very stable and abundant mRNA encoding the cell surface proteins of the procyclic life cycle stage (the stage that resides in the tsetse fly midgut) (Bühlmann et al., Reference Bühlmann, Walrad, Rico, Ivens, Capewell, Naguleswaran, Roditi and Matthews2015). The reason for this massive stabilization of this group of mRNAs upon block in RNA export is not fully understood, but stabilization depends on the mRNAs conserved 5′ UTR and is independent on transcription or translation (Bühlmann et al., Reference Bühlmann, Walrad, Rico, Ivens, Capewell, Naguleswaran, Roditi and Matthews2015). The likeliest explanation is that the block in mRNA export prevents the mRNAs to reach their cytoplasmic destiny of degradation.
Do trypanosomes have two alternative pathways to export mRNAs? MEX67 RNAi is lethal, indicating that the XPO1/NMD3 system cannot compensate for the absence of MEX67. It is therefore more likely that MEX67-Mtr2 and XPO1/NDM3 export pathways interact and depend on each other, in a way that needs to be established.
Nuclear pores and NUPs
Many NUPs of the NPC play active roles in regulating or mediating mRNA export (Ashkenazy-Titelman et al., Reference Ashkenazy-Titelman, Shav-Tal and Kehlenbach2020). In vertebrates, five FG Nups have direct interactions with the C-terminal region of NXT1, namely Nup62 (cytoplasmic), Nup98 (outer ring), Nup153 (nucleoplasmic), Nup214 (cytoplasmic) and Nup358 (cytoplasmic outer ring) (Bachi et al., Reference Bachi, Braun, Rodrigues, Panté, Ribbeck, von Kobbe, Kutay, Wilm, Görlich, Carmo-Fonseca and Izaurralde2000; Forler et al., Reference Forler, Rabut, Ciccarelli, Herold, Köcher, Niggeweg, Bork, Ellenberg and Izaurralde2004); if present, the respective homologues in yeast and trypanosomes are indicated by black asterisks in Fig. 2. At the nuclear basket Nup1 (NUP153 in vertebrates) binds the export competent RNP via the TREX-2 complex; in vertebrates TPR (the homologue to yeast Mlp1) contributes to this interaction (Ullman et al., Reference Ullman, Shah, Powers and Forbes1999; Soop et al., Reference Soop, Ivarsson, Björkroth, Fomproix, Masich, Cordes and Daneholt2005; Rajanala and Nandicoori, Reference Rajanala and Nandicoori2012; Umlauf et al., Reference Umlauf, Bonnet, Waharte, Fournier, Stierle, Fischer, Brino, Devys and Tora2013; Jani et al., Reference Jani, Valkov and Stewart2014; Aksenova et al., Reference Aksenova, Smith, Lee, Bhat, Esnault, Chen, Iben, Kaufhold, Yau, Echeverria, Fontoura, Arnaoutov and Dasso2020). In yeast, the basket NUP Mlp1 acts as a gatekeeper to prevent the export of immature mRNAs, in particular of unspliced mRNAs (Green et al., Reference Green, Johnson, Hagan and Corbett2003; Galy et al., Reference Galy, Gadal, Fromont-Racine, Romano, Jacquier and Nehrbass2004; Vinciguerra et al., Reference Vinciguerra, Iglesias, Camblong, Zenklusen and Stutz2005) and this function appears conserved in Metazoans (Coyle et al., Reference Coyle, Bor, Rekosh and Hammarskjold2011; Rajanala and Nandicoori, Reference Rajanala and Nandicoori2012). In yeast, the Mlp1/Mlp2 interacting protein Pml39 is equally essential for retention of unspliced transcripts and may work as an upstream regulator of Mlp1 (Palancade et al., Reference Palancade, Zuccolo, Loeillet, Nicolas and Doye2005). These interactions of the RNP with proteins of the nuclear basket dock the export-competent RNP to the pore, in preparation for export. The human β-actin mRNA resides on average 80 ms at the basket (Grünwald and Singer, Reference Grünwald and Singer2010). Translocation through the export channel is fast (5–20 ms for β-actin (Grünwald and Singer, Reference