1. Introduction
Strictly speaking, taking quantitative biology approach means that we use numbers, and typically also mathematics, to describe biological processes (Figure 1). However, this is not merely a nice-to-have extra or a technological increment; it actually revolutionises knowledge production. Quantitation leads to identifying and modelling dependencies between different measurements, which is the way to form new hypotheses. Statistical approaches measure the strength of such dependencies. Modelling then allows a preliminary test of the hypothesis in silico, with predictions tested again in experiments with quantitative results, while all along probability theory is used to make sure that we draw sound inferences, and account for noise and robustness. This is what a quantitative approach in the multiple senses used in this review means—an iterative approach of measurement, statistical analyses, hypothesis testing in silico, in vitro and in planta, and back to the next cycle with the gained knowledge we have just obtained.
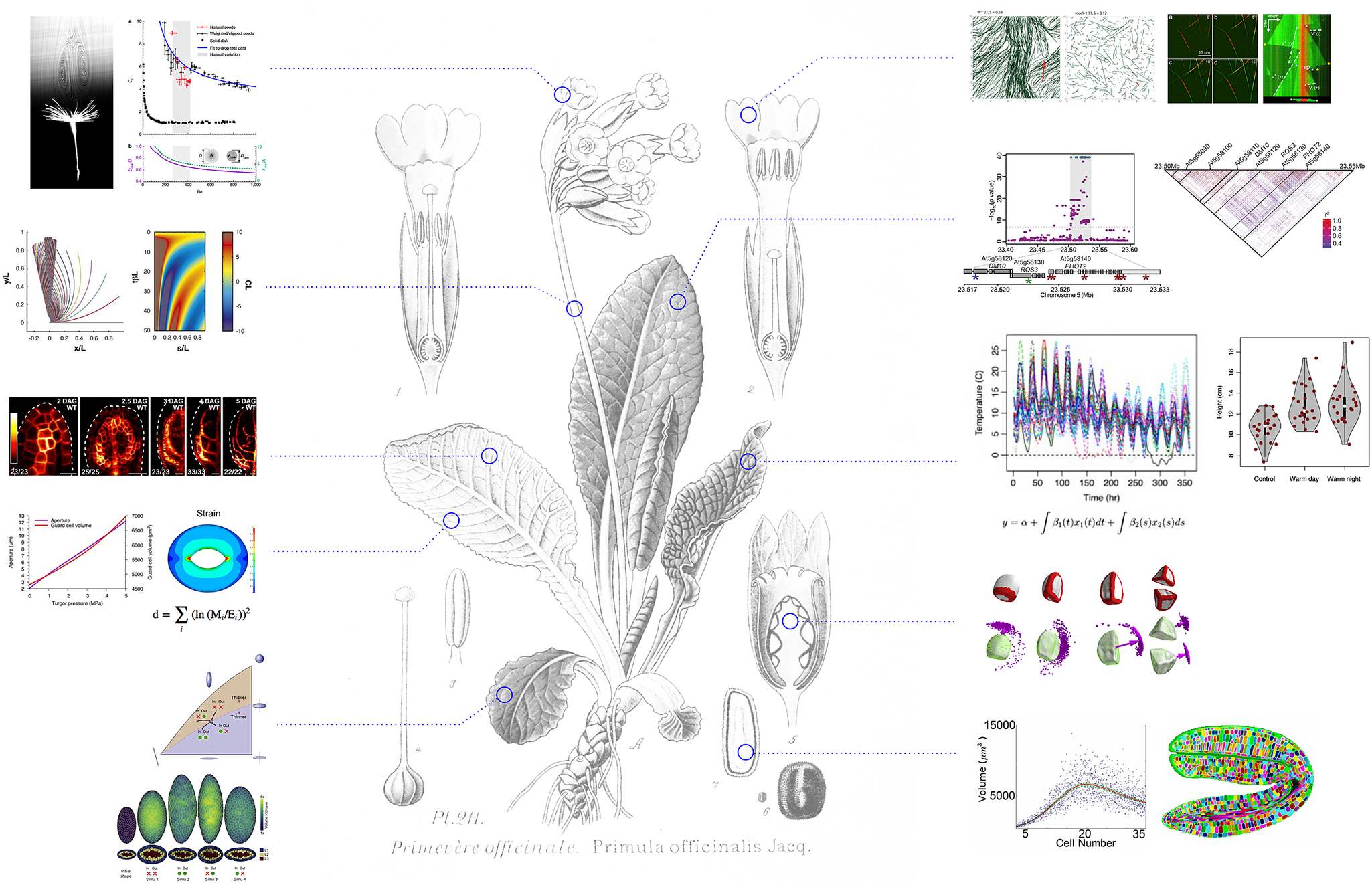
Fig. 1. A quantitative revolution in plant science. Whereas molecular insights in plant biology could simply provide a molecular catalogue of plant ontology, the integration of mathematics and computational modelling has instead helped to identify new questions with the aim to unravel the principles of plant life. Hypotheses are formalised and tested in computational models, and results from simulations fuel further experimental analysis. Assessment of the validity of the results, from molecules to ecosystems, involves statistical validation and further quantitative exploration. Background image (Primula officinalis) taken from Atlas des plantes de France, A. Masclef, Paul Klincksieck Ed., Paris, 1890. Quantitative examples extracted from Verna et al., Reference Verna, Sree, Sawchuk, Nguyen and Enrico2019 eLife; Chakrabortty et al., Reference Chakrabortty, Willemsen, Zeeuw, Liao, Weijers, Mulder and Scheres2018 Curr. Biol.; Bastien et al., Reference Bastien, Bohr, Moulia and Douady2013 PNAS; Brestovitsky et al., Reference Brestovitsky and Ezer2019 Plant Direct; Zhao et al., Reference Zhao, Du, Oliveri, Zhou, Ali, Chen, Feng, Wang, Lü, Long, Schneider, Sampathkumar, Godin, Traas and Jiao2020 Curr. Biol.; Woolfenden et al., Reference Woolfenden, Bourdais, Kopischke, Miedes, Molina, Robatzek and Morris2017 Plant J; Cummins, Reference Cummins, Seale, Macente, Certini, Mastropaolo, Maria Viola and Nakayama2018 Nature; Allard, Reference Allard, Wasteneys and Cytrynbaum2010 Mol. Biol. Cell. and Fache et al., Reference Fache, Gaillard, Van Damme, Geelen, Neumann, Stoppin-Mellet and Vantard2010 Plant Cell.
Furthermore, such an interdisciplinary approach also fuels creativity and triggers new questions for two reasons: (a) being able to formalise questions within a defined mathematical framework also means that hypotheses become truly testable and interoperable. This is key to understand plants as multiscale (both in space and in time) systems, (b) because interdisciplinary settings imply that not everybody is an expert in all techniques, the focus remains on the question at hand, and not the techniques and their development. This means that quantitative biology is one of the best ways to open new avenues of research, and identify new questions (Figure 2). This is what we are attempting to elaborate on in this review.
