Introduction
The Argasidae family consists of soft-bodied ticks that parasitize semi-terrestrial and terrestrial vertebrate hosts (Guglielmone et al., Reference Guglielmone, Robbins, Apanaskevich, Petney, Estrada Peña, Horak, Shao and Barker2010; Manzano-Román et al., Reference Manzano-Román, Díaz-Martín, de la Fuente and Pérez-Sánchez2012). Soft ticks have a global distribution and are typically found in microclimates near their hosts (Vial, Reference Vial2009; Dietrich et al., Reference Dietrich, Gómez-Díaz and McCoy2011; Estrada-Peña et al., Reference Estrada-Peña, Kleinerman and Baneth2017; Kleinerman and Baneth, Reference Kleinerman and Baneth2017). They primarily feed at night for short periods but can endure extended periods without hosts (Estrada-Peña et al., Reference Estrada-Peña, Kleinerman and Baneth2017; Kleinerman and Baneth, Reference Kleinerman and Baneth2017). They possess a ventrally located gnathostoma and lack a chitinous dorsal scutum (Vial, Reference Vial2009; Estrada-Peña et al., Reference Estrada-Peña, Kleinerman and Baneth2017). Argasidae includes approximately 218 tick species in 15 genera, 1 of which is the genus Ornithodoros (Guglielmone et al., Reference Guglielmone, Robbins, Apanaskevich, Petney, Estrada Peña, Horak, Shao and Barker2010; Mans et al., Reference Mans, Featherston, Kvas, Pillay, de Klerk, Pienaar, de Castro, Schwan, Lopez, Teel and de León2019; Mans, Reference Mans2023).
Ornithodoros ticks are distinguished from other soft ticks by the absence of a ‘sutural line’ at the border between their dorsal and ventral surfaces (Estrada-Peña et al. Reference Estrada Pena, Mangold, Nava, Venzal, Labruna and Guglielmone2010, Reference Estrada-Peña, Kleinerman and Baneth2017; Mans, Reference Mans2023). The genus Ornithodoros is diverse, encompassing around 134 species distributed among 4 subgenera (Estrada-Peña et al., Reference Estrada Pena, Mangold, Nava, Venzal, Labruna and Guglielmone2010, Reference Estrada-Peña, Kleinerman and Baneth2017; Muñoz-Leal et al., Reference Muñoz-Leal, Dias, Abrahão and Labruna2017; Mans et al., Reference Mans, Featherston, Kvas, Pillay, de Klerk, Pienaar, de Castro, Schwan, Lopez, Teel and de León2019, Reference Mans, Kelava, Pienaar, Featherston, de Castro, Quetglas, Reeves, Durden, Miller, Laverty and Shao2021; Dantas-Torres and Otranto, Reference Dantas-Torres, Otranto and Rezaei2022). One of these subgenera is the Pavlovskyella (Supplementary Table 1), characterized by a combination of characters in adults: transverse postanal groove intersected by the median postanal groove, absence of large cheeks covering the capitulum, presence of dorsoventral groove and humps in tarsus I (Clifford et al., Reference Clifford, Kohls and Sonenshine1964; Filippova, Reference Filippova1966; Pospelova-Shtrom, Reference Pospelova-Shtrom1969; Estrada Peña et al., Reference Estrada Pena, Mangold, Nava, Venzal, Labruna and Guglielmone2010, Reference Estrada-Peña, Kleinerman and Baneth2017; Muñoz-Leal et al., Reference Muñoz-Leal, Dias, Abrahão and Labruna2017; Mans et al., Reference Mans, Featherston, Kvas, Pillay, de Klerk, Pienaar, de Castro, Schwan, Lopez, Teel and de León2019; Reference Mans, Kelava, Pienaar, Featherston, de Castro, Quetglas, Reeves, Durden, Miller, Laverty and Shao2021).
Ornithodoros tholozani is the type species of the subgenus and has a wide geographic distribution (Desportes and Campana, Reference Desportes and Campana1946). Three subspecies of O. tholozani have been proposed: O. tholozani sensu stricto (Iran, Iraq), O. tholozani pavlovskyi (Central Asia) and O. tholozani crossi (India) (Desportes and Campana, Reference Desportes and Campana1946). The latter subspecies, originally described as Ornithodoros crossi, was collected in Punjab, India (Brumpt, Reference Brumpt1922), near the border with Pakistan. Although Sapre (Reference Sapre1944) redescribed O. crossi using specimens collected in the type locality, he was unable to examine the type series that was lost. Additionally, the validity of this species had been questioned earlier by Leeson (Reference Leeson1953). However, Sapre's redescription of O. crossi clearly indicates that O. tholozani crossi is not a synonym because of marked differences in the chaetotaxy of tarsus IV. Therefore O. tholozani crossi sensu Desportes & Campana should neither be considered a synonym of O. crossi nor of any other closely related species. We consider O. crossi a nomen nudum because the type series is missing. Noteworthily, O. tholozani was considered a synonym of Ornithodoros papillipes by the North American group of taxonomists (Hoogstraal, Reference Hoogstraal1985). We consider O. papillipes valid because it has a denticulated tuft of setae arising from the hood, in front of the camerostome (Pavlovsky, Reference Pavlovsky1930), and this character is absent in O. tholozani (Sapre, Reference Sapre1944). Ornithodoros species exhibit a lack of critical morphological features, which has historically led to confusion in species identification, emphasizing the necessity for molecular-based identification methods.
There are notable disagreements regarding the systematic classification of argasid ticks among 4 primary schemes: the Soviet scheme (Pospelova-Shtrom, Reference Pospelova-Shtrom1946; Filippova, Reference Filippova1966), the American scheme (Clifford et al., Reference Clifford, Kohls and Sonenshine1964; Hoogstraal, and Kohls, Reference Hoogstraal and Kohls1966; Hoogstraal, Reference Hoogstraal1985), the French scheme (Camicas and Morel, Reference Camicas and Morel1977; Camicas et al., Reference Camicas, Hervy, Adam and Morel1998) and the morphological cladistic scheme (Klompen and Oliver, Reference Klompen and Oliver1993). A more advanced classification scheme has recently been established based on mitochondrial genomes (Mans et al., Reference Mans, de Klerk, Pienaar, de Castro and Latif2012, Reference Mans, Featherston, Kvas, Pillay, de Klerk, Pienaar, de Castro, Schwan, Lopez, Teel and de León2019, Reference Mans, Kelava, Pienaar, Featherston, de Castro, Quetglas, Reeves, Durden, Miller, Laverty and Shao2021), which is the one adopted in this article. Additionally, studies have indicated that the genus Ornithodoros, along with some of its subgenera, including Pavlovskyella, are paraphyletic (Labruna et al., Reference Labruna, Terassini, Camargo, Brandão, Ribeiro and Estrada-Peña2008; Barros-Battesti et al., Reference Barros-Battesti, Landulfo, Onofrio, Faccini, Marcili, Nieri-Bastos, Venzal and Labruna2011; Mans et al., Reference Mans, Featherston, Kvas, Pillay, de Klerk, Pienaar, de Castro, Schwan, Lopez, Teel and de León2019, Reference Mans, Kelava, Pienaar, Featherston, de Castro, Quetglas, Reeves, Durden, Miller, Laverty and Shao2021; Muñoz-Leal et al., Reference Muñoz-Leal, Venzal, Kneubehl, Lopez, Martins and Labruna2023). The controversial phylogenetic relationships among argasid ticks could be addressed through mitogenomic and nuclear 18S–28S rDNA-based analysis of both the established and newly discovered argasid tick species.
The soft tick fauna of Pakistan has received limited attention. Given its zoogeographical location, which falls between the Palearctic and Oriental regions, Pakistan is expected to host a diverse range of soft tick species. Approximately, 9 soft tick species have been reported in this country, including 5 species (Alveonasus lahorensis, Argas sp. ‘rousetti’, Argas persicus, Carios vespertilionis and Ornithodoros sp.) identified through molecular analysis and 4 species identified based on morphological characteristics (Argas abdussalami, O. papillipes, Argas reflexus and O. tholozani) (Rao and Kalra, Reference Rao and Kalra1949; Doss et al., Reference Doss, Farr, Roach and Anastos1978; Hoogstraal, Reference Hoogstraal1985; Karim et al., Reference Karim, Budachetri, Mukherjee, Williams, Kausar, Hassan, Adamson, Dowd, Apanskevich, Arijo and Sindhu2017; Zahid et al., Reference Zahid, Muñoz-Leal, Khan, Alouffi, Labruna and Ali2021, Reference Zahid, Alouffi, Almutairi, Ateeq, Tanaka, Chang, Chen and Ali2023; Ali et al., Reference Ali, Numan, Khan, Aiman, Muñoz-Leal, Chitimia-Dobler, Labruna and Nijhof2022; Mans et al., Reference Mans, Chitimia-Dobler, Pienaar, de Castro, Khan, Almutairi, Alouffi and Ali2024). The objective of this study was to thoroughly investigate the status of the previously undetermined Ornithodoros sp. (Ali et al., Reference Ali, Numan, Khan, Aiman, Muñoz-Leal, Chitimia-Dobler, Labruna and Nijhof2022) through detailed morphological and molecular analyses.