Grünwald and Singer2010)) and the contributing mRNA-specific Nups are less well-known, possibly because central channel Nups are structurally too essential to test specific roles. In vertebrates, Nup98, Nup133 and Nup160 have suspected roles in intermediate mRNA export (Powers et al., Reference Powers, Forbes, Dahlberg and Lund1997; Vasu et al., Reference Vasu, Shah, Orjalo, Park, Fischer and Forbes2001; Blevins et al., Reference Blevins, Smith, Phillips and Powers2003) (red asterisks in Fig. 2). The final steps of RNA export at the cytoplasmic filaments (80 ms for β-actin (Grünwald and Singer, Reference Grünwald and Singer2010)) are better understood. Central is the DEAD-box RNA helicase Dbp5 (DDX19 in vertebrates) that remodels the RNP complex by separating double-stranded RNA regions and RNA−protein interactions, to release export factors, including Mex67 (Lund and Guthrie, Reference Lund and Guthrie2005; von Moeller et al., Reference von Moeller, Basquin and Conti2009; Lin et al., Reference Lin, Correia, Cai, Huber, Jette and Hoelz2018). The ATP dependency of this process ensures directionality of mRNA export. The second key-player in this cytoplasmic remodelling process is the NUP Gle1 (Murphy and Wente, Reference Murphy and Wente1996; Watkins et al., Reference Watkins, Murphy, Emtage and Wente1998), which is required to activate Dbp5 (Alcázar-Román et al., Reference Alcázar-Román, Tran, Guo and Wente2006; Weirich et al., Reference Weirich, Erzberger, Flick, Berger, Thorner and Weis2006). In yeast, inositol hexakisphosphate as an essential co-activator of Dbp5 (Alcázar-Román et al., Reference Alcázar-Román, Tran, Guo and Wente2006; Weirich et al., Reference Weirich, Erzberger, Flick, Berger, Thorner and Weis2006); whether this small molecule is also needed in vertebrates is still debated (Adams et al., Reference Adams, Mason, Glass, Aditi and Wente2017; Lin et al., Reference Lin, Correia, Cai, Huber, Jette and Hoelz2018). Both Dbp5 and Gle1 have direct interactions with NUPs of the cytoplasmic filaments: Dbp5 binds Nup159 (Nup214 in vertebrates) and Gle1 binds Nup42 (hCG1 in human) (Murphy and Wente, Reference Murphy and Wente1996; Strahm et al., Reference Strahm, Fahrenkrog, Zenklusen, Rychner, Kantor, Rosbach and Stutz1999; Kendirgi et al., Reference Kendirgi, Rexer, Alcázar-Román, Onishko and Wente2005; Alcázar-Román et al., Reference Alcázar-Román, Bolger and Wente2010).
The architecture of the yeast NPC is well known (Alber et al., Reference Alber, Dokudovskaya, Veenhoff, Zhang, Kipper, Devos, Suprapto, Karni-Schmidt, Williams, Chait, Rout and Sali2007a, Reference Alber, Dokudovskaya, Veenhoff, Zhang, Kipper, Devos, Suprapto, Karni-Schmidt, Williams, Chait, Sali and Rout2007b), in subnanometer resolution (Kim et al., Reference Kim, Fernandez-Martinez, Nudelman, Shi, Zhang, Raveh, Herricks, Slaughter, Hogan, Upla, Chemmama, Pellarin, Echeverria, Shivaraju, Chaudhury, Wang, Williams, Unruh, Greenberg, Jacobs, Yu, de la Cruz, Mironska, Stokes, Aitchison, Jarrold, Gerton, Ludtke, Akey, Chait, Sali and Rout2018) and the structure of the trypanosome NPC was modelled based on homology studies and affinity capture/mass spectrometry interactomic studies (DeGrasse et al., Reference DeGrasse, Chait, Field and Rout2008, Reference DeGrasse, DuBois, Devos, Siegel, Sali, Field, Rout and Chait2009; Obado et al., Reference Obado, Brillantes, Uryu, Zhang, Ketaren, Chait, Field and Rout2016, Reference Obado, Field and Rout2017) (Fig. 2). A comparison shows that structure and composition of the NPCs are in principle conserved between yeast and trypanosomes, in particular within the inner