Transmissible spongiform encephalopathies (TSEs) are fatal neurological diseases which can occur in a range of species including humans [Reference Manson1]. The archetypal form of TSE is classical scrapie in small ruminants, which was first reported nearly 300 years ago. Central to the disease process is the conversion of the host cellular prion protein, PrP, into a disease-specific isoform called PrPSc [Reference Soto2]. In small ruminants, there are many polymorphisms in the gene encoding for prion protein (the PRNP gene), which affect disease susceptibility, and this has been widely employed as a disease control measure for classical scrapie strains in sheep populations [3]. It has been proposed that the polymorphic nature of the goat PRNP gene could be similarly exploited, but this is made more complicated by the greater geographical and breed-related heterogeneity of goat genetics [Reference Vaccari4], and the relative absence of data on the biological and geographical diversity of caprine scrapie strains that may exist. Polymorphisms at codon 146 have been reported to affect natural susceptibility to classical scrapie in Cypriot goats. Specifically the presence of asparagine (N) at codon 146 appears to be associated with susceptibility, whereas its replacement with aspartic acid (D) or serine (S) has shown association with genetic resistance to classical scrapie in goats [Reference Papasavva-Stylianou5–Reference Ortiz-Pelaez7]. The possibility of using the D146 and S146 alleles as a nationwide strategy for breeding for resistance in the Cypriot goat population has been repeatedly described [Reference Papasavva-Stylianou6–Reference Gonzalez8]. The objective of this study is to assess whether the risk of scrapie in non-NN goats is different from that of NN goats under the same natural conditions.
Healthy home-bred goats with at least one putatively resistant allele (S or D) at codon 146, that had been exposed to a scrapie-contaminated environment for at least 6 years, were pre-selected from herds which had had confirmed cases of caprine scrapie since 2004 or earlier. A number of goats with the N146N genotype, matched by age and herd, were equally pre-selected as putative non-resistant. This ensured that all pre-selected animals had been born into an infected environment, and had remained in such throughout their lifetime. Although it was not possible to quantify precisely the level of exposure, there is clear evidence that the scrapie agent can persist for long periods in the environment [Reference Konold9–Reference Georgsson11]. When these pre-selected animals were culled for management reasons, at the end of their productive life, testing was undertaken to look for any evidence of disease using the presence of PrPSc as a marker.
The genotyping of whole herds and the identification of the genotype of pre-selected goats encouraged herd owners to voluntarily extend the productive lifespan of any S and/or D allele-carrying goats in order to increase their capacity to breed for presumptive resistance within their herds. As a result, it was difficult to sample a sufficient number of goats carrying those alleles and those with the matching criteria (N146N) in some of the recruited herds.
Tissue samples were collected post-mortem from a total of 246 putative resistant and 310 putative non-resistant goats aged between 6 and 13 years, from 39 herds, covering all districts where the disease is present, over the period August 2009 to October 2013.
Fresh brainstem was tested using the BioRad TeSeE ELISA (Bio-Rad, USA), according to the manufacturer's instructions. Previous studies in which parallel testing was undertaken in goats indicated that brainstem ELISA alone is not very sensitive for detecting infected animals. Consequently, brainstem at the level of the obex, and retropharyngeal lymph node were also sampled into 10% formal saline, processed into wax and screened by immunohistochemistry (IHC) using the anti-PrP monoclonal antibody 2G11 (ABDSerotec) as described in detail elsewhere [Reference Simmons12]. In a parallel study it was established, using intracerebrally challenged animals ([13]; S. Georgiadou & M. M. Simmons, unpublished data), that IHC using this antibody can detect positive cases in animals polymorphic at codon 146.
An animal was considered infected if it was positive in either tissue by either of the tests conducted.
Of the 556 goats with results available from both IHC (obex and retropharyngeal lymph node) and the rapid test, 117 animals (all N146N genotype) were positive by at least one of them. This represented 37·7% of all the NN animals tested. Eighteen (46·1%) out of the 39 herds participating in the study had positive cases among the pre-selected goats for the study with a median prevalence of 20·2% [interquartile range (IQR)Q1-Q3 10·8–37·8%]. However the sampling at herd level was very variable during the period of the cull (median 7, IQRQ1-Q3 2–22) and the level of detection was very much dependent on the number of goats sampled in each herd.
A total of 114 goats were positive by IHC. Eighty-eight (77·2%) of these were positive in both the brainstem and retropharyngeal lymph node, with six (5·3%) being positive only in the brainstem and the remaining 20 (17·5%) animals IHC positive only in the retropharyngeal lymph node. Fifty-eight (61·7%) of the goats positive by IHC in the brainstem were also positive by ELISA. A further three cases were positive only in the ELISA test, and could not be confirmed by IHC. The risk difference when comparing the two groups of interest was equal to 37·7% (95% confidence interval 32·3–43·1), with the probability of the risk difference being zero (null hypothesis) lower than 5% (P < 0·001) [package ‘fmsb’, R Development Core Team (http://www.R-project.org]. The overall results (the total numbers of tested and positive goats by age and genotype) are given in Table 1.
Table 1. Number of goats tested by age, and genotype at codon 146
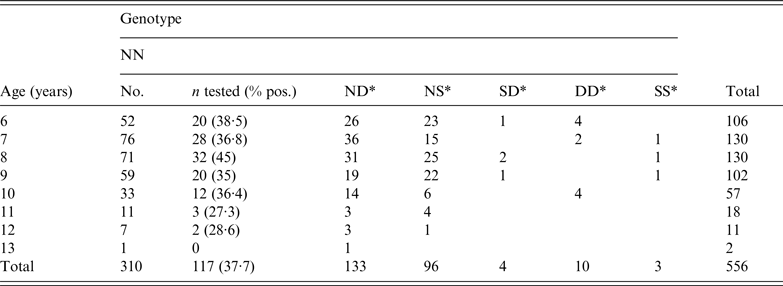
* All animals of this genotype tested negative.
The results of this study confirm and extend those of previous studies that have shown that the susceptibility of Cypriot goats to scrapie in natural conditions is associated mostly with the N146N genotype [Reference Papasavva-Stylianou5–Reference Ortiz-Pelaez7]. Although the objective of this study was to conduct in parallel both the rapid and confirmatory tests to maximize the detection ability of infected animals and not test the test performance, the results are aligned with the low ability of the rapid test to detect positive cases to IHC as previously reported at the herd [Reference Ortiz-Pelaez7, Reference Gonzalez8] and population [Reference Corbiere14] levels. They also further strengthen the assertion that animals with at least one aspartic acid (D) or serine (S) at codon 146 are resistant to classical scrapie, despite a lifetime natural exposure to the infective agent, and indicate that the introduction of a breeding programme [3] using codon 146 would have a substantial beneficial effect on disease prevalence in Cyprus.
ACKNOWLEDGEMENTS
This study was partly funded by the Cypriot Government and the European Commission through the EU Reference Laboratory for TSEs. The authors are grateful to the owners of the participating herds, staff of the District Veterinary Offices, the Histopathology and TSE laboratories of the Cypriot Veterinary Services, and the staff of the Pathology Department, APHA Weybridge for logistical and technical support.
DECLARATION OF INTEREST
None.