Materials and methods
Tick specimens and research area
Ornithodoros specimens for this study were obtained from previously reported specimens from 5 districts of Khyber Pakhtunkhwa, Pakistan: Shangla (34°47′19.9″N, 72°41′30.6″E), Bajaur (34°46′04.7″N, 71°29′49.1″E), Dir Upper (35°11′37.7″N, 71°54′17.2″E), Dir Lower (34°52′25.0″N, 71°46′49.2″E) and Orakzai (33°41′13.0″N, 70°59′48.7″E) (Ali et al., Reference Ali, Numan, Khan, Aiman, Muñoz-Leal, Chitimia-Dobler, Labruna and Nijhof2022). Additionally, a new collection from these districts was made, with the addition of specimens from the new district Swat (34°44′18.7″N, 72°20′50.6″E). These districts, situated in the northern province of Pakistan, are characterized by their mountainous landscapes, with elevations ranging from approximately 1500 to 3500 m. They experience a temperate climate, with cold winters and moderate summers. The environmental conditions support agriculture and forestry. The global positioning system or Google Maps (https://www.google.com/maps/) was used to determine the geographic coordinates of the collection sites, and ArcGIS v. 10.3.1 was used to design the study map (Fig. 1).

Figure 1. Map of Khyber Pakhtunkhwa province, Pakistan, showing the locations where Ornithodoros ticks were collected for this study.
Specimen collection and preservation
The Ornithodoros ticks were collected during 2019 and 2023 from cracks, crevices, burrows and debris in animal shelters (that housed small or large ruminants) throughout 6 monitoring locations/districts: Shangla, Bajaur, Dir Upper, Dir Lower, Orakzai and Swat in KP, Pakistan. The Ornithodoros specimens were collected in 15 mL Falcon tubes, and sent to the Department of Zoology, Abdul Wali Khan University, Mardan, Garden Campus, KP, Pakistan. To remove any contaminants from the body surface, the tick specimens were washed in distilled water and then with 70% ethanol. They were subsequently preserved in 100% absolute ethanol in Eppendorf tubes for further molecular analysis.
Morphological analysis
The collected Ornithodoros specimens were identified morphologically using a stereo-zoom microscope (StereoBlue-euromex SB.1302-1, Arnhem, Netherlands) using the taxonomic key of Filippova (Reference Filippova1966), because it includes the species of Ornithodoros occurring in Pakistan and other geographically related species. Late-instar nymphs and adult specimens were prepared for scanning electron microscopy (SEM). Briefly, specimens were sonicated in a solution of dish soap for 30 min, and dehydrated in a 70–100% ethanol battery to undergo critical point drying and metallization. Micrographs were captured with a JEOL JMS-5900 and a Hitachi SU35000 scanning electron microscope. The nomenclature of this study followed Cooley and Kohls (Reference Cooley and Kohls1944), Filippova (Reference Filippova1966) and Muñoz-Leal et al. (Reference Muñoz-Leal, Martins, Nava, Landulfo, Simons, Rodrigues and Labruna2020). Specimens were deposited in the following tick collections: Colección Chilena de Garrapatas ‘Daniel González-Acuña’ (CCG) and the United States National Tick Collection (USNTC).
Genomic DNA isolation and next-generation sequencing
DNA was extracted from Ornithodoros specimens using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. NanoDrop (Nano-Q, Optizen, Daejeon, South Korea) was used to quantify the genomic DNA, which was then stored for further examinations at −20°C. Genomic DNA was processed using the MGIEasy Universal DNA Library Prep kit (MGI, Shenzhen, China) and sequenced on the MGI DNBSEQ-G400 sequencing instrument using the PE150 (paired-end 2 × 150 bp) format (Agricultural Research Council-Biotechnology Platform, South Africa) to obtain ~10 Gb of data per sample. The data quality and assembly protocol are similar to that obtained using Illumina paired-end short read sequencing previously used for the assembly of tick mitochondrial genomes (Mans et al., Reference Mans, de Klerk, Pienaar, de Castro and Latif2015, Reference Mans, Featherston, Kvas, Pillay, de Klerk, Pienaar, de Castro, Schwan, Lopez, Teel and de León2019, Reference Mans, Kelava, Pienaar, Featherston, de Castro, Quetglas, Reeves, Durden, Miller, Laverty and Shao2021). Specimens prepped for sequencing included Shangla, Bajaur and Dir Lower.
Next-generation sequence assembly and evolutionary analyses
Paired-end sequence data were quality trimmed (0.001 quality limit) and MGI adapters removed using CLC Genomics Workbench v. 20.1 software (Qiagen Digital Insights, Aarhus, Denmark). Standard assembly parameters (mismatch cost-2, insertion cost-3, deletion cost-3, length fraction-0.9, similarity-0.9, minimum contig length-200 and automatic bubble size) were used and assembly performed using a kmer size of 49 in CLC Genomics Workbench v. 20.1 software (Qiagen Digital Insights). Contigs were identified as mitochondrial, 18S or 28S rDNA using BLASTn analysis (Altschul et al., Reference Altschul, Gish, Miller, Myers and Lipman1990). Final contigs were obtained by mapping data back to the contigs using CLC Genomics Workbench v. 20.1 (mismatch cost-2, insertion cost-3, deletion cost-3, length fraction-0.5 and similarity-0.9), to obtain consensus sequences and final coverage values. The mitochondrial genome was annotated using the MITOS and ARWEN servers to identify tRNA genes (Laslett and Canbäck, Reference Laslett and Canbäck2008; Bernt et al., Reference Bernt, Donath, Jühling, Externbrink, Florentz, Fritzsch and Stadler2013). Protein coding genes were identified using the Expasy Translation Server (https://web.expasy.org/translate/) and BLASTp analysis (Altschul et al., Reference Altschul, Gish, Miller, Myers and Lipman1990). Following BLAST, the maximum identity sequences were retrieved in the FASTA format from the NCBI for each gene used in the analysis including the cox1 (cytochrome c oxidase subunit 1), cox2 (cytochrome c oxidase subunit 2), atp8 (ATP synthase F0 subunit 8), atp6 (ATP synthase F0 subunit 6), cox3 (cytochrome c oxidase subunit 3), nad3 (NADH dehydrogenase subunit 3), nad5 (NADH dehydrogenase subunit 5), nad4 (NADH dehydrogenase subunit 4), nad4L (NADH dehydrogenase subunit 4L), nad6 (NADH dehydrogenase subunit 6), cytb (cytochrome b), nad1 (NADH dehydrogenase subunit 1), nad2 (NADH dehydrogenase subunit 2), 12S rDNA (12S ribosomal DNA), 16S rDNA (16S ribosomal DNA), 18S rDNA (18S ribosomal DNA) and 28S rDNA (28S ribosomal DNA) sequences. The BioEdit Sequence Alignment Editor v. 7.0.5 was used for the alignment of the acquired sequences with the downloaded sequences using ClustalW multiple alignments (Hall et al., Reference Hall, Biosciences and Carlsbad2011). The obtained coding sequences were aligned by using the integrated MUSCLE algorithm (Edgar, Reference Edgar2004). The alignments of DNA (12S–16S rDNA and 18S–28S rDNA) and amino acid (cox1, cox2, atp8, atp6, cox3, nad3, nad5, nad4, nad4L, nad6, cytb, nad1 and nad2) sequences were subjected to Molecular Evolutionary Genetics Analysis (MEGA-11) (Tamura et al., Reference Tamura, Stecher and Kumar2021) for concatenation into datasets. The concatenated alignment files were exported in the FASTA format and converted to the RELAXPHYLIP format via NCL converter v. 2.1 tool (Lewis, Reference Lewis2003), in the Cyberinfrastructure for Phylogenetic Research (CIPRES) Science Gateway v. 3.3 (CIPRES; https://www.phylo.org/portal2/) (Miller et al., Reference Miller, Pfeiffer and Schwartz2010). The maximum-likelihood phylogenetic trees were constructed through the IQ-Tree v. 2.2.2.7 tool run on XSEDE in CIPRES (Miller et al., Reference Miller, Pfeiffer and Schwartz2010) using default options with 1000 ULTRAFAST bootstrap replicates (Hoang et al., Reference Hoang, Chernomor, Von Haeseler, Minh and Vinh2018). The output tree file was visualized by the FigTree v. 1.4.4 tool (Rambaut, Reference Rambaut2018). The concatenated datasets were also subjected to MEGA-11, using the maximum composite likelihood model (Tamura et al., Reference Tamura, Nei and Kumar2004), to determine pairwise genetic distances. The number of base substitutions per site between pairwise gene sequences was determined. These analyses involved 46, 48, 41 and 32 sequences for protein coding amino acid sequences, protein coding nucleotide sequences, 12S–16S rDNA and 18S–28S rDNA sequences, respectively.