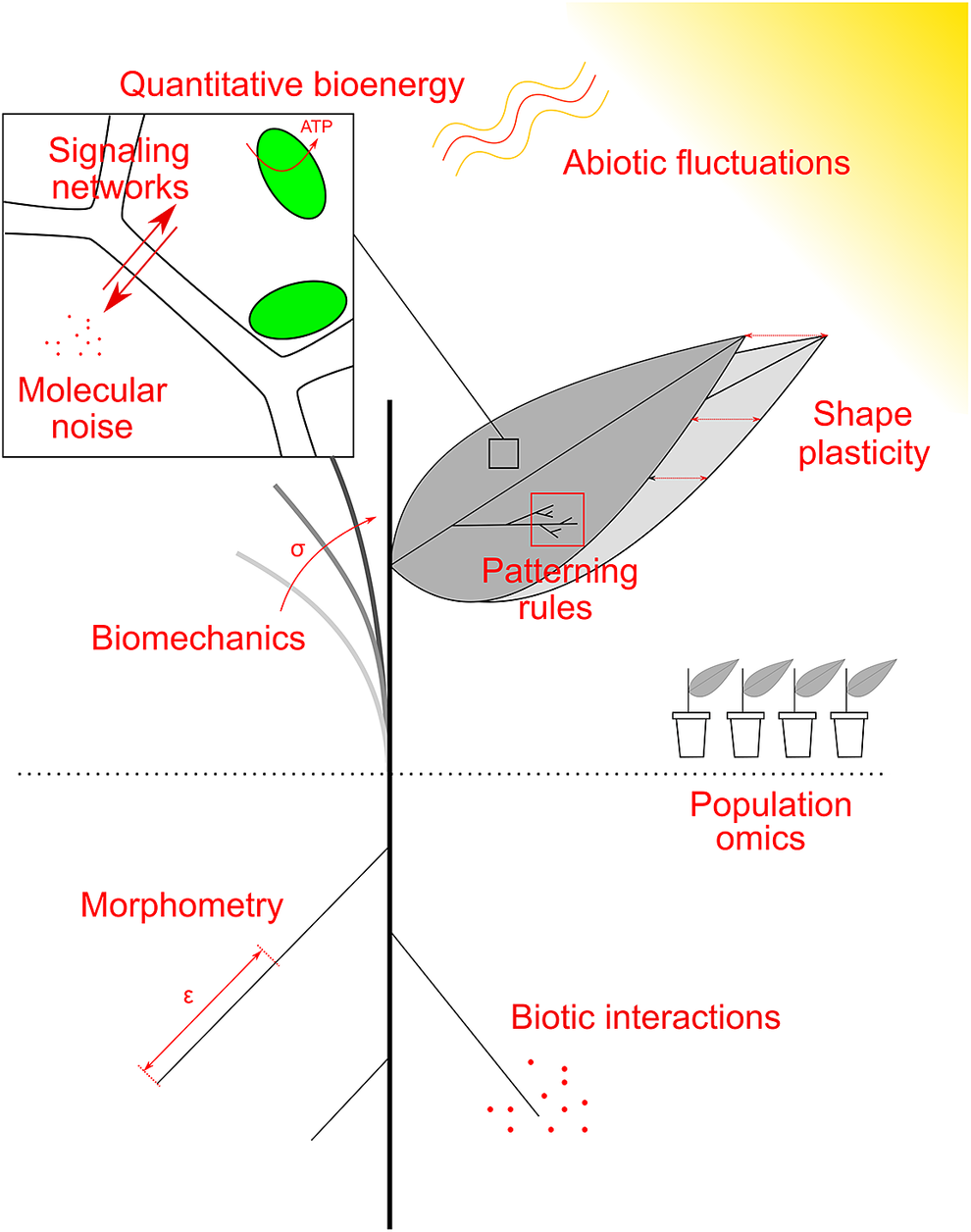
Fig. 2. Examples of research topics at the crossroad between plant biology, physics and maths. Quantitative plant biology explores these topics, and the common mathematical framework allows their integration, across spatial and temporal scales. See main text for details.
2. Signalling networks
Signalling networks primarily act to process and integrate information perceived through a multitude of receptor systems, and relay this information to cellular effectors, which, in turn, enact a response tailored to the conditions. This definition implies that information is encoded and decoded, and thus can be formalised. This is a thriving field of study in computational modelling (Long et al., Reference Long, Brady and Benfey2008). In terms of identifying complex relationships between inputs and outputs, machine learning approaches are becoming increasingly popular. Furthermore, exciting developments in technologies coupled with statistical techniques now allow for the inference signalling networks from large genomic datasets which are becoming readily available (Carré et al., Reference Carré, Mas and Krouk2017).
Most studies on signalling networks have emphasised the identification of the molecular components critical for signal transduction pathways in a mostly binary manner (‘on’ vs. ‘off’), often considering a reduced set of actors (minimally a receptor and its ligand), under controlled-lab growth conditions. To expand our knowledge beyond the description of pathway architecture and understand how integrated signalling networks behave in varying conditions, quantitative biology approaches are required. For example, how can signals be discriminated from each other when they simultaneously occur? How can priorities be established and an integrated response achieved when a cell is challenged with multiple inputs, beyond the so-called ‘hormonal cross-talk’? How are thresholding and noise filtering mechanisms molecularly encoded to prevent spurious pathway activation? Answering these questions requires accepting a number of technical challenges, for example, concerning the quantification of signalling molecules and graded responses.
A major achievement in plant signalling has been the identification of the core components of ‘receptor–ligand’ pairs for many signal transduction pathways, through the isolation of mutants that have modified responses to a particular signal, that either fail to respond, or respond constitutively even in the absence of that signal. Genetics further uncovered the hierarchy and interactions among transduction pathway components and showed how complex the networks involved in the integration of signalling inputs and outputs are. This includes feedback mechanisms, where signalling sometimes provides unexpected compensatory responses. For instance, a dwarf cellulose synthase mutant is partially rescued when it bears another mutation in the wall integrity pathway: the plant becomes unable to detect the damages in its cell wall and thus does not trigger growth arrest mechanisms (Hématy et al., Reference Hématy, Sado, Van Tuinen, Rochange, Desnos, Balzergue, Pelletier, Renou and Höfte2007).
Compared to other systems, the contribution of signalling dynamics, that is, the duration, frequency and amplitude of a signal for downstream responses, has been somewhat neglected in plants (Purvis & Lahav, Reference Purvis and Lahav2013). For example, in mammalian cells, transient activation of extracellular signal-regulated kinase (ERK) through epidermal growth factor can result in cell proliferation, whereas sustained activation by nerve growth factor can lead to cell differentiation (Avraham & Yarden, Reference Avraham and Yarden2011). It has been predicted and experimentally validated that modulation of feedback strength in a single inhibitory loop from ERK to one of its upstream kinases (RAF) can result in a variety of stable output states ranging from a sustained monotone response to a transient adapted output, to oscillation, or to bi-stable, switch-like responses (Kholodenko et al., Reference Kholodenko, Hancock and Kolch2010). In plants, our understanding of this temporal dimension of information encoding is far less developed and thus opens many avenues for pioneering studies in quantitative plant biology.