ring of the pore (Fig. 2). However, there are some striking differences: (a) Trypanosome nuclear pores are highly symmetrical, with the only exception of the trypanosome-specific proteins NUP110 and NUP92, which are exclusively found at the nuclear basket. In contrast, yeast (and also metazoan) NPCs contain several nuclear pore proteins that specifically localize to either the nuclear basket or the cytoplasmic site of the pore. This asymmetry is crucial for the directionality of mRNP export in opisthokonts (Hurwitz et al., Reference Hurwitz, Strambio-de-Castillia and Blobel1998; Schmitt et al., Reference Schmitt, von Kobbe, Bachi, Panté, Rodrigues, Boscheron, Rigaut, Wilm, Séraphin, Carmo-Fonseca and Izaurralde1999; Folkmann et al., Reference Folkmann, Noble, Cole and Wente2011) and it remains unknown, how directionality of transport is achieved in trypanosomes. (b) Among the proteins that are asymmetrically distributed in yeast and absent in trypanosomes are many proteins with important and well-characterized functions in mRNP export, namely Gle1, Dbp5 and Nup159 at the cytoplasmic filaments. Moreover, whether the only trypanosome proteins with asymmetric distribution, NUP92 and NUP110, are orthologues of the Opisthokont Mlp proteins is not certain, as evidence indicates independent ancestry (Holden et al., Reference Holden, Koreny, Obado, Ratushny, Chen, Chiang, Kelly, Chait, Aitchison, Rout and Field2014). Function of NUP92 in chromosome segregation appears conserved, albeit a knock-out is viable and can adapt to normal growth over time (Holden et al., Reference Holden, Koreny, Obado, Ratushny, Chen, Chiang, Kelly, Chait, Aitchison, Rout and Field2014). (c) Of the five FG NUPs that in vertebrates have direct interactions with NXF1, trypanosomes only have two (NUP62 and NUP158) while yeast only lack the metazoan specific protein NUP358 (Fig. 2). (d) In opisthokonts, mRNP export is thriven by ATP hydrolysis that is used by the RNA helicase Dbp5 for remodelling the mRNP complex at the cytoplasmic site of the pore. Trypanosomes have no Dbp5 and the interaction of Mex67 with importin1, Ran, RanBP1 and the corresponding GAP (see section ‘Mex67-Mtr2 (NXF1-NXT1 or TAP-p15): the major mRNA export factor’) indicates that mRNA export is GTP dependent instead: another fundamental difference between trypanosomes and yeast mRNA export.
The nuclear exosome, the TRAMP complex and the NNS complex
Faulty RNAs and all processing by-products that accumulate in the nucleus are degraded by the nuclear exosome. The core of the eukaryotic RNA exosome is a barrel-shaped structure out of six RNAse PH-like proteins (that are enzymatically inactive) with three S1/KH RNA-binding-domain containing proteins positioned at the top of the barrel (Schmid and Jensen, Reference Schmid and Jensen2019). RNA degradation activity is provided by the processive 3′–5′ exonuclease and endonuclease Dis3 (also called Rrp44) at the bottom of the barrel and the distributive 3′–5′ exonuclease Rrp6 (EXOSC10 in humans) localized at the top (Schmid and Jensen, Reference Schmid and Jensen2019). Two further proteins are found at the top of the barrel: Lrp1 (also Rrp47, C1D in human) and Mpp6 (MPP6 in human). This 13-subunit nuclear exosome (also called Exo13, Table 7) is already active, but requires further subunits for efficient and target-specific RNA degradation.
Table 7. Exosome and TRAMP complex subunits in trypanosomes and yeast

a The six trypanosome RNAse PH subunits cannot be clearly assigned to the yeast orthologues.
b It is not yet established, which of these proteins (if any) is the functional orthologue to Air1.