Results
Morphological description of Ornithodoros pakistanensis sp. nov.
Order: Ixodida Leach, 1815
Family: Argasidae Canestrini, 1890
Genus: Ornithodoros Koch, 1837, 1844
Ornithodoros pakistanensis sp. nov. Ali, Chitimia-Dobler, Muñoz-Leal & Mans
Type locality: Khyber Pakhtunkhwa (KP), Pakistan.
Type specimens: Holotype: 1 female in ethanol (USNTC, in progress). Allotype: 1 male in ethanol at USNTC, same accession number. Paratypes: 5 males, 14 females, 46 nymphs in ethanol (CCG-86), 11 males, 9 females and 24 nymphs in ethanol at the Gertrud Theiler Tick Museum (ARC-OVR) and 202 males, 277 females, 207 nymphs in ethanol at the Department of Zoology, Abdul Wali Khan University, Mardan, KP, Pakistan.
Etymology: The species epitet ‘pakistanensis’ is in allusion to the geographical origin of this newly discovered species.
Hosts: Collected in cracks, crevices, burrows and debris of animal shelters that housed small or large ruminants (goats, sheep, cattle, buffaloes) and equids.
Known infectious agents: Rickettsia sp. belong to the limoniae group (Ali et al., Reference Ali, Numan, Khan, Aiman, Muñoz-Leal, Chitimia-Dobler, Labruna and Nijhof2022).
Disease relationships: Unknown.
Zoobank registration number: LSID: 0FC991C3-6B96-49D6-B23C-CB939D7BBFC7.
Female (measurements based on 14 specimens, Fig. 2)
Body: Sub-oval, 5.775 ± 0.025 mm (5.750–5.800 mm) long, 3.340 ± 0.080 mm (3.260–3.420 mm) wide. Lateral margins subparallel, curving posteriorly, converging to a round apex anteriorly.
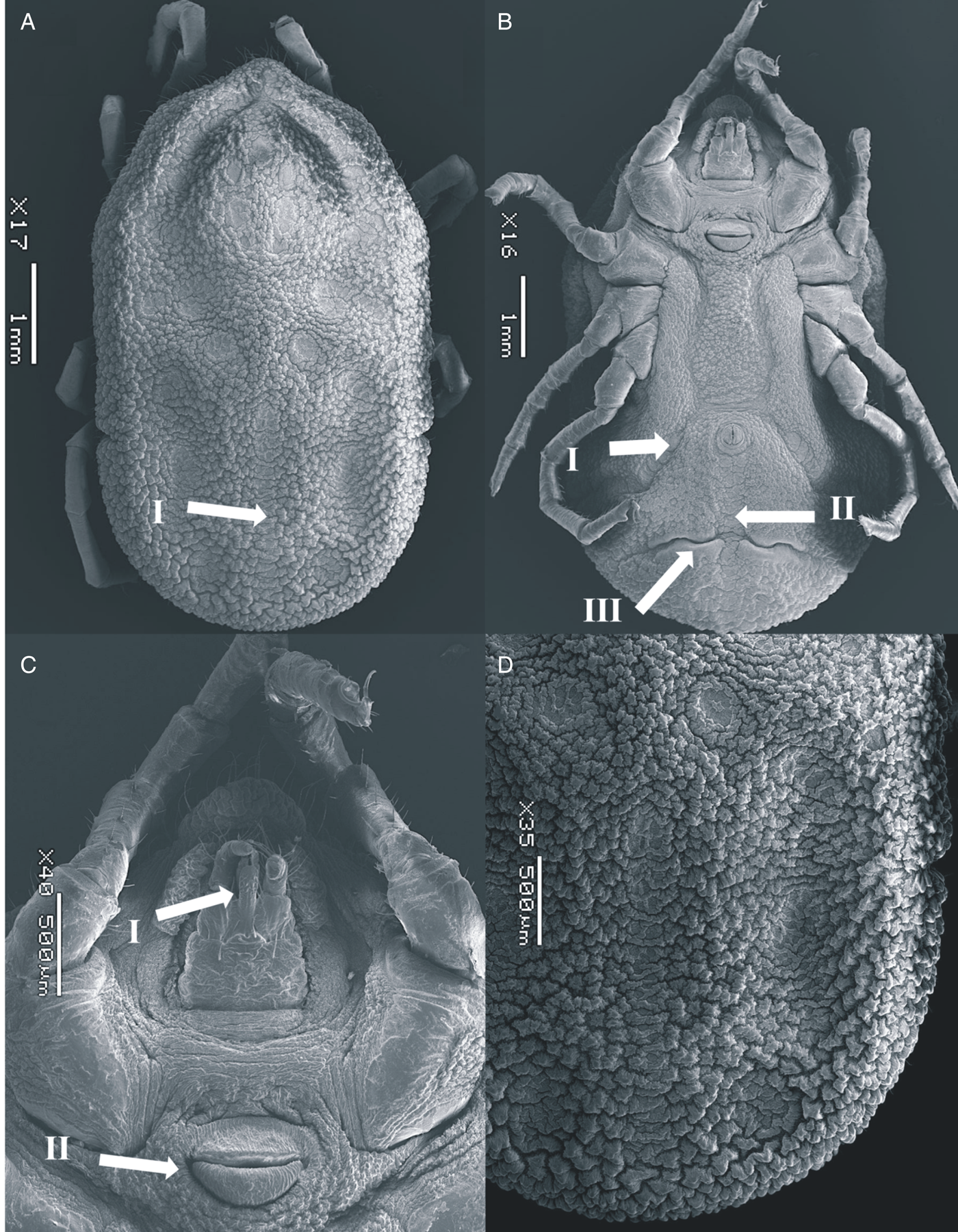
Figure 2. SEM of Ornithodoros pakistanensis female: (A) idiosoma dorsal view (I: posterior mammillae), (B) idiosoma ventral view (I: preanal grooves, II: medium postanal grooves and III: transverse postanal grooves), (C) ventral capitulum (I: hypostome and II: genital aperture) and (D) idiosoma dorsal/posterior mammillae collected in this study.
Dorsum: Covered by irregularly shaped mammillae, with a smooth surface top; tightly interlocked with irregular buttresses; larger in body margins; some with a small seta. Eyes absent. Dorsoventral groove present. Dorsal discs faint, pebbled with thick marginal ridges. Anteromedian disc present, anterior to centrolateral discs; anterolateral discs merged into a curved line forming a groove in the anterior margins of idiosome. Intermedian anterior and intermedian central discs merged; intermedian central and intermedian posterior discs not merged, separated diagonally. Median disc merged with the posteromedian file of discs, reaching the posterior margin of idiosome; posterolateral file begins posterior to median disc and intersected by mammillae.
Venter: Covered by small mammillae in the middle, larger mammillae in the posterior part. Discs present along preanal and median postanal grooves. Transverse postanal groove intersecting perpendicularly the median postanal groove slightly below the medial point from its origin in anal ring. Coxal folds devoid of glabrous patches of integument. Spiracular plates located between coxae III and IV. Genital area surrounded by a striated ring merging with the anterior lip that lacks a pore on its surface. Anterior and posterior lips similarly sized.
Capitulum: Located below a large hood. The hood devoid of setae. Cheeks small, not covering the capitulum. Basis capitulum rectangular, wrinkled, with 1 postpalpal and 1 posthypostomal pair of setae, similar in length. Three to 4 short basal setae laterally. Palpi elongated, provided with abundant setae. Palpal article I large, with a large ridge-like extension along its internal margin. Hypostome reaching the second palpal article in length; with crenulations at the tip and below the denticles. Two rows of 3–5 denticles present in the distal half, notched apically.