Despite increased efforts, the availability of quantitative data on signalling events remains the major constraint for approaches aiming at a deeper understanding and modelling of signalling networks. An ever-expanding set of biosensors, which allow for the in vivo visualisation and quantification of signalling molecules with cellular or even subcellular resolution, are among the most promising remedies for this lingering ailment. In fact, very recently, biosensor-based approaches have brought breakthroughs to the field of plant signalling. For example, a study by Toyota and colleagues has elegantly elucidated ‘Rapid, long-distance signalling in plants’ showing that when injured on one leaf by a nibbling insect, a plant can alert its other leaves to begin anticipatory defence responses (Toyota et al., Reference Toyota, Spencer, Sawai-Toyota and Gilroy2018). This development went hand in hand with quantitative approaches and mathematical modelling that led to the proposal of a propagation mechanism (Evans et al., Reference Evans, Choi, Gilroy and Morris2016). In addition, emerging systems biology tools may provide new ways to perturb signalling network components in a spatially and temporally controlled manner to illustrate network behaviour.
Finally, beyond the computational models of networks, validation with molecular genetics and biosensors, quantitative approaches on signalling also need to deal with numerous players, their redundancy, synergy and antagonism. Crucial gene activities are often shared by several redundant homologs, and thus single mutations in such genes cause only partial defects. It is also challenging to identify the primary defect from all the secondary defects. These difficulties can be solved by extensive phenotype quantification. For example, the roles of various miRNAs, which were identified by small RNA sequencing of Arabidopsis embryos, were clarified based on their mutant phenotypes on each embryonic tissue and developmental stage (Plotnikova et al., Reference Plotnikova, Kellner, Schon, Mosiolek and Nodine2019). Such quantification can be further combined with CRISPR/Cas9-based genome editing techniques that enable tissue-specific and conditional gene manipulation of target gene (Decaestecker et al., Reference Decaestecker, Buono, Pfeiffer, Vangheluwe, Jourquin, Karimi, Van Isterdael, Beeckman, Nowack and Jacobs2019; Wang et al., Reference Wang, Ye, Lyu, Ursache, Löytynoja and Mähönen2020). With the help of such systems, one can knockout genes in specific cell types at will, thus enabling the identification of the distinct roles of the identified genes.
3. Noise and robustness
Another layer of complexity is brought about by a prevalent factor in biology: noise. Stochastic, or random, effects pervade biology across scales (Lestas et al., Reference Lestas, Vinnicombe and Paulsson2010; Tsimring, Reference Tsimring2014), from cells where molecules are constantly buffeted by thermal noise, to the robust formation of organs by collections of cells, to the environmental fluctuations experienced by crops in the field. Noise also invades our efforts to measure the biological world, leading to technical variation and requiring transparent quantitative methods for responsible analysis. Unavoidable, multiscale noise in biology provides both challenges and opportunities for plants, and a large and growing body of work seeks to elucidate how plants attempt to filter out, or exploit, randomness. We cannot hope here to give a comprehensive survey of the many ways stochastic influences shape plant biology, but hope that a few classic examples across scales will illustrate the ubiquity, and importance, of stochasticity in plant biology (Abley et al., Reference Abley, Locke and Leyser2016).
At the most fundamental level, stochasticity in the form of spontaneous mutations and other events underlies all plant evolution (e.g., Rose et al., Reference Rose, Rees and Grubb2002) studying the interplay of stochastic variation and fitness in thistle populations. With the view of neutral theory of molecular evolution, stochastic events continually shape plant population structure (e.g., Menges, Reference Menges2014, modelling the impact of stochastic extinction events on plant populations).
Within plants, cellular noise impacts vital processes across scales including the circadian clock (Guerriero et al., Reference Guerriero, Pokhilko, Fernández, Halliday, Millar and Hillston2012), gene expression (Araújo et al., Reference Araújo, Pietsch, Keizer, Greese, Balkunde, Fleck and Hülskamp2017; Wang et al., Reference Wang, Tian, Madlung, Lee, Chen, Lee, Watson, Kagochi, Comai and Chen2004), internal signalling (Trewavas, Reference Trewavas2012), tropisms (Meroz & Bastien, Reference Meroz and Bastien2014), patterning (Meyer et al., Reference Meyer, Teles, Formosa-Jordan, Refahi, San-Bento, Ingram, Jönsson, JCW and Roeder2017) organ shape plasticity (Hong et al., Reference Hong, Dumond, Zhu, Tsugawa, Li, Boudaoud, Hamant and Roeder2018) and seed germination (see below). The cytoskeletal polymer networks formed by actin and microtubules are prime examples of why quantitative approaches are inevitable: they are far-out of equilibrium, stochastic, interacting many-particle systems with a high number of spatial degrees of freedom. Emergent properties from such behaviour include the formation of parallel arrays or cell division plane orientations. As such, they are at the cutting edge of statistical physics, a vibrant field of highly quantitative research in its own right (Deinum & Mulder, Reference Deinum and Mulder2013; Wasteneys & Ambrose, Reference Wasteneys and Ambrose2009).
Stochastic modelling can provide a powerful framework to understand the interactions of plants with their environments (e.g., Katul et al., Reference Katul, Porporato and Oren2007). Elegant modelling work coupling mechanical and stochastic influences has been used to describe whole-plant development (e.g., Costes et al., Reference Costes, Smith, Renton, Guédon, Prusinkiewicz and Godin2008; for apple trees). The explosion of multiomics technology has allowed genetic features shaping noise levels in transcripts and metabolites to be discovered (Jimenez-Gomez et al., Reference Jimenez-Gomez, Corwin, Joseph, Maloof and Kliebenstein2011).
Some of these stochastic influences constitute challenges for plants—for example, cellular noise in signalling pathways means that plants need to invest extra resources in maintaining the fidelity of signals. However, noise can also be beneficial, providing a useful source of variability in plants (Muller et al., Reference Muller, Guédon, Passot, Lobet, Nacry, Pagès, Wissuwa and Draye2019). Bet-hedging in seeds provides a compelling example of plants exploiting noise: a generation of seeds that germinate at different times will be more robust to unpredictable environmental change than one that germinates synchronously. Johnston and Bassel (Reference Johnston and Bassel2018) identified a network motif encoding a variability enabling bet-hedging in seeds. In this system, noisy positive feedback onto both ABA synthesis and ABA degradation can result in significantly varying final hormone levels. Noise can even help signals to become detectable, because noise on top of a weak signal can make the signal detectable, a phenomenon called ‘stochastic resonance’ (Rué et al., Reference Rué, Domedel-Puig, Garcia-Ojalvo and Pons2012).
Variability in environmental inputs is also leveraged by dormant seeds. Topham et al. (Reference Topham, Taylor, Yan, Nambara, Johnston and Bassel2017) identified a system whereby seeds make preferential use of fluctuating ambient and low temperatures over constant low temperature to break their dormancy. This mechanism indicates that the perception of low temperature in seeds is not a matter of the linear accumulation of cold, but rather complex processing of these fluctuating inputs. The adaptive significance of this may relate to daily temperature fluctuations being greater in the spring and autumn, with these variable signals acting as indicators of the changing seasons.
The relationship between the variability in single cells and their collective contribution towards robust organ shape and size has also been investigated. It is proposed that noise across molecular and cellular scales can be amplified to prime organogenesis (Uyttewaal et al., Reference Uyttewaal, Burian, Alim, Landrein, Borowska-Wykręt, Dedieu, Peaucelle, Ludynia, Traas, Boudaoud, Kwiatkowska and Hamant2012) or filtered in order to ultimately result in the formation of organs having consistent morphologies (Hervieux et al., Reference Hervieux, Tsugawa, Fruleux, Dumond, Routier-Kierzkowska, Komatsuzaki, Boudaoud, Larkin, Smith, Li and Hamant2017).