One is the TRAMP complex (Trf4-Air2-Mtr4 polyadenylation), that consists of the RNA helicase Mtr4 (MTR4 (SKIV2L2) in human), the poly(A) polymerase Trf4 (PAPD5 (TRF4-2) in human) and the RNA binding protein Air1 (ZCCHC7 (AIR1) in human) (Schmid and Jensen, Reference Schmid and Jensen2019) (Table 7). Trf4 is thought to add short A-tails to 3′ ends of exosome targeted RNAs; these tails are believed to facilitate loading of the RNA substrate to Mtr4, which resides at the top of the exosome barrel and probably unwinds the RNA substrate prior to presenting it either to Rrp6 or injecting it into the barrel for degradation by Dis3 (Schmid and Jensen, Reference Schmid and Jensen2019). The Zn-finger containing Air1 protein likely provides RNA binding activity to the TRAMP complex. The TRAMP complex is engaged in multiple functions, including the decay of highly structured RNAs that would be insensitive to exosomal digestions without the Mtr4 helicase.
Substrate recognition of the TRAMP complex can occur via its RNA binding protein Air1, however, RNA polymerase II products are often recognized by the NNS complex (Schmid and Jensen, Reference Schmid and Jensen2019). In yeast, this complex consists of the RNA binding proteins Nrd1 and Nab3 and the RNA helicase Sen1. Nrd1 and Nab3 have sequence-specific RNA binding domains involved in exosome substrate recognition. Next to its interaction with the exosome, Nrd1 also interacts with serine phosphorylated CTD of RNA polymerase II, linking early transcription with decay (Schmid and Jensen, Reference Schmid and Jensen2019). One outstanding question is how the exosome distinguishes faulty RNAs from correctly processed RNAs destined for export. The model that currently fits best to the available data is that nuclear RNA degradation is not very selective but rather the default pathway (Schmid and Jensen, Reference Schmid and Jensen2018; Tudek, Reference Tudek2019). The turn-over rate of nuclear RNAs is in general high ((Wyers et al., Reference Wyers, Rougemaille, Badis, Rousselle, Dufour, Boulay, Régnault, Devaux, Namane, Séraphin, Libri and Jacquier2005; Preker et al., Reference Preker, Nielsen, Kammler, Lykke-Andersen, Christensen, Mapendano, Schierup and Jensen2008) and RNAs prevented from nuclear export are therefore more likely to be degraded than RNAs that exit fast. For example, in yeast, the spliceosome and the exosome compete for intron-containing RNAs and more than half are degraded instead of spliced (Gudipati et al., Reference Gudipati, Xu, Lebreton, Séraphin, Steinmetz, Jacquier and Libri2012). After splicing, cap and poly(A) tail appear to provide a certain protection, but if nuclear export is inhibited these mature transcripts are doomed to degradation too (Tudek et al., Reference Tudek, Schmid, Makaras, Barrass, Beggs and Jensen2018). Consistently, long-lived nuclear RNAs (such as snRNAs or snoRNAs) require specific protective measures to escape the default RNA decay pathway in the nucleus (Schmid and Jensen, Reference Schmid and Jensen2018).