Legs: Without micromammillae. Coxae anteriorly elongated, with the anterior two-thirds sclerotized. Coxa I with a patch of micromammillae laterally. Coxae I–IV decreasing on size; coxae I and II separated, II–IV contiguous. Tarsus I with 5 small dorsal humps, equally protruding, tarsus II with 4; tarsus III and IV with an apical protuberance, without humps. Tarsus IV with 6–7 pairs of setae ventrally. Bifid claws and pulvillus present.
Male (measurements based on 5 specimens, Fig. 3)
Body sub-oval 4.687 ± 0.042 mm (4.645–4.729 mm) long, 2.757 ± 0.052 mm (2.705–2.810 mm) wide, similar to female. Cheeks smaller than female, not covering the capitulum. Dorsum with mammillae slightly more separated than the female. Dorsal discs visible, topology as female. Hypostome short, reaching the first palpal article in length. Genital flap chiefly straight, with crenulations at the base. Legs similar to female.
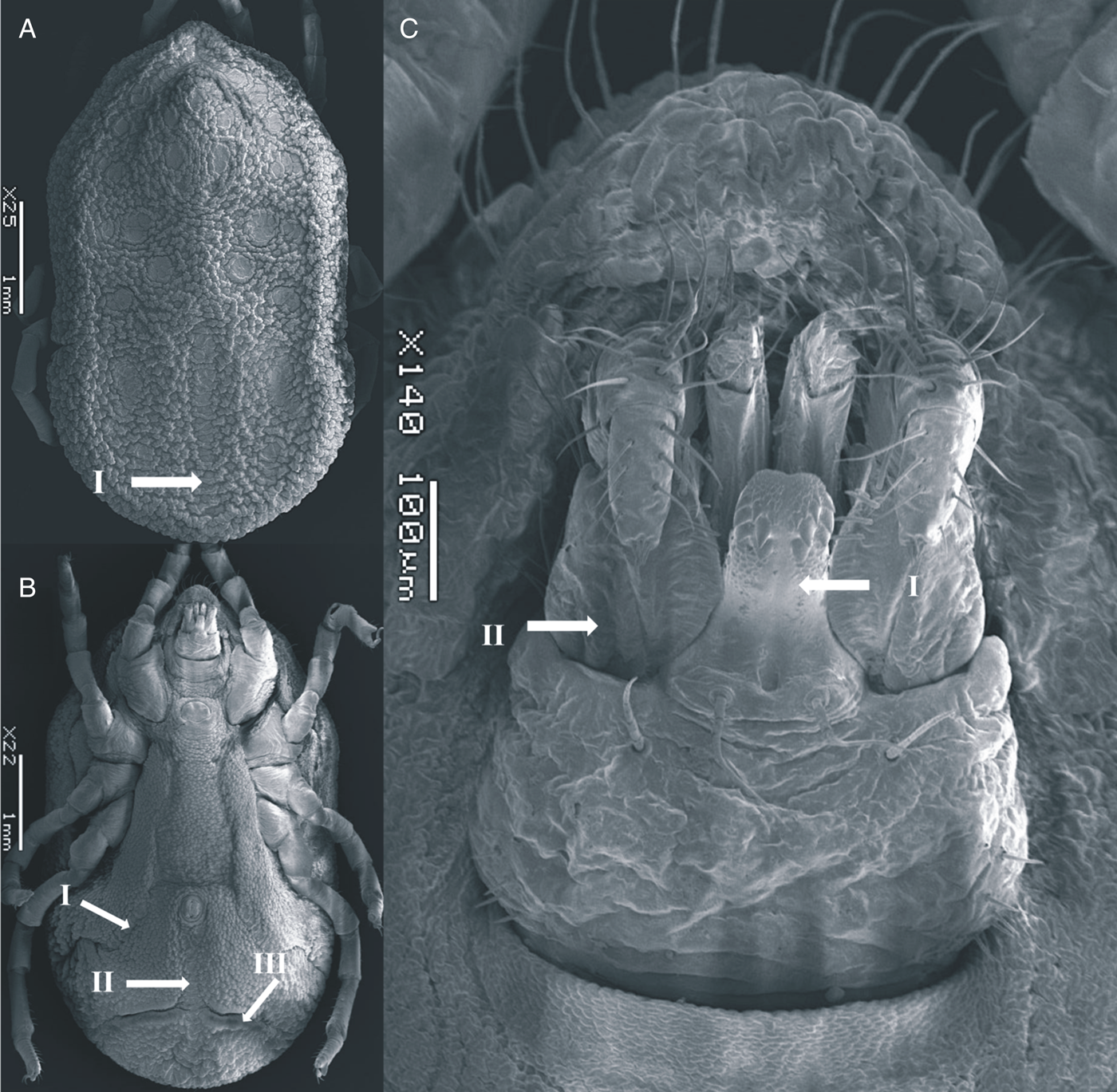
Figure 3. SEM of O. pakistanensis male: (A) idiosoma dorsal view (I: posterior mammillae), (B) idiosoma ventral view (I: preanal grooves, II: medium postanal grooves and III: transverse postanal grooves) and (C) ventral capitulum (I: hypostome and II: palps) collected in this study.
Late-instar nymph (measurements based on 20 specimens, Fig. 4)
Body sub-oval 5.381 ± 0.168 mm (5.213–5.550 mm) long, 2.775 ± 0.025 mm (2.750–2.800 mm) wide. Morphologically similar to female but lacking genital apron. Tarsus IV provided with 4–6 pairs of ventral setae.
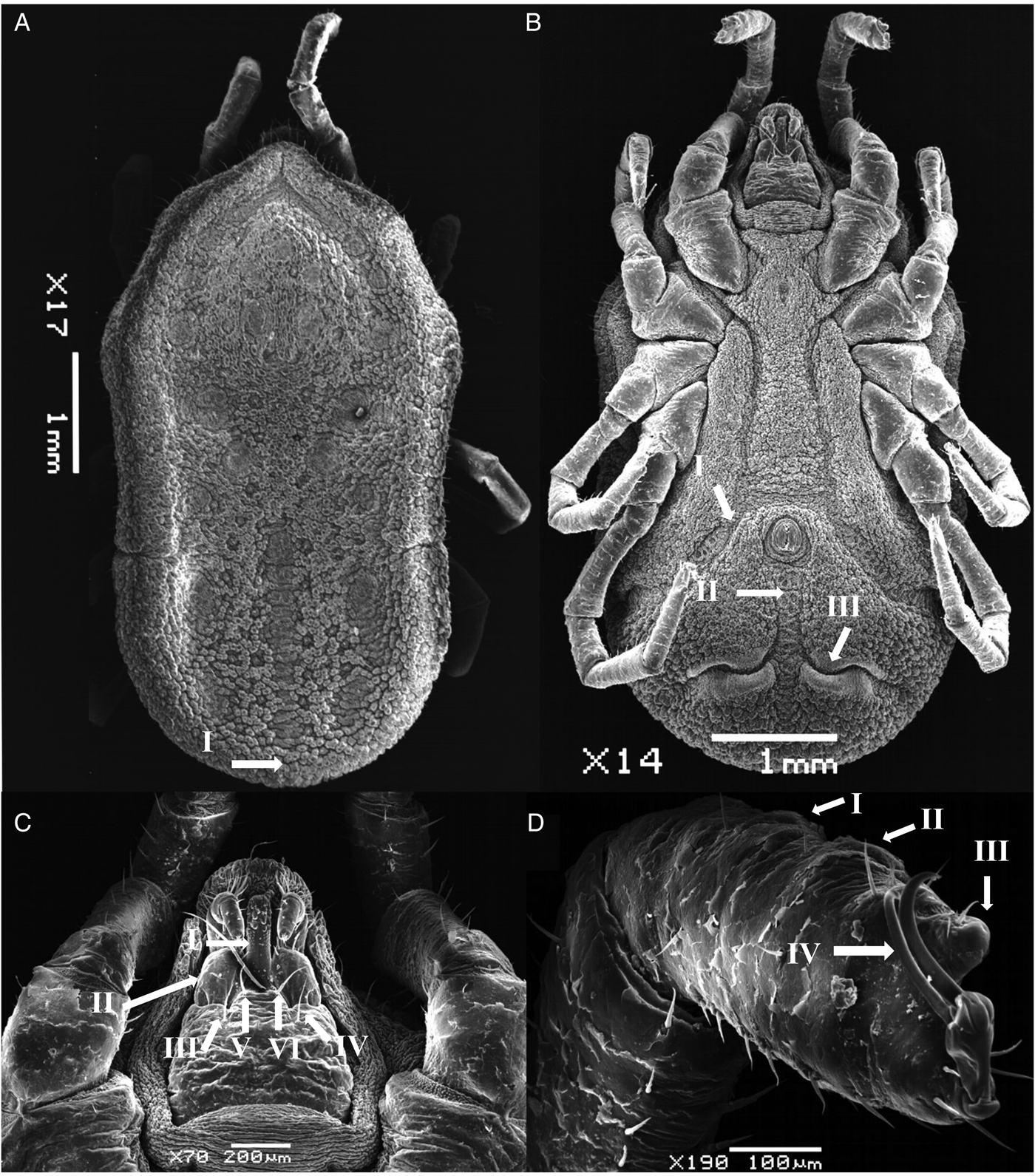
Figure 4. SEM of nymph stage for O. pakistanensis: (A) idiosoma dorsal view (I: posterior mammillae), (B) idiosoma ventral view (I: preanal grooves, II: medium postanal grooves and III: transverse postanal grooves), (C) ventral capitulum (I: hypostome, II: palps, III and IV: setae on capitulum, V and VI: postpalpal setae) and (D) legs (I–III: tarsus/metatarsus and IV: pair of spur) collected in this study.