The study of these vital influences is quantitative in essence. Statistical measures to quantify variability and heterogeneity across scales have been developed. The characterisation of noise requires these quantitative measures (for example, a measure of dispersion like the coefficient of variation or Fano factor; Tsimring, Reference Tsimring2014). Large numbers of quantitative observations are required for accuracy in these measurements, and to distinguish mechanistic hypotheses when noise is involved. It is only through suitably coupled quantitative models, experiments and statistical methods that we can hope to unravel the sources and effects of stochasticity in plant biology, and the mechanisms by which plants deal with, and exploit, the resulting variability. There is much at stake since robust agrosystems increasingly build on genetic heterogeneity (agroforestry, agroecology and mixed varieties) while facing increasing environmental fluctuations. The future of food security will involve careful assessment of uncertainties and better ways to build on the added values of stochasticity.
Another issue related to noise exists in concert—perhaps less biologically exciting but equally, if not more, societally important. Statistical misunderstandings of the role noise plays in experiments has led to most published research findings being wrong, at least in some scientific fields (Ioannidis, Reference Ioannidis2005). To combat this, we need both to embrace rigorous (but not necessarily complicated) statistical methods and a shift in quantitative philosophy, where noise and uncertainty are more transparently and openly addressed and critically analysed. The p<.05 paradigm, and accompanying focus on statistical rather than scientific significance, has been much criticised (Ziliak & McCloskey, Reference Ziliak and McCloskey2008). Even within this paradigm, statistical tests are frequently misused. A common example is using a t-test for integers or non-negative entities when the standard deviation (not the standard error!) is of similar magnitude to the mean. Clearly, such a sample cannot be normally distributed, and other tests (like Mann–Whitney) should be favoured (Saxon, Reference Saxon2015). The series of articles in Saxon (Reference Saxon2015) highlights several other statistical misuses that confound scientific progress. As the transition towards more quantitative and data-rich plant biology continues, we urge researchers to adapt their statistical approaches to ensure this exciting world is understood as accurately as possible.
4. Tissue topology as an instructive cue
Signalling and noise occur in cells that reside in tissues. An individual cell’s function is thus highly biased by that of its neighbours. Beyond the molecular aspects, plant biology thus embeds another strong quantitative component: topology.
Cell-to-cell communication is central to plants. Coordination between cells is needed to orchestrate growth and development, as well as responses to their environment. Plant cells can transmit information using chemical, mechanical and electrical signals. Chemical signals typically move between cells through a secreted peptide sensed by a transmembrane receptor kinase, through symplastic channels called ‘plasmodesmata’ or via efflux and influx transporters. Mechanical signals are likely to be transduced via the connecting cell wall surfaces, although the precise mechanisms for this remain to be elucidated. Electrical signals could be transferred through membrane voltage through plasmodesmata or the transport of ions. All these mechanisms are dependent on either symplastic connectivity or common surface areas between cells. Topology is thus an essential part of plant biology.
As cells divide, the daughter cells may lose connections to some of the previously neighbouring cells. Growth can cause movement or deformation of cells, thus leading to changes in the contact surface area with neighbouring cells and potentially new connections. One would therefore expect that growth and development would lead to changes in communication between cells. How could plant cells then know, in this ever-changing environment, how to act and whether they should divide or differentiate (Jackson et al., Reference Jackson, Duran-Nebreda, Kierzkowski, Strauss, Xu, Landrein, Hamant, Smith, Johnston and Bassel2019)? How is this dynamic cell-to-cell communication achieved and coordinated (Bassel, Reference Bassel2018)?
One possibility is spatial restriction acting to modulate the expression of key proteins, whose flux in concentration gradients create unique microenvironments. The microenvironment of a cell is established by a combination of signals within the original cell, known as cell autonomous signalling, along with external signals known as non cell autonomous signals. These signals originate from neighbouring cells, such as phytohormones or mobile proteins, executing non cell autonomous functions in plants. The importance of regulatory mobile proteins in establishing a unique microenvironment has been shown to be central for the proper cellular organisation of the root stem cell niche, and thus cell-to-cell communication in this region. For example, non cell autonomous signalling of SHORTROOT (SHR) and its binding partner SCARECROW (SCR) guide the timing of cell division and determine cell fate of quiescent centre (QC) cells and cortex-endodermis initial (CEI) cells in the Arabidopsis root stem cell niche (Cruz-Ramírez et al., Reference Cruz-Ramírez, Díaz-Triviño, Blilou, Grieneisen, Sozzani, Zamioudis, Miskolczi, Nieuwland, Benjamins, Dhonukshe, Caballero-Pérez, Horvath, Long, Mähönen, Zhang, Xu, JAH, Benfey, Bako and Scheres2012).
Studying the communication between cells is critical both to address complex problems at the whole-organism level, and to understand systemwide cellular behaviours. To accommodate the study of cell-to-cell communication, several exciting approaches have been used. Using scanning fluorescence correlation spectroscopy, one can quantify protein characteristics within a specific cell type, such as complex stoichiometry, as well as molecular dynamics between different cells, such as protein movement (Clark et al., Reference Clark, Hinde, Winter, Fisher, Crosti, Blilou, Gratton, Benfey and Sozzani2016). SHR and SCR expression activity differs in QC and CEI cells, as they form complexes in each cell type with different stoichiometric proportions. Methods like raster image correlation spectroscopy, pair correlation function and number and brightness allow mobile transcription factor motility and expression levels to be analysed, thus quantifying how they contribute to developmental processes. Computational modelling can then be used to predict cellular behaviour, such as cell division or differentiation timing, as well as expression dynamics of key regulatory proteins in specific QC and CEI cells. The latter can be accomplished through mathematical methods such as ordinary differential equations, which can take into consideration the number and concentration of stoichiometric complexes of SHR and SCR in each cell type, the difference in SHR and SCR expression between cell types QC and CEI and relevant upstream regulatory elements (Clark et al., Reference Clark, Fisher, Berckmans, den Broeck, Nelson, Nguyen, Bustillo-Avendaño, Zebell, Moreno-Risueno, Simon, Gallagher and Sozzani2020).
Mathematical modelling also has a key role to play in developing and testing hypotheses on communication through plasmodesmata. Our understanding of how this process works is still relatively poor, but two mathematical models have recently offered new insights into how this may work. Deinum et al. (Reference Deinum, Mulder and Benitez-Alfonso2019) developed a model of diffusion from cell to cell through plasmodesmata. The authors built a detailed multilevel model based on realistic plasmodesmatal geometries and investigated the impact of different geometrical parameters and plasmodesmatal distributions. This model allows for wall permeabilities, as a function of geometrical parameters, to be inferred from experimental data. Park et al. (Reference Park, Knoblauch, Oparka and Jensen2019) modelled cell-to-cell communication via plasmodesmatal flux as a function of turgor pressure. They hypothesised a plasmodesmata closing mechanism based on mechanosensing. This model offers an explanation for rapid closing for which the alternate model of callose deposition seems too slow. Further work, both theoretical and experimental, is needed to elucidate the mechanisms underpinning the functioning of these important communication channels.