Trypanosomes have orthologues to all RNA exosome subunits and most co-precipitate with each other (Estevez et al., Reference Estevez, Kempf and Clayton2001, Reference Estevez, Lehner, Sanderson, Ruppert and Clayton2003) (Table 7). The lack of co-precipitation of the Rrp44 orthologue questioned whether this subunit is part of the complex (Estevez et al., Reference Estevez, Kempf and Clayton2001; Clayton and Estevez, Reference Clayton and Estevez2010), but given that Rrp44 and Rrp6 have identical, characteristic localization patterns to the nucleoplasm and to the periphery of the nucleolus (Kramer et al., Reference Kramer, Piper, Estevez and Carrington2016) and both are involved in 5.8S rRNA processing (Estevez et al., Reference Estevez, Kempf and Clayton2001) the lack of co-precipitation is likely to reflect a weak interaction rather than none. All evidence points towards trypanosomes having a conserved RNA exosome with mostly or entirely nuclear localization and with mostly conserved and essential function in rRNA processing (Estevez et al., Reference Estevez, Kempf and Clayton2001), snoRNA processing (Fadda et al., Reference Fadda, Färber, Droll and Clayton2013) and removal of unspliced mRNAs (Kramer et al., Reference Kramer, Piper, Estevez and Carrington2016). Importantly, all these exosomal functions were concluded from accumulation of the respective RNA species upon depletion of exosome components; thus, whether the exosome specifically targets these RNAs, or, whether these RNAs are degraded because they have an extended exposure time to the exosome is not known. The later model, which is in agreement to the current model in opisthokonts (Schmid and Jensen, Reference Schmid and Jensen2018; Tudek, Reference Tudek2019), is supported by a simulation of trypanosome mRNA decay pathways that predicts co-transcriptional degradation of mRNA precursors by the exosome: accordingly, mRNA processing and degradation compete and longer mRNAs are more likely degraded than short mRNAs simply because processing time and thus exosomal exposure is longer (Fadda et al., Reference Fadda, Ryten, Droll, Rojas, Färber, Haanstra, Merce, Bakker, Matthews and Clayton2014). These data explain the negative correlation between mRNA abundance and mRNA size (Fadda et al., Reference Fadda, Ryten, Droll, Rojas, Färber, Haanstra, Merce, Bakker, Matthews and Clayton2014). The model of a rather unspecific exosome is supported by the findings that several short-lived RNA species are stabilized, when nuclear export is inhibited in various ways ((Bühlmann et al., Reference Bühlmann, Walrad, Rico, Ivens, Capewell, Naguleswaran, Roditi and Matthews2015) and see section ‘mRNA export by the rRNA transporters NMD3 and XPO1?’) and that developmentally regulated mRNAs are enriched in nuclear fractions in the related parasite T. cruzi (Pastro et al., Reference Pastro, Smircich, Di Paolo, Becco, Duhagon, Sotelo-Silveira and Garat2017): in both cases, mRNA levels appear controlled by transcript-specific cytoplasmic RNA degradation systems rather than by the exosome.
Trypanosomes have nuclear-localized orthologues to at least two of the three subunits of the TRAMP complex, MTR4 and NPAPL (also called ncPAP1) and both are essential for growth (Cristodero and Clayton, Reference Cristodero and Clayton2007; Etheridge et al., Reference Etheridge, Clemens, Gershon and Aphasizhev2009) (Table 7). MTR4 and ncPAP1 can be co-isolated together from trypanosome extracts using either protein as a bait, and both respective purified complexes exhibit PAP activity (Etheridge et al., Reference Etheridge, Clemens, Gershon and Aphasizhev2009). MTR4 is involved in 5.8S rRNA processing and controls RNA quality by a process that involves polyadenylation (Cristodero and Clayton, Reference Cristodero and Clayton2007). Three further proteins with nuclear localization were co-purified with ncPAP1 (Etheridge et al., Reference Etheridge, Clemens, Gershon and Aphasizhev2009). Two have zinc-knuckles and could theoretically be Air1 orthologues: the nucleolar protein NOP47 and the SR protein RRM1 (Table 7). However, neither is the closest homologue to yeast Air1, NOP47 associates with the spindle during mitosis (Zhou et al., Reference Zhou, Lee, Kurasawa, Hu, An and Li2018) and HA-RRM1 does not co-purify MTR4 or ncPAP1, at least not in amounts resulting in detectable bands on a Coomassie gel (Naguleswaran et al., Reference Naguleswaran, Gunasekera, Schimanski, Heller, Hemphill, Ochsenreiter and Roditi2015); whether either is the functional orthologue to Air1 remains to be investigated. Interestingly, the third protein co-purified with ncPAP1 is PUF10, a Pumilio domain protein with a function in 5.8S rRNA processing (Schumann Burkard et al., Reference Schumann Burkard, Käser, de Araújo, Schimanski, Naguleswaran, Knüsel, Heller and Roditi2013) and perhaps a trypanosome-specific TRAMP complex subunit. No exosome subunits were co-purified with ncPAP1 (Etheridge et al., Reference Etheridge, Clemens, Gershon and Aphasizhev2009). Thus, trypanosomes are likely to have a TRAMP-like complex that awaits further characterization. Trypanosomes have no homologues to the proteins of the NNS complex.