Genetic profiling and evolutionary analysis
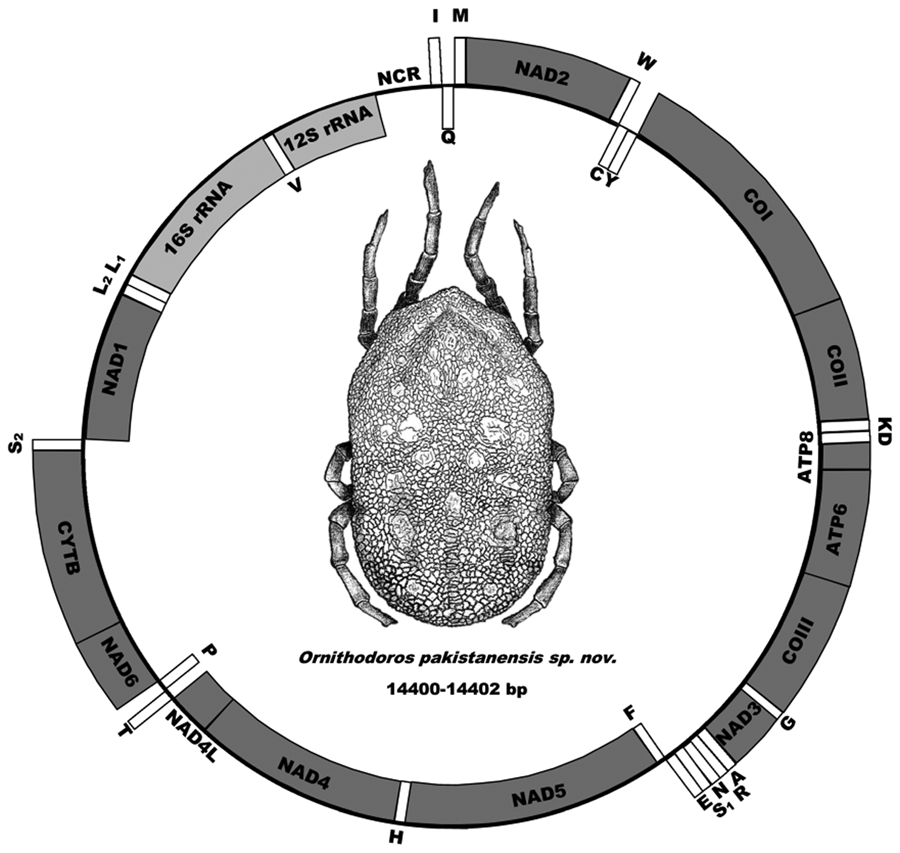
The mitochondrial genome has the standard arrangement of 13 protein genes (cox1, cox2, atp8, atp6, cox3, nad3, nad5, nad4, nad4L, nad6, cytb, nad1 and nad2), 2 ribosomal RNA genes (12S rDNA and 16S rDNA) and 22 transfer RNA genes as well as the gene structure observed for argasid ticks (Fig. 5) (Shao et al., Reference Shao, Aoki, Mitani, Tabuchi, Barker and Fukunaga2004; Mans et al., Reference Mans, de Klerk, Pienaar, de Castro and Latif2012, Reference Mans, Featherston, Kvas, Pillay, de Klerk, Pienaar, de Castro, Schwan, Lopez, Teel and de León2019, Reference Mans, Kelava, Pienaar, Featherston, de Castro, Quetglas, Reeves, Durden, Miller, Laverty and Shao2021; Burger et al., Reference Burger, Shao, Labruna and Barker2014). Sequence alignment of the 3 mitochondrial genomes indicates pairwise identities of 99% and a mitochondrial genome size of 14 400–14 402 bp.

Figure 5. Mitochondrial genome arrangement of O. pakistanensis sp. nov. Arrangements of genes on the forward strand (outside, clockwise) and reverse strand (inside, anti-clockwise) is indicated.
The results of BLASTp for protein coding amino acid sequences of the mitogenome of O. pakistanensis sp. nov. are provided in Table 1. All concatenated protein coding amino acid sequences showed the lowest pairwise genetic distance of 0.128 in comparisons with Ornithodoros verrucosus, followed by 0.129 with O. tholozani and 0.133 with Ornithodoros tartakovskyi (Supplementary Table 2). Based on their maximum identities and concatenated phylogenetic analysis, the obtained protein coding amino acid sequences for O. pakistanensis sp. nov. clustered with species of the subgenus Pavlovskyella, namely O. tholozani, O. verrucosus and O. tartakovskyi (Fig. 6).
Table 1. Outcomes of BLASTp and BLASTn analyses for protein coding genes in the mitogenome of Ornithodoros pakistanensis sp. nov.

O. verrucosus (Ukraine), O. tholozani (Israel) and O. tartakovskyi (Rocky Mountain Laboratory colony).
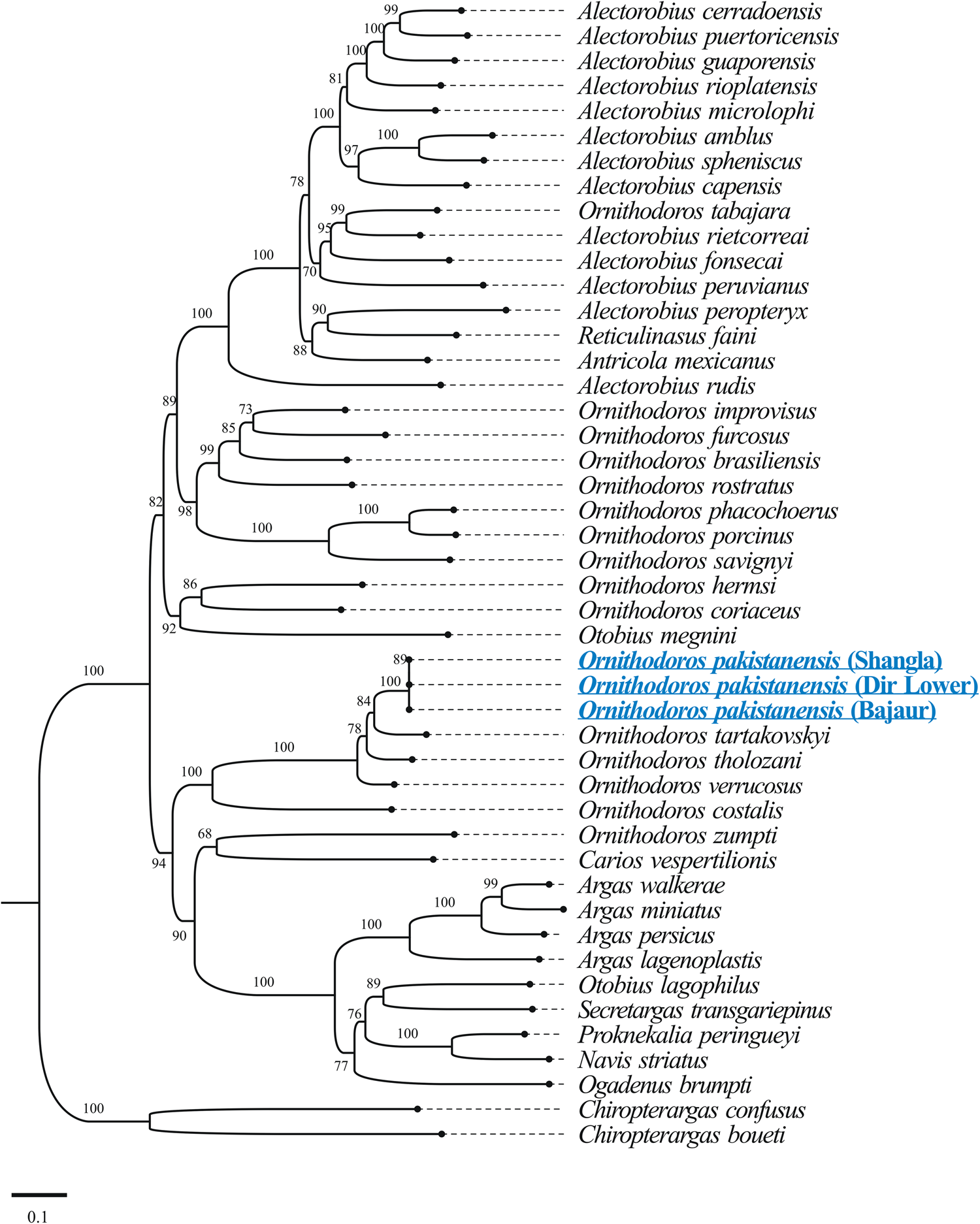
Figure 6. Maximum-likelihood tree for the concatenated 13 mitochondrial protein sequences (cox1, cox2, atp8, atp6, cox3, nad3, nad5, nad4, nad4L, nad6, cytb, nad1 and nad2). The sequence of Chiropterargas confusus and Chiropterargas boueti was used as outgroup. The bootstrap values (1000 replicates) are shown at each node. The obtained sequences for O. pakistanensis are underlined and presented in blue.