An interdisciplinary approach is required for another key aspect of this topic: ‘decoding’ the information involved in cell communication. Increasingly, detailed experiments and bioinformatics are revealing the mechanisms of production, and dynamics, of different signals (molecular and biophysical). But how such complex signals are used by the plant to convey information is an open question. What processing converts an intracellular combination of hormone concentrations into an actionable signal, for example? Over what length and timescales can signals of different physical forms be sent and received through the plant? How is the fidelity of such signals retained in the face of inevitable noise (Lestas et al., Reference Lestas, Vinnicombe and Paulsson2010)? Quantitative answers to these questions have tremendous potential for basic biology and agronomical resilience, but will require a cross-disciplinary approach using information theory, modelling and statistics to harness exciting new data.
5. Morphometric atlas for morphogenesis
Scaling up from tissue topology, another quantitative aspect of plant biology is shape, which can be complex in case of many organs, and morphogenesis, that is, shape changes in time or its maintenance despite organ growth. At the tissue scale, quantitative plant biology can take the form of growth kinematics, which is growth description of high spatiotemporal resolution that is necessary to understand plant growth dynamics, that is, how the plants develop (Silk, Reference Silk1984). The most influential proponents of quantitative studies of plant growth and development were Ralph O. Erickson (1914–2006) and his students or followers, the late Zygmunt Hejnowicz (1929–2016) and Paul B. Green (1931–1998), as well as Wendy K. Silk (Meicenheimer & Silk, Reference Meicenheimer and Silk2006). These scientists have used quantitative and interdisciplinary approaches despite the average biologists’ prejudice against math and statistics at that time (Erickson, Reference Erickson1988). In his review devoted to modelling of plant growth, Erickson (Reference Erickson1976) summed up their approach writing that ‘in any attempt at modelling it should be possible to relate the experimental data to the differential equations which represent the process being modelled’. Experimental data thus must have to be extensive in terms of precision and robust, through large sample size. In such endeavours, the required quantitative analysis is often complemented by modelling.
Kinematics of plant growth has been studied from two biophysical perspectives: fluid dynamics (Silk, Reference Silk1984) and solid body mechanics (Hejnowicz & Romberger, Reference Hejnowicz and Romberger1984). The first perspective is based on the analogy between plant growth and fluid flow: individual cells ‘flow’ through a growing plant organ that maintains an almost steady shape. The second perspective focuses on the continuous character of the symplastic growth, typical for plant tissues, which is cell growth coordinated at tissue and organ levels. The symplastic growth of plant organs, which is often also anisotropic, is the irreversible tissue deformation that observes the continuum condition of solid body mechanics. Both perspectives, fluid dynamics and solid body mechanics, put forward the tensorial nature of plant organ growth, which implies elaborate quantifications and modelling (Hejnowicz & Romberger, Reference Hejnowicz and Romberger1984; Silk, Reference Silk1984).
Empirical studies of plant growth and development as well as its complex regulation are technically challenging, because growth is often unsteady and inhomogeneous. Therefore, critical for the progress of our understanding of plant morphogenesis are techniques enabling the acquisition of high spatiotemporal resolution, high quality and well-quantified imaging data and its subsequent analysis. Recently, various techniques were developed to support live imaging, such as autotracking of moving samples and in vitro tissue cultivation under microscopes. These techniques were further combined with minimally invasive microscopy, including light sheet and two-photon excitation systems, and enabled long-term time-lapse imaging to visualise the four-dimensional dynamics during pattern formation. For example, the combination of Arabidopsis ovule cultivation and two-photon excitation microscopy revealed the 4D atlas of cell lineages during embryo patterning (Gooh et al., Reference Gooh, Ueda, Aruga, Park, Arata, Higashiyama and Kurihara2015). Furthermore, high-resolution live imaging enabled to monitor the intracellular behaviour of developing cells, such as cytoskeletal rearrangement during lateral root initiation and vacuolar shape change during zygote polarisation (Kimata et al., Reference Kimata, Kato, Higaki, Kurihara, Yamada, Segami, Morita, Maeshima, Hasezawa, Higashiyama, Tasaka and Ueda2019; Vilches Barro et al., Reference Vilches Barro, Stöckle, Thellmann, Ruiz-Duarte, Bald, Louveaux, von Born, Denninger, Goh, Fukaki, J. E. M. and Maizel2019).
The advanced imaging technologies provide a vast amount of spatiotemporal data, and this can mask important information. Therefore, image quantification is essential to extract key properties from the 4D big data. For example, cell volume and division orientation were quantified at various stages of embryogenesis, and these values were utilised for simulation modelling, which revealed the importance of geometric cell division rules and its modification by plant hormone auxin in embryo patterning (Yoshida et al., Reference Yoshida, Barbier de Reuille, Lane, Bassel, Prusinkiewicz, Smith and Weijers2014; Moukhtar et al., Reference Moukhtar, Trubuil, Belcram, Legland, Khadir, Urbain, Palauqui and Andrey2019). Thus, the combination of 4D imaging and detailed quantification can provide a powerful tool to identify fundamental rules underlying plant morphogenesis. As shown next, such morphometric analyses are now integrated with gene networks and cell identities in comprehensive 4D atlases to identify patterning rules.
6. Patterning spatial and temporal information
It has long been recognised that plants exploit algorithm-like patterns in their development (Prusinkiewicz & Hanan, Reference Prusinkiewicz and Hanan1989). We are only now beginning to uncover the complex mechanisms based on long-range transport and sensing of small metabolites, such as the ubiquitous auxin, in some sense the charge-carrier of the analogue ‘electronics’ steering plant development.
To integrate 4D imaging-based quantitative information of dynamic transcriptional or signalling networks, developmental atlases are arising as functional tools, complementary to computational modelling (Refahi et al., Reference Refahi, Zardilis, Michelin, Wightman, Leggio, Legrand, Faure, Vachez, Armezzani, Risson, Zhao, Das, Prunet, Meyerowitz, Godin, Malandain, Jönsson and Traas2021). For instance, this coupling of approaches, allowing the precise spatial registration of auxin maxima and signalling in the shoot apical meristem, uncovered a novel mechanistic framework to explain phyllotaxis (Galvan-Ampudia et al., Reference Galvan-Ampudia, Cerutti, Legrand, Brunoud, Martin-Arevalillo, Azais, Bayle, Moussu, Wenzl, Jaillais, Lohmann, Godin and Vernoux2020).
Other examples include the formation of trichomes, the division profile of the stomatal lineage, the emergence of lateral roots and the positioning of root hairs. Such patterning processes often involve local signalling modules, with positive and negative feedback loops. Interestingly, while many of these networks build on biochemical interactions, as, for example, in reaction-diffusion Turing-like patterns, there is increasing interests in more holistic models that also include mechanics. For instance, lateral root emergence from the pericycle, and through cortical tissues, involves adjacent cells and their mechanical accommodation to such an invasion (Ditengou et al., Reference Ditengou, Teale, Kochersperger, Flittner, Kneuper, van der Graaff, Nziengui, Pinosa, Li, Nitschke, Laux and Palme2008; Lucas et al., Reference Lucas, Kenobi, von Wangenheim, Voβ, Swarup, De Smet, Van Damme, Lawrence, Péret, Moscardi, Barbeau, Godin, Salt, Guyomarc’h, EHK, Maizel, Laplaze and Bennett2013; Vermeer et al., Reference Vermeer, von Wangenheim, Barberon, Lee, EHK, Maizel and Geldner2014). Similarly, the stomatal lineage patterning involves a tight control of polarity, which also involves large-scale patterns of tissue tension across the leaf (Bringmann & Bergmann, Reference Bringmann and Bergmann2017). Finally, some of the feedbacks involved in patterning can be geometrical. For instance, it has been proposed that the positioning and size of the WUS-expressing domain at the shoot apical meristem depends on cytokinins diffusing from the epidermis, and thus scales with meristem shape (Gruel et al., Reference Gruel, Landrein, Tarr, Schuster, Refahi, Sampathkumar, Hamant, Meyerowitz and Jönsson2016).