Co-transcriptional initiation of RNA export indicates the lack of major mRNA export checkpoints in trypanosomes
The major differences in nuclear mRNA metabolism between trypanosomes and opisthokonts detailed above, in particular the absence of many factors involved in mRNA export control raise the question, whether and how trypanosomes regulate mRNA export. With only two introns present in trypanosomes, the main question is how and whether trypanosomes prevent the export of polycistronic mRNA precursors that have not undergone trans-splicing and polyadenylation. It is established that mRNA export control is at least not tight in trypanosomes: Polycistronic mRNAs were detected in the cytoplasm by fractionations (Jäger et al., Reference Jäger, De Gaudenzi, Cassola, D'Orso and Frasch2007; Kramer et al., Reference Kramer, Marnef, Standart and Carrington2012) and also by single molecule RNA FISH: ¼ of all tubulin dicistronic RNAs were in the cytoplasm (Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019).
We have recently studied nuclear export in trypanosomes using three-colour intramolecular single molecule fluorescence in situ hybridization (smFISH) (Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019). For this, a large endogenous mRNA (FUTSCH, >22 000 nts long) is stained in three colours by hybridization with three nucleotide probe-sets: red at the 5′ end, infrared (pink false-colour) in the middle part, and green at the 3′ end (Fig. 3A). This allows the simultaneous detection and classification of multiple mRNA metabolism intermediates, based on colour combinations (Kramer, Reference Kramer2017; Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019). Using this approach, we observed mRNAs with their 5′ end (red dot) already in the cytoplasm, the middle part (infrared dot) still in the nucleus, and no 3′ end (no green dot) (Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019) (Fig. 3B), suggestive of co-transcriptional nuclear export. Further experiments (using orthogonal methods and different mRNAs) confirmed the presence of co-transcriptional mRNA export in trypanosomes (Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019). Importantly, not all mRNAs are exported co-transcriptionally: only about half of the very long transcripts leave the nucleus while still in transcription. Instead, what the data show is that trypanosomes lack a quality control checkpoint that prevents unprocessed mRNAs from starting export. Given that long mRNAs have longer transcription and processing times than short mRNAs, they are more likely to reach the pore while still in transcription. Average-sized mRNAs, in contrast, have fast processing times and, in addition, are probably too short to reach the pore from their site of transcription. The data do not exclude the presence of a checkpoint that prevents the completion of mRNA export, if these are unprocessed, for example by recognizing the absence of a poly(A) tail and/or associated factors.

Fig. 3. Trypanosomes can initiate mRNA export co-transcriptionally. (A) Principle of three-colour intramolecular single molecule FISH: a large transcript is simultaneously probed with smFISH probe sets in three different colours, covering the 5′ end, the middle part and the 3′ end. A rare example of an mRNA with all three colours visible as separate dots is shown on the right. (B) Detection of co-transcriptional mRNA export in trypanosomes by three-colour intramolecular single molecule FISH: the mRNA 5′ end (red) is already in the cytoplasm, while the middle part (pink false colour) is still in the nucleus and the 3′ end (green colour) has not yet been transcribed. (C) Trypanosome cells that express DHH1-eYFP as a marker for RNA granules are shown untreated (left) and treated with sinefungin to inhibit trans-splicing (right). DHH1 localizes to nuclear periphery granules (NPGs) at the outside of the nucleus (yellow arrow). (D) Model of co-transcriptional mRNA export in untreated cells (left) and in cells treated with sinefungin to inhibit trans-splicing (right). In both conditions, transcripts leave the nucleus while still in transcription. When trans-splicing is inhibited, progress of export is slowed or prevented by an unknown mechanism (anchor), resulting in the formation of granules at the cytoplasmic site of the nuclear pore. These granules contain unspliced transcripts stuck in export as well as cytoplasmic RNA binding proteins. (E) NPGs are visible by transmission electron microscopy as dense areas, connected to the nuclear pores via electron dense fibre-like structures (yellow arrow). All microscopy images of this figure are taken from Goos et al. (Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019).