The outcomes of BLASTp for protein coding nucleotide sequences of the mitogenome of O. pakistanensis sp. nov. are presented in Table 1. All concatenated protein coding nucleotide sequences showed lowest pairwise genetic distance of 0.306 in comparisons with O. verrucosus, followed by 0.345 with O. tholozani and 0.393 with O. tartakovskyi (Supplementary Table 3). In the concatenated phylogenetic tree based on protein coding nucleotide sequences, O. pakistanensis sp. nov. clustered with O. tholozani, O. verrucosus and O. tartakovskyi of the subgenus Pavlovskyella (Fig. 7).
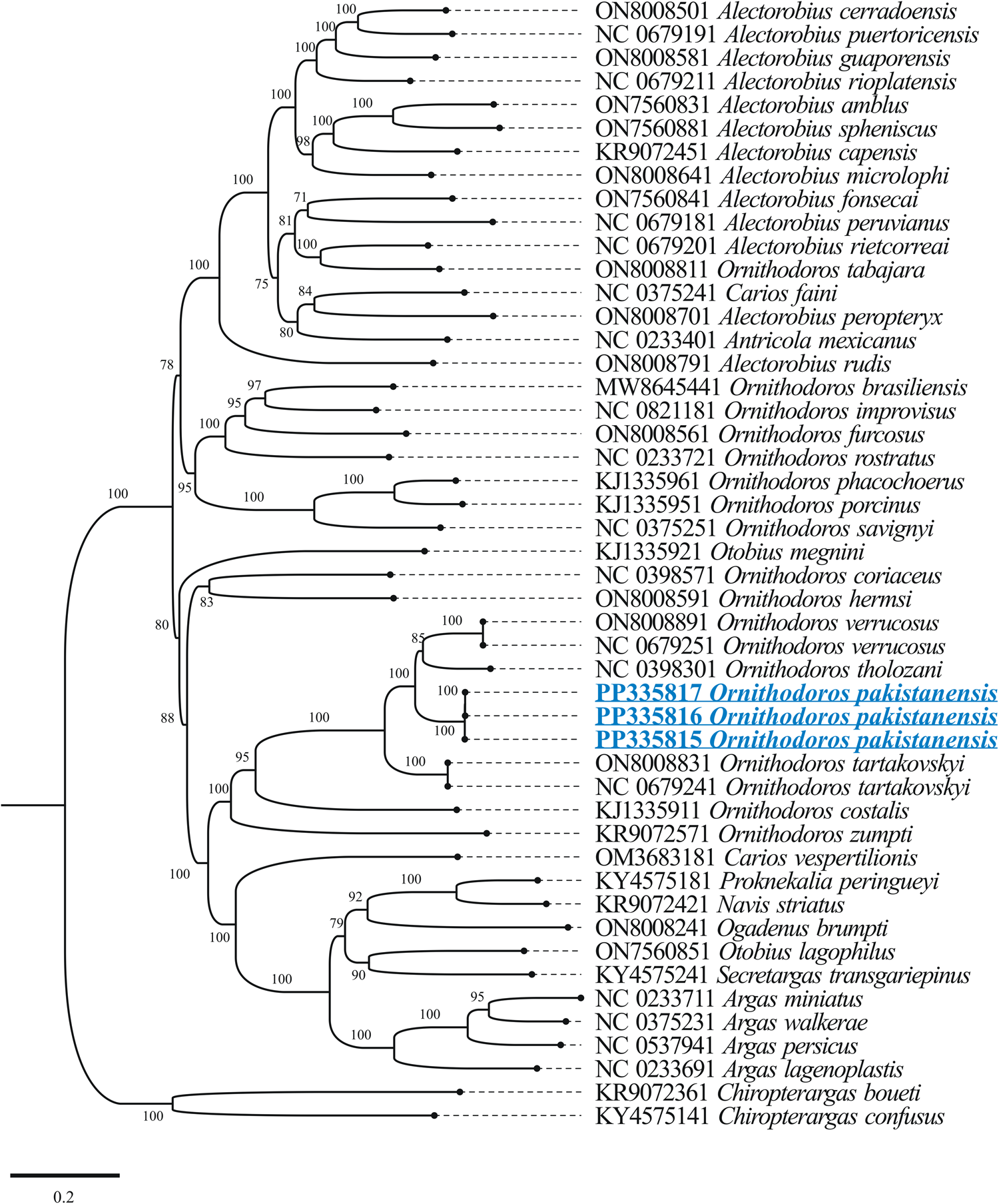
Figure 7. Maximum-likelihood phylogenetic tree 13 concatenated protein coding nucleotide sequences (cox1, cox2, atp8, atp6, cox3, nad3, nad5, nad4, nad4L, nad6, cytb, nad1 and nad2). Chiropterargas confusus and C. boueti were used as an outgroup. The bootstrap values (1000 replicates) are shown at each node. The obtained sequences for O. pakistanensis are underlined and coloured in blue.
The BLASTn analysis of the obtained 12S rDNA sequence (684 bp) for O. pakistanensis sp. nov. showed 88.47% maximum identity with O. tartakovskyi (ON800883) followed by 87.65% with O. verrucosus (ON800889), and 87.54% with O. tholozani (NC039830). The obtained 16S rDNA sequence (1022 bp) of O. pakistanensis sp. nov. showed 88.69% maximum identity with O. tholozani (NC039830) followed by 86.91% with O. verrucosus (ON800889), and 85.27% with O. tartakovskyi (ON800883). The 12S–16S rDNA sequences showed the lowest 0.125 pairwise genetic distance in comparisons with O. tholozani (NC039830) followed by 0.144 with O. verrucosus (ON800889) and 0.151 with O. tartakovskyi (ON800883) (Supplementary Table 4). Based on their concatenated phylogenetic tree, the 12S–16S rDNA sequences were clustered with the O. tartakovskyi, O. tholozani and O. verrucosus reported from Rocky Mountain Laboratory colony, Israel and Ukraine, respectively (Fig. 8).