Importantly, integrated quantitative approaches also allow us to address how stereotypical patterns and organ shapes are. Defining stereotypes, in turn, paves the way to quantify the variability of morphogenesis, which is likely a crucial component per se of development (Waddington, Reference Waddington1942). In plants, mechanisms controlling specifically plasticity and robustness of development are starting to be explored (Hong et al., Reference Hong, Dumond, Zhu, Tsugawa, Li, Boudaoud, Hamant and Roeder2018), and will shed new lights on plant growth and form.
7. Shape plasticity
Whereas many animals can developmentally rest on their laurels after reaching maturity, plant morphogenesis is by definition an unfinished project. As such, postembryonic development is central to plant developmental biology. Organogenesis can be plastic and nonuniform, and organ shapes can respond to environmental cues. While in animals, symmetry, repeatability and scalability are key due to the importance of motion for animal survival, in plants instead plastic development in response to environmental conditions is central to plant fitness. It is this plasticity that enables plants, for instance, to generate new lateral roots where nutrients or water is found, yet save valuable resources by not investing in growth elsewhere.
Heterogeneity in growth, ranging from organ shape deformations to tropisms, are key to plant adaption. Organ or architecture plasticity is observed not only across plant evolution between species, but also within single individuals during development, or in response to changing environments. Such plasticity can also be predictable, meaning that quantitative approaches can measure, describe and help to understand its features. Toward this aim, robust shape descriptors have been developed. For instance, leaf shape plasticity has been assessed quantitatively in grapevine, using more than 5,500 leaves representing 270 vines from more than 11 species, to understand environmental impact (Chitwood & Sinha, Reference Chitwood and Sinha2016). Similarly, at the cellular level, careful cell shape description in 3D identified geometric cues predicting specific formative asymmetric division, shifting organism growth from 2D to 3D in a basal plant, the moss Physcomistrella patens (Tang et al., Reference Tang, Duijts, Bezanilla, Scheres, Vermeer and Willemsen2020).
Cell growth orientation and anisotropy, defining final cell shape, clearly determine asymmetric cell divisions, but also symmetric cell divisions in proliferative tissues, as cell division plane depends on geometrical and mechanical rules (von Wangenheim et al., Reference von Wangenheim, Fangerau, Schmitz, Smith, Leitte, EHK and Maizel2016; Willis et al., Reference Willis, Refahi, Wightman, Landrein, Teles, Huang, Meyerowitz and Jönsson2016; Yoshida et al., Reference Yoshida, Barbier de Reuille, Lane, Bassel, Prusinkiewicz, Smith and Weijers2014). As such, cell shape defines cell division patterns, which are key in various aspect of plant morphogenesis, such as stomata, root development or early embryogenesis in Arabidopsis. However, mutants displaying random cell division patterns, like tonneau, are still able to pattern normal fates territories (Traas et al., Reference Traas, Bellini, Nacry, Kronenberger, Bouchez and Caboche1995), and embryo patterning occurs in monocots despite highly variable cell division patterns (Zhao et al., Reference Zhao, Begcy, Dresselhaus and Sun2017). These examples suggest that supracellular mechanisms operate to control morphogenesis, as predicted by the organismal theory (Kaplan, Reference Kaplan1992).
Shape robustness entails a control of organ shape plasticity to ensure reproducible final shapes while facing external and internal perturbations. This implies the coordination of heterogeneous cellular behaviours. This coordination involves various mechanisms ranging from mechanical stress to mobile morphogens controlled by cell-to-cell communications (e.g., Hervieux et al., Reference Hervieux, Tsugawa, Fruleux, Dumond, Routier-Kierzkowska, Komatsuzaki, Boudaoud, Larkin, Smith, Li and Hamant2017). Importantly, local cellular growth variability can be as well used by the plant to buffer shape variations at the organ or tissue level, as shown, for instance, during Arabidopsis sepal development (Hong et al., Reference Hong, Dumond, Tsugawa, Sapala, Routier-Kierzkowska, Zhou, Chen, Kiss, Zhu, Hamant, Smith, Komatsuzaki, Li, Boudaoud and Roeder2016), or leaf primordia initiation at the shoot apical meristem (Uyttewaal et al., Reference Uyttewaal, Burian, Alim, Landrein, Borowska-Wykręt, Dedieu, Peaucelle, Ludynia, Traas, Boudaoud, Kwiatkowska and Hamant2012), with a primary role of the epidermis (Malivert et al., Reference Malivert, Hamant and Ingram2018; Zhou et al., Reference Zhou, Du, Feng, Hu, Lü, Long and Jiao2020). Finally, shape definition during plant development is a multiscale and integrated process, which is plastic by nature. Indeed, the gradual production of new cells during iterative development imposes continuously new geometric, topological and mechanical constraints, which, in turn, canalise individual cells growth and shapes.
Plastic cellular patterns can rely on local fluctuations in gene expression, establishing thresholds determining cell fate trajectories. Indeed, the coupling of variations in gene expression and in cell fate has been observed for individual genes, such as ATML1 in leaf and sepal epidermal cells. In epidermal cells arrested in G2 phase of the cell cycle, when ATML1 level exceeds a certain threshold, endoreduplication starts, allowing differential cell growth (Meyer et al., Reference Meyer, Teles, Formosa-Jordan, Refahi, San-Bento, Ingram, Jönsson, JCW and Roeder2017). Recent years have seen the rise of single cell transcriptome techniques. To obtain separate cells for single cell analysis, cell walls need to be removed (i.e., generation of protoplasts) quickly from plant tissues, whose effect on gene expression remains to be comprehensively assayed. Protoplasts are easy to extract from the root meristem of Arabidopsis, and therefore several papers describing the single cell transcriptome of that tissue have recently been published. These pioneering studies confirmed the identities of different cell types in the root meristem, but most interestingly, with the help of bioinformatics, they also revealed cell differentiation trajectories (Efroni et al., Reference Efroni, Mello, Nawy, Ip, Rahni, DelRose, Powers, Satija and Birnbaum2016). Differentiation of the stomatal lineage have also been addressed recently (Lee et al., Reference Lee, Wengier and Bergmann2019), showing that more and more plant tissues can be amenable to single cell transcriptomic approaches. Interestingly, differentiation trajectories can be mapped on pseudotime curves, by coupling single cell techniques to careful tissue staging, as recently shown for male germline precursors in maize (Nelms & Walbot, Reference Nelms and Walbot2019). Through spatial transcriptomics (Duncan et al., Reference Duncan, Olsson, Hartley, Dean and Rosa2016; Giacomello et al., Reference Giacomello, Salmén, Terebieniec, Vickovic, Navarro, Alexeyenko, Reimegård, LS, Mannapperuma, Bulone, Ståhl, Sundström, Street and Lundeberg2017), we will obtain information on transcriptional programs during morphogenesis at a temporal and spatial resolution we thought impossible just a while ago. Such high-resolution transcriptome technology would allow us to understand how individual cells dynamically determine their cell fates in response to diverse inputs, such as nutrient amount mechanical stress and the dynamics of neighbouring cells, to actualise plastic plant development.