To investigate any possible mRNA quality control mechanism further, we have massively increased the amount of polycistronic mRNAs by inhibiting trans-splicing. This can be done equally well in two independent ways, either using sinefungin (a drug that inhibits cap methylation (McNally and Agabian, Reference McNally and Agabian1992)) or by transfecting a morpholino antisense to the U2 snRNA (Matter and König, Reference Matter and König2005; Kramer et al., Reference Kramer, Marnef, Standart and Carrington2012). When trans-splicing is blocked, we observed a large proportion of polycistronic tubulin mRNAs in the cytoplasm (Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019), confirming the absence of a rigid mRNA export control machinery. To our surprise, inhibition of trans-splicing also correlated with many RNA binding proteins localizing to granular structures at the outside of the nuclear pores and we named these granules NPGs (nuclear pore granules) (Kramer et al., Reference Kramer, Marnef, Standart and Carrington2012; Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019) (Fig. 3C). NPG-like structures were not observed after inhibition of splicing in HeLa cells or inhibition of trans-splicing in C. elegans (Kramer et al., Reference Kramer, Marnef, Standart and Carrington2012; Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019), indicating that they may be unique to trypanosomes. We determined the proteome of purified NPGs and found that the granules contain the full set of cytoplasmic RNA binding proteins (Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019). Proteins involved in nuclear mRNA processing steps, such as splicing (Lsm5, SmE), capping (CGM1) and export (XPO1, MEX67, NUP96, RANBP1) were absent (Kramer et al., Reference Kramer, Marnef, Standart and Carrington2012). Also, most translation initiation factors were absent, with the exception of some isoforms of the eIF4F complex (eIF4E3, eIF4E1 and possibly eIF4E5, eIF4G1 and eIF4G2) (Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019), consistent with the granules being insensitive to translational inhibitors (Kramer et al., Reference Kramer, Marnef, Standart and Carrington2012). Moreover, we could detect polycistronic mRNAs in these granules by smFISH (Goos et al., Reference Goos, Dejung, Wehman, M-Natus, Schmidt, Sunter, Engstler, Butter and Kramer2019). The easiest explanation consistent with all data is that these granules are newly exported 5′ ends of polycistrons, bound to their natural set of RNA binding proteins that have not yet started translation (Fig. 3D). At least part of the polycistron is still stuck inside the pore and possibly extending into the nucleus: in electron microscopy images an electron-dense string-like structure is often visible that connects the pore with the NPGs (Fig. 3E). The major, still unanswered question is: why is this structure visible? Perhaps, export is somewhat slowed and otherwise transient structures of export accumulate. Export could be slowed because large polycistrons physically block the pores, perhaps because some remain attached to the transcription site and fail to exit completely. Alternatively, a quality control checkpoint could act at the nuclear basket that recognizes an mRNA as not fully processed, slowing or preventing export. Interestingly, while most cytoplasmic RNA binding proteins relocalize to NPGs upon inhibition of trans-splicing, we found four cytoplasmic RNA binding proteins that relocalized fully or partially to the nucleus. Most (ZFP1, ZFP2, ZC3H29) are CCCH-type zinc-finger proteins and most (ZFP1, ZFP2, Tb927.11.6600) function in trypanosome life cycle regulation (Hendriks et al., Reference Hendriks, Robinson, Hinkins and Matthews2001; Hendriks and Matthews, Reference Hendriks and Matthews2005; Paterou et al., Reference Paterou, Walrad, Craddy, Fenn and Matthews2006; Mony et al., Reference Mony, MacGregor, Ivens, Rojas, Cowton, Young, Horn and Matthews2014). Importantly, this relocalization was not detected when transcription was blocked by actinomycin D, indicating that it is not the absence of mature mRNA, but rather the presence of polycistronic RNA that causes relocalization. One further protein, XPO-5, moved from the nucleoplasm to the nuclear pores upon inhibition of trans-splicing. The function of this putative transporter protein is unknown and it is not essential in procyclic cells (Hegedűsová et al., Reference Hegedűsová, Kulkarni, Burgman, Alfonzo and Paris2019). Whether either of these five proteins acts in RNA export control remains to be investigated.