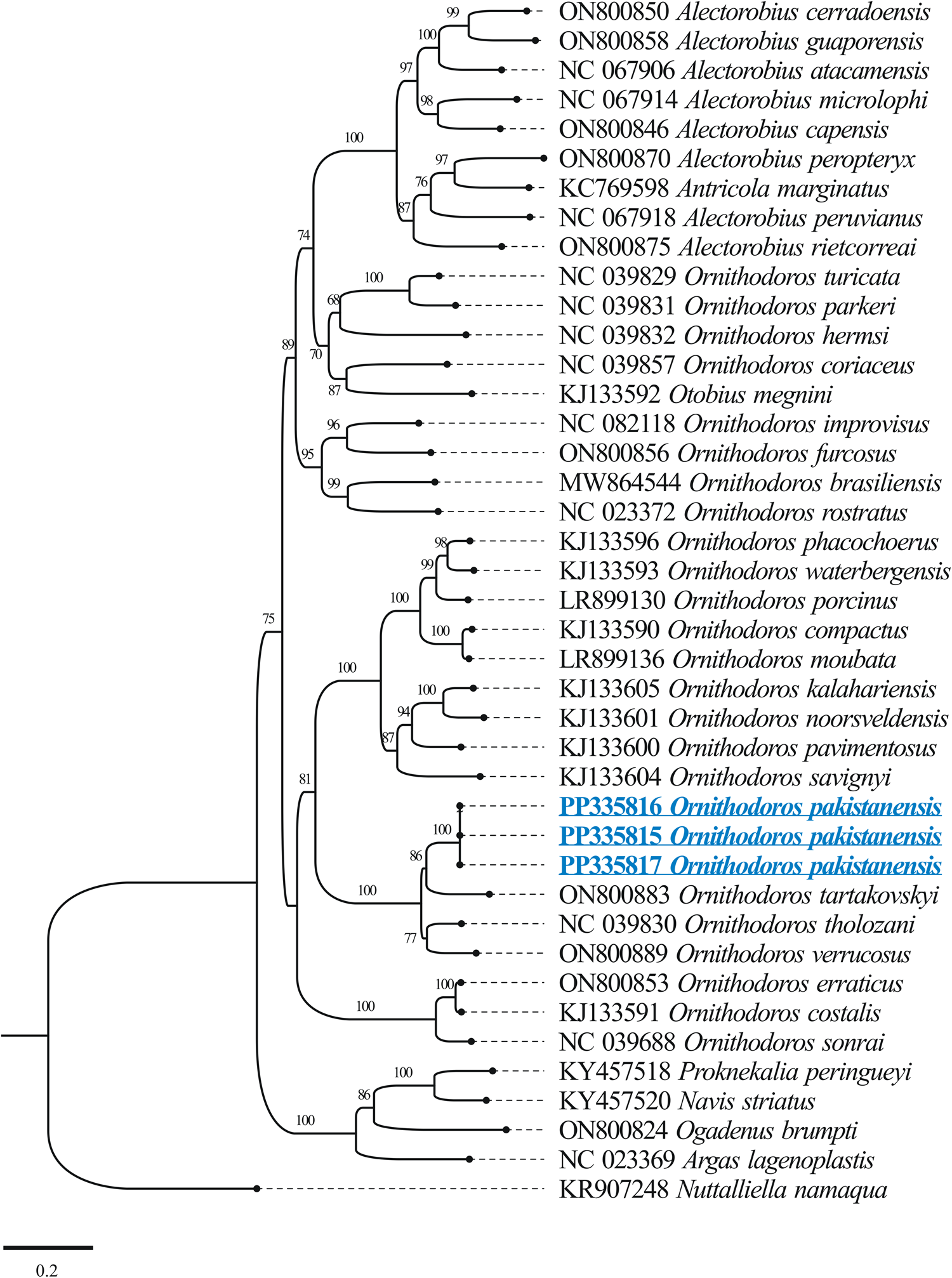
Figure 8. Maximum-likelihood phylogenetic tree based on 12S (684 bp)–16S (1022 bp) rDNA sequences The sequence of Nuttalliella namaqua was used as an outgroup. The bootstrap values (1000) are shown at each node. The obtained sequences for the present study are underlined and coloured in blue.
The BLASTn analysis of the obtained 18S rDNA sequence (1539 bp) and 28S rDNA sequence (1377 bp) for O. pakistanensis sp. nov. showed 99.09% (MF818025) and 99.27% (MF818024), maximum identity with O. tholozani, respectively. Matrix of evolutionary differences on pairwise comparisons of the mitochondrial 18S–28S rDNA sequences revealed small genetic divergence (0.009) with O. tholozani (MF818025/MF818024), followed by 0.022 with Ornithodoros sonrai (MF818028/MF818027) (Supplementary Table 5). Based on their concatenated phylogenetic tree, the 18S–28S rDNA sequences were clustered with the O. tholozani sequence reported from Israel (Fig. 9).

Figure 9. Maximum-likelihood phylogenetic tree based on the concatenated 18S (1539 bp)–28S (1377 bp) rDNA sequences. The sequence of N. namaqua was used as an outgroup. The bootstrap values (1000 replicates) are shown at each node. The obtained sequences for the present study are underlined and coloured in blue.
The obtained full-length mitochondrial genome (PP335815, PP335816, PP335817), 18S rDNA (PP333199, PP333200, PP333201) and 28S rDNA (PP333202, PP333203, PP333204) sequences for O. pakistanensis sp. nov. were deposited in GenBank.
Discussion
The morphological based identification of Ornithodoros spp. has caused some historical confusions (Bakkes et al., Reference Bakkes, De Klerk, Latif and Mans2018). Conducting comprehensive morphological and molecular analyses across a broad geographic range is essential for the accurate identification of both the established and newly discovered Ornithodoros ticks. This approach not only contributes to a more precise understanding of systematics of the genus Ornithodoros but also facilitates an accurate assessment of the epidemiology of the infectious agents they transmit. Herein, O. pakistanensis sp. nov. was described as belonging to the subgenus Pavlovskyella based on morphology, the mitochondrial genome and the nuclear 18S–28S rDNA. This species demonstrated close morphological and molecular resemblances to species within the subgenus Pavlovskyella, such as O. tholozani, while maintaining distinct characteristics that validate it as a new or separate species.
Morphological species relationship
Adults and late-instar nymphs of O. pakistanensis sp. nov. are similar to other representatives of the Pavlovskyella subgenus. However, the novel species can be separated from O. papillipes because it lacks a tuft of setae on the ventral surface of the hood (Pavlovsky, Reference Pavlovsky1930). Ornithodoros pakistanensis sp. nov. separates from Ornithodoros cholodkovskyi because in the latter species the transverse postanal groove intersects the median postanal groove clearly before the median point from its origin in the anal ring (Pavlovsky, Reference Pavlovsky1930). Ornithodoros tartakovskyi is also similar to O. pakistanensis sp. nov., but the transverse postanal groove of the former forms a sharp angle in its intersection with the median postanal groove (Filippova, Reference Filippova1966). Moreover, O. tartakovskyi has 3 bulky humps on tarsus I compared to 5 small humps in O. pakistanensis sp. nov. The novel species is also similar to O. verrucosus; however, in the latter species the third of 5 humps is larger and higher than the others (Filippova, Reference Filippova1966). In O. pakistanensis sp. nov. all humps on tarsus I are evenly protruding. The comparison of closely related O. tholozani subspecies and O. crossi with O. pakistanensis sp. nov. needs to take into account the ventral pairs of setae on tarsus IV (Table 2). However, the holotype and original description of O. crossi are missing (Leeson, Reference Leeson1953) and O. crossi is considered a nomen nudum. We therefore based our comparisons with the redescription of the species, which states (in drawings) that O. crossi has 2 pairs of ventral setae in tarsus IV (Sapre, Reference Sapre1944). Instead, O. pakistanensis sp. nov. has 4–7 pairs in nymphs and 5–7 pairs in adults. The comparison of the chaetotaxy of tarsus IV has been previously studied in O. tholozani from different localities and supported the designation of 3 subspecies; particularly, O. tholozani crossi was proposed for the specimens collected in Punjab, India (Descamps and Campana, 1946). Considering this character, and the geographical proximity where the collections were carried out in Pakistan, O. tholozani crossi should be now regarded as Ornithodoros pakistanensis sp. nov. (Table 2).
Table 2. Chaetotaxy of tarsus IV of O. tholozani subspecies sensu Descamps & Campana (1946) and O. pakistanensis sp. nov.
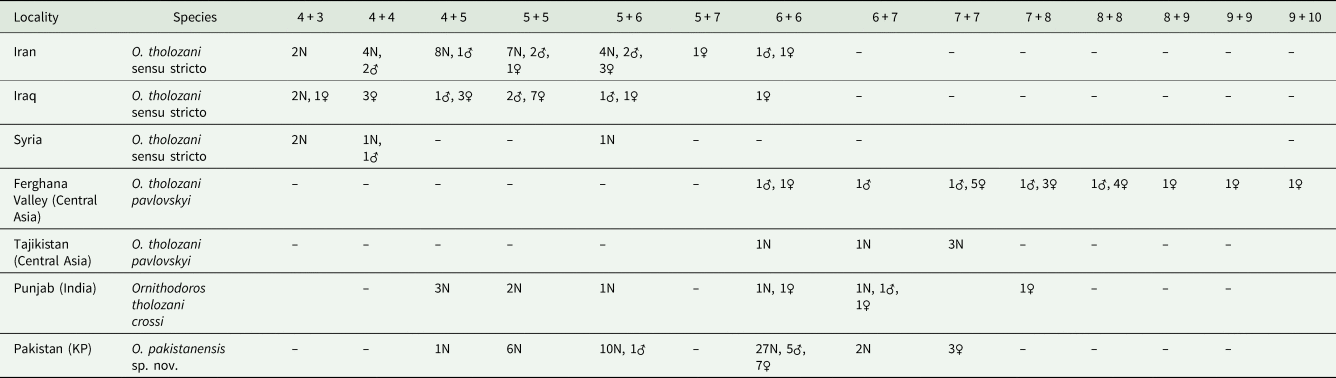
The number of setae is separated by ‘+’ because not all the setae were paired.