8. Mechanics behind growth and motion
A quantitative approach is also indispensable in the field of plant biomechanics. Cell growth is a classical and enlightening example of how quantitative descriptions and mechanical reasoning can shed light on a key biological process. It has been quantitatively studied since the 19th century, with experiments on osmosis by Wilhelm Pfeffer (1845–1920). Many control factors are involved (temperature, hormones, osmole concentration, oxygen etc.). Although turgor pressure is understood to be the force behind cell growth, its action appears contradictory: On the one hand, turgor pushes on the cell wall, promoting growth; on the other hand, it raises the water potential, inhibiting water entry into the cell. By modelling the cell wall as a viscoplastic material, Lockhart (Reference Lockhart1965) was able to combine these two opposite tendencies into a single equation, from which turgor is absent. In Lockhart’s equation, the relative growth rate becomes nonzero when the difference in osmotic pressure Δπ rises above a yield pressure PY :
 $$\begin{align*}\frac{1}{V}\frac{dV}{dt}=\frac{\varphi L}{\varphi +L}\left(\varDelta \pi -{P}_Y\right),\end{align*}$$
$$\begin{align*}\frac{1}{V}\frac{dV}{dt}=\frac{\varphi L}{\varphi +L}\left(\varDelta \pi -{P}_Y\right),\end{align*}$$
where φ is the extensibility of the cell wall and L is its relative hydraulic conductance. As argued by Green (Reference Green and King1996), these two physical parameters enter the equation in a nonlinear way, which could not have been deduced from co-variation studies. Biophysical modelling, supported by quantitative data, was therefore a necessary step. Ortega (Reference Ortega1985) added an elastic component to the equation, accounting for reversible changes in cell volume, which can be significant, for instance, in diurnal variations in tree stem diameter.
Another biophysical approach to cell growth is based on the principle of minimum energy (Hejnowicz, Reference Hejnowicz2011). However, neither this nor the Lockhart–Ortega equation directly accounts for the effects of temperature, hormones or other factors. Actually, these factors act on the extensibility of the cell wall. Investigating the corresponding relationships requires more detailed models of the chemo-rheological processes occurring in the cell wall (e.g., Ali & Traas, Reference Ali and Traas2016), assessed against quantitative data (Proseus et al., Reference Proseus, Zhu and Boyer2000).
Upscaling mechanical properties from a single cell to an organ(ism) is nontrivial. The whole structure has to be closely considered. Because cell walls are stiffer than protoplasts and are under tensile stress generated by turgid protoplasts, the apoplast is the load bearing and load transmitting part of the organ. The green organ can thus be regarded as made of a pressurised cellular solid, where the contribution of various organ tissues into its mechanical properties/stiffness depends on tissue distribution and structure. One of the first plant morphologists who studied biophysical aspects of plant structure and development was Hofmeister, who also noted that cell walls are under tension. His ‘ability to combine exceptional observational detail with an emphasis on experimental methodology’ (Kaplan & Cooke, Reference Kaplan and Cooke1996) was an early manifestation of quantitative plant biology. The structure of plant organs was studied from a mechanical perspective and looked at as a supportive system also by other pioneers of plant anatomy, such as Sachs and Simon Schwendener (1829–1919), although later these aspects of plant structure were generally neglected for decades (Romberger & Hejnowicz, Reference Romberger and Hejnowicz1993).
Mechanics also explains plant movements, such as the operation of contractile roots, the catapult-like action of fern sporangia (Noblin et al., Reference Noblin, Rojas, Westbrook, Llorens, Argentina and Dumais2012) or the closing of the Venus flytrap (Forterre et al., Reference Forterre, Skotheim, Dumais and Mahadevan2005). Some of these movements, including nastic ones, have important morphogenetic implications, for instance, to explain how leaves flatten as they grow (Derr et al., Reference Derr, Bastien, Couturier and Douady2018). All these phenomena in living organisms have to be studied observing empirical rigors of physics, and thus contribute to quantitative plant biology.
As for biochemistry, mechanics is not only an output of the gene network, but also an input. Growth and geometry changes cause mechanical tension and stress during morphogenesis, and this, in turn, feeds back to tissue patterning (Heisler et al., Reference Heisler, Hamant, Krupinski, Uyttewaal, Ohno, Jönsson, Traas and Meyerowitz2010; Nakayama et al., Reference Nakayama, Smith, Mandel, Robinson, Kimura, Boudaoud and Kuhlemeier2012). A growing number of studies report that both short- and long-distance signalling between plant cells is accompanied by tension/stress sensing mechanisms enabling correct morphogenetic processes (Trinh et al., Reference Trinh, Alonso-Serra, Asaoka, Colin, Cortes, Malivert, Takatani, Zhao, Traas, Trehin and Hamant2021). Because forces are invisible in essence, and as these regulatory mechanisms take place in three dimensions and at different timescales, computational modelling has become a vital tool to understand patterning processes ranging from the tissue, organ to whole plant scale.
Because plants develop ‘in-place’, they might be more geometry-aware than many other organisms. This is apparent even at the level of individual plant cells, which like plants themselves largely develop ‘in-place’. The unique properties of the semirigid plant cell wall, means that plant cells need to maintain and hence be able to sense their geometry often at a size scale of tens of microns. It is remarkable that Paul Green in a landmark paper (Green, Reference Green1962; Green & King, Reference Green and King1966) basically predicted the existence of cellulose microfibril reorientation by mechanical stress on the basis of his observations of the plant cell wall, even before microtubules were discovered (Ledbetter & Porter, Reference Ledbetter and Porter1963). Microtubules were later on found to guide the synthesis and orientation of cellulose microfibrils, which determine cell growth orientation, through live imaging (Paredez et al., Reference Paredez, Somerville and Ehrhardt2006).
Not only the role of mechanical stress in development is well established (Green, Reference Green1999; Hamant et al., Reference Hamant, Heisler, Jonsson, Krupinski, Uyttewaal, Bokov, Corson, Sahlin, Boudaoud, Meyerowitz, Couder and Traas2008; Lynch & Lintilhac, Reference Lynch and Lintilhac1997), but it is also widely accepted that mechanics is at the basis of the supportive functional system of plant organs (Romberger et al., 1993). Noteworthy, plant organs are often prestressed constructions. Namely, an important role in this system is played by tissue stresses (Hejnowicz & Sievers, 1996; Kutschera, Reference Kutschera1989), that is, the tensegrity at the organ level. The structural tissue stresses, an indirect result of the turgor pressure, exist in plant organs that are composed of turgid tissues that differ in cell size as well as thickness and mechanical properties of the cell walls.