Summary and outlook
A fully processed trypanosome mRNA bears no major differences to an mRNA from opisthokonts and mRNA processing appears conserved in its mains features. However, with the exception of Mex67-Mtr2, all complexes and proteins involved in regulating mRNA export in opisthokonts are either absent (TREX, TREX-2, RES, DBP5, probably Mlp1-2) or have no reported functions in mRNA export (non-classical CTD, SR proteins except perhaps RRM1, EJC, TRAMP complex). Moreover, trypanosomes evolved several unique complexes and pathways. For example, mRNA export in trypanosomes is likely thriven by GTP using the RanGTP system instead of ATP and it may also use the XPO1-NMD pathway in addition or together with Mex67-Mtr2. The missing mRNA export control elements in trypanosomes may explain the leakage of unspliced mRNAs into the cytoplasm and the fact that export can start co-transcriptionally, rather than being dependent on the completion of all processing steps. In the near absence of introns, a leakage of unspliced (usually dicistronic) mRNAs may be tolerable to the parasite, with the worst damage being a misregulation in gene expression, but no production of faulty proteins. To keep the leakage of unprocessed mRNAs to a sufficiently low level, it may be sufficient to ensure fast, efficient and mostly co-transcriptional mRNA processing, perhaps supported by preferential cytoplasmic degradation of faulty mRNAs (the latter has not been shown).
A comparison of RNA export pathways throughout the tree of life came to the conclusion that RanGTP-dependent RNA export pathways (exporting rRNA, tRNA and snRNA) are relatively well conserved, while the RanGTP-independent export pathway of mRNA is not (Serpeloni et al., Reference Serpeloni, Vidal, Goldenberg, Avila and Hoffmann2011b). The Apicomplexa Toxoplasma gondii, for example, also lacks the TREX complex with the exception of the Sub2 helicase, has no Mex67 (albeit an unrelated C2H2 zinc-finger protein may act as a functional orthologue) and whether the mRNA export is RanGTP dependent is not certain (albeit a Dbp5 homologue is present in the genome) (Avila et al., Reference Avila, Cabezas-Cruz and Gissot2018). Plants have a TREX complex and a TREX-2 complex with some plant-specific adaptations and also a Dbp5 homologue, but homologues to Mex67 are absent (Ehrnsberger et al., Reference Ehrnsberger, Grasser and Grasser2019). It is likely that the highly conserved RanGTP-dependent transport system was the export system that has evolved first and was originally used for all RNA and protein transport processes. Later, export systems became more specialized to serve the specific needs of the eukaryotes. Trypanosomes may have experienced little pressure to evolve a sophisticated mRNA export control system and it will be highly interesting to investigate mRNA export in other protozoa with mostly intron-less transcripts.
Acknowledgements
I like to thank Jack Sunter (Oxford Brookes University, Oxford, UK), Richard Wheeler (University of Oxford, Oxford, UK) and Sam Dean (University of Oxford, Oxford, UK) for providing the highly valuable source of TrypTag, that has helped to speculate about the functions of many putative mRNA metabolism proteins discussed in this review. Christine Clayton (University of Heidelberg, Heidelberg, Germany) is acknowledged for her highly useful comments in TriTrypDB and Martin Zoltner (Charles University in Prague, Prague, Czech Republic) for critical proofreading of the manuscript. My apologies go to the many scientists whose work I have not been able to cite and discuss, due to space constraints.
Financial support
This work was supported by the Deutsche Forschungsgemeinschaft [Kr4017_3-1].
Conflicts of interest
None.
Ethical standards
Not applicable.