Phylogenetic species relationship
Particular focus on mitochondrial genome sequences offers the potential to elucidate disputed phylogenetic relationships within soft ticks (Burger et al., Reference Burger, Shao, Labruna and Barker2014; Mans et al., Reference Mans, Featherston, Kvas, Pillay, de Klerk, Pienaar, de Castro, Schwan, Lopez, Teel and de León2019, Reference Mans, Kelava, Pienaar, Featherston, de Castro, Quetglas, Reeves, Durden, Miller, Laverty and Shao2021; Mohamed et al., Reference Mohamed, Moustafa, Kelava, Barker, Matsuno, Nonaka, Shao, Mans, Barker and Nakao2022). Consequently, ticks identified based on their morphology in this study were subjected to molecular characterization through mitogenome sequencing. Utilizing molecular analysis of various genes, O. pakistanensis sp. nov. was determined to be most closely related to O. tholozani sensu stricto, O. verrucosus and O. tartakovskyi. In contrast to the minimal divergence observed in 18S rDNA and 28S rDNA (less than 1%), 12S rDNA, 16S rDNA and the mitogenome displayed a substantial divergence of more than 12%. The former indicates high levels of conservation between species for the nuclear rDNA genes, the latter confirms the distinct nature of different species (Labruna et al., Reference Labruna, Terassini, Camargo, Brandão, Ribeiro and Estrada-Peña2008; Mans et al., Reference Mans, Featherston, Kvas, Pillay, de Klerk, Pienaar, de Castro, Schwan, Lopez, Teel and de León2019, Reference Mans, Kelava, Pienaar, Featherston, de Castro, Quetglas, Reeves, Durden, Miller, Laverty and Shao2021; Khan et al., Reference Khan, Islam, Khan, Islam, Muñoz-Leal, Labruna and Ali2022; Ali et al., Reference Ali, Khan, Alouffi, Almutairi, Paguem, Chitimia-Dobler, Pienaar, de Castro and Mans2024). In the phylogenetic analysis using these genes, O. pakistanensis sp. nov. clustered within a monophyletic clade alongside O. tholozani, either independently (based on 18S rDNA and 28S rDNA), or in combination with other species such as O. verrucosus and O. tartakovskyi (based on cox1, 12S rDNA and 16S rDNA). Although there may be a common ancestor for these species, as suggested by sharing the subgenus Pavlovskyella (Kneubehl et al., Reference Kneubehl, Muñoz-Leal, Filatov, De Klerk, Pienaar, Lohmeyer, Bermúdez, Suriyamongkol, Mali, Kanduma and Latif2022; Carnero-Morán et al., Reference Carnero-Morán, Oleaga, Cano-Argüelles and Pérez-Sánchez2023), they exhibit significant variations in morphology, host preference and habitat use.
On the presence of O. tholozani in Pakistan
Climate is considered to have a significant influence, when compared to biological factors such as host availability and vegetation patterns, on the distribution of Ornithodoros ticks (Cumming, Reference Cumming2002; Vial et al., Reference Vial, Ducheyne, Filatov, Gerilovych, McVey, Sindryakova, Morgunov, de León, Kolbasov and De Clercq2018). While confirming the climatic suitability of central Asia for the establishment of Ornithodoros ticks, Vial et al. (Reference Vial, Ducheyne, Filatov, Gerilovych, McVey, Sindryakova, Morgunov, de León, Kolbasov and De Clercq2018) were unable to confirm the climate suitability of Pakistan due to poor evidence of any Ornithodoros species in the country, particularly O. tholozani sensu stricto. However, the region meets essential climate criteria for Ornithodoros feeding activity, including a spring temperature surpassing 10°C, a 3-month summer temperature exceeding 20°C, annual precipitation ranging between 60 and 750 mm, dry seasons interspersed by small rain showers and availability of residual water from perennial rivers near habitats.
Ornithodoros spp. typically thrive in microclimates within arid regions, infesting single or multiple vertebrate hosts during nighttime (Hoogstraal, Reference Hoogstraal1985; Oliver, Reference Oliver1989; Guglielmone et al., Reference Guglielmone, Robbins, Apanaskevich, Petney, Estrada Peña, Horak, Shao and Barker2010). Ornithodoros tholozani, known for infestations in both domestic and wild animals, is widely distributed in the deserts and semi-desert regions of Asia (Parola and Raoult, Reference Parola and Raoult2001; Estrada-Peña et al., Reference Estrada-Peña, Kleinerman and Baneth2017). Without any substantial evidence, this species has been considered to exist in Pakistan (Vial et al., Reference Vial, Ducheyne, Filatov, Gerilovych, McVey, Sindryakova, Morgunov, de León, Kolbasov and De Clercq2018). Herein, O. pakistanensis sp. nov., a morphologically closely related species, is reported in the northwestern mountainous terrain of Pakistan, which comprises a range of semi-arid to humid regions, characterized by significant vegetation (Ali et al., Reference Ali, Numan, Khan, Aiman, Muñoz-Leal, Chitimia-Dobler, Labruna and Nijhof2022; Khan et al., Reference Khan, Almutairi, Alouffi, Tanaka, Chang, Chen and Ali2023; Tila et al., Reference Tila, Khan, Almutairi, Alouffi, Ahmed, Tanaka, Tsai and Ali2023). Moreover, based on specific collection sites, it is suggested that domestic hosts such as cattle and small ruminants are the hosts of O. pakistanensis sp. nov.
Conclusion
This study described O. pakistanensis sp. nov., a new species from the northwestern part of Pakistan using morphological and molecular analyses. Given that the genus Ornithodoros and the subgenus Pavlovskyella are paraphyletic, this study adds a novel species that fits Pavlovskyella sensu stricto morphology and genetics, closely related to O. tholozani, the type species of the subgenus. Ornithodoros pakistanensis can be morphologically distinguished from O. tholozani by the presence of 4–7 setal pairs in tarsus IV ventral chaetotaxy in nymphs, and 5–7 pairs in adults. Due to the overlap in several aspects of these 2 tick species, including morphology and distribution, the reliability of certain previous reports describing O. tholozani in the region, especially in Pakistan, may be questionable.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182024000982
Data availability statement
All data in the study is available in the public databases as listed by their accession numbers.
Acknowledgements
We are thankful for the financial support offered by the Pakistan Science Foundation (PSF) and Higher Education Commission (HEC) of Pakistan.
Author contributions
A. Ali: conceptualization, data curation, funding acquisition, investigation, methodology, project administration, supervision, writing original draft, writing – review and editing. M. N.: investigation, data curation, software, visualization, writing – original draft, writing – review and editing. M. K.: investigation, formal analysis, software, writing – original draft, writing – review and editing. A. Alouffi: investigation, methodology, data curation, resources, writing – original draft, writing – review and editing. M. M. A.: investigation, methodology, project administration, resources, writing – original draft, writing – review and editing. R. P.: methodology, visualization, writing – original draft, writing – review and editing. M. H. C.: investigation, methodology, visualization, writing – original draft, writing – review and editing. L. C.-D.: methodology, visualization, writing – review and editing. S. M.-L.: investigation, methodology, visualization, morphology, microscopy, writing original draft, writing – review and editing. B. J. M.: investigation, methodology, project administration, mitogenome analysis, funding acquisition, visualization, writing original draft, writing – review and editing.
Financial support
The researchers supporting project number (RSP2024R494), King Saud University, Riyadh, Saudi Arabia. This study was supported by the National Research Foundation of South Africa (grant number: 137966). S. M.-L. was funded by Fondecyt Iniciación no. 11220177.
Competing interests
None.
Ethical standards
This research was approved by the Advanced Studies Research Board (ASRB: Dir/A&R/AWKUM/2022/9396) committee members of Abdul Wali Khan University, Mardan, Khyber Pakhtunkhwa, Pakistan. During the observation and tick collecting process, the owners of the animal shelters provided their oral permission.