Due to their size and perennity, trees are of special interest for plant biomechanics. As slender structures, they lend themselves well to engineering approaches like beam theory (Niklas, Reference Niklas1992). Prompted by the pioneering works of Schwendener (Schwendener, Reference Schwendener1874), biomechanicians started to investigate the quantitative constraints put on tree growth by their own weight (Greenhill, Reference Greenhill1881) or external loads like wind (Metzger, Reference Metzger1893). Trees greatly differ from engineered structures as they change their mass and size by large factors during their lifetime and experience a wide range of mechanical loads. In addition, they explore their environment and adapt to it (see, e.g., Alonso-Serra et al., Reference Alonso-Serra, Shi, Peaucelle, Rastas, Bourdon, Immanen, Takahashi, Koivula, Eswaran, Muranen, Help, Smolander, Su, Safronov, Gerber, Salojärvi, Hagqvist, Mähönen, Helariutta and Nieminen2020; Eloy et al., Reference Eloy, Fournier, Lacointe and Moulia2017). Defining integrative biomechanical traits makes it possible to quantify how well a tree is adapted to its environment and to infer which ecological strategy it follows (Fournier et al., Reference Fournier, Dlouhá, Jaouen and Almeras2013). This also illustrates how physics, engineering and plant ecology can work hand in hand.
9. Bioenergetics
As is the case for all living organisms, plant growth, development and physiology involve continuous dynamic energy conversion, abiding the laws of thermodynamics. Photosynthesis is the most important energy-harvesting process on Earth. By driving the flow of electrons extracted from water molecules through several highly organised photosynthetic complexes, sun energy is momentarily converted into two chemical energy currencies of the cells, ATP and NADPH. At the same time, it drives the assimilation of carbon, nitrogen and sulphur. The chemical energy generated from photosystems then fuels many anabolic metabolisms that promote the mechanisms behind plant growth described above.
As shown for other themes, cell bioenergetics is examined by quantitative approaches, such as absorption spectrometry, chlorophyll fluorescence, irradiance measurement, optical microscopy, gas analysis, electrodes or isotopic labelling. These methods allow the estimation of photosynthetic parameters, carbon assimilation rate, electron flows, metabolic fluxes, stomatal conductance or respiration rate (Fernandez-Jaramillo et al., Reference Fernandez-Jaramillo, Duarte-Galvan, Contreras-Medina, Torres-Pacheco, de J Romero-Troncoso, Guevara-Gonzalez and Millan-Almaraz2012).
The acquired data are integrated into metabolic networks and flux-balance analysis models to describe energy metabolism in photosynthetic organisms (Cheung et al., Reference Cheung, Poolman, Fell, Ratcliffe and Sweetlove2014; Rügen et al., Reference Rügen, Bockmayr and Steuer2015). One example is the Farquhar, von Caemmerer and Berry model for predicting net CO2 uptake (A) in C3 plants by linking A to the carboxylation rate of ribulose 1,5-bisphosphate (RuBP), oxygenation rate of RuBP and mitochondrial respiration in the light (Farquhar et al., Reference Farquhar, von Caemmerer and Berry1980). This model, which has been cited more than 7,500 times since its publication 40years ago, has been applied to a wide range of studies, from investigating C3 bioenergetics to predicting photosynthetic fluxes of ecosystems on a global scale.
In fact, the importance of modelling and quantitative studies of photosynthesis goes beyond the cellular, tissue or organismal levels. Field and global measurements of solar-induced vegetation fluorescence by recent remote sensing technologies using airborne sensors or satellite systems allow scientists to monitor the temporal and seasonal changes of vegetation and the fluxes of carbon, water and energy on a regional or a global scale, and to study the interaction between vegetation, primary productivity, environmental stresses and climate changes (Wu et al., Reference Wu, Albert, Lopes, Restrepo-Coupe, Hayek, Wiedemann, Guan, Stark, Christoffersen, Prohaska, Tavares, Marostica, Kobayashi, Ferreira, Campos, da Silva, Brando, Dye, Huxman and Saleska2016). Coupled with data gathered in the field, this opens many opportunities to bridge scales and revisit the essential role of plant productivity for our civilisation and ecosystem.
10. Integrating fluctuating and diverse abiotic environmental factors
Within a single day, plants may experience all kinds of challenges, including fluctuating light intensity and/or quality, temperature changes, wind, rain or snow. Each of such fluctuations in the surrounding environment factors may represent a threat to plants, unless dealt with properly.
The information contained in abiotic signals needs to be processed and integrated in order to arrive to developmental decisions. To elucidate this process, a quantitative systems approach is required, because the intricate interplay between the several parts of the signalling networks often escapes intuitive reasoning (Boer et al., Reference Boer, Santos Teixeira and Ten Tusscher2020). There are numerous quantitative challenges related to: (a) learning the structure of environmental signal integration networks, (b) understanding the dynamic properties of these networks and (c) designing genetic changes to the network that would cause the plant to respond to environmental parameters in a specific way.
One of the most thoroughly studied abiotic response systems in plants is light and temperature signal integration. Plants sense the quantity and quality of light, for which a variety of photosensors are used ranging from the UV to the red part of the spectrum (Möglich et al., Reference Möglich, Yang, Ayers and Moffat2010; Paik & Huq, Reference Paik and Huq2019). For instance, measuring the duration of the day by sensing dawn and dusk is important for the entrainment of the circadian clock (Seaton et al., Reference Seaton, Toledo-Ortiz, Ganpudi, Kubota, Imaizumi and Halliday2018; Wenden et al., Reference Wenden, Kozma-Bognár, Edwards, Hall, Locke and Millar2011). But also other responses, such as germination, de-etiolation, regulation of flowering and responses to canopy shade, are regulated by light sensing networks (Chen et al., Reference Chen, Chory and Fankhauser2004; Galvão & Fankhauser, Reference Galvão and Fankhauser2015; Kami et al., Reference Kami, Lorrain, Hornitschek and Fankhauser2010; Paik & Huq, Reference Paik and Huq2019). Plants respond differently to different spectral distributions of the incident light, which is achieved not by the light sensors alone, but by an interplay between the sensors and their interacting factors, that is, by the light sensing network (Galvão & Fankhauser, Reference Galvão and Fankhauser2015; Klose et al., Reference Klose, Venezia, Hussong, Kircher, Schäfer and Fleck2015; Paik & Huq, Reference Paik and Huq2019; Rausenberger et al., Reference Rausenberger, Hussong, Kircher, Kirchenbauer, Timmer, Nagy, Schäfer and Fleck2010). Due to this, the plant’s response to environmental light is a system property and cannot be understood by analysing the properties of the photoreceptors alone, besides simple cases.
A prominent example of this is phytochrome A (phyA) in Arabidopsis thaliana. The spectral response of phyA depends on the light intensity; for low intensities, phyA responds to red (660nm) light, whereas for high intensities, it responds to far-red light (720nm; Franklin et al., Reference Franklin, Allen and Whitelam2007). This intensity-dependent change of the spectral response without a change of the physical properties of phyA can only be understood, if one analyses the signalling network (Possart et al., Reference Possart, Fleck and Hiltbrunner2014; Rausenberger et al., Reference Rausenberger, Tscheuschler, Nordmeier, Wüst, Timmer, Schäfer, Fleck and Hiltbrunner2011; Schäfer, Reference Schäfer1975).
The understanding of the light signalling networks and their different responses to the light spectrum is very challenging and requires joining the forces of experiments and mathematical modelling. This is even more true if one aims to unravel the integration and processing of light and temperature signals in plants. Plants are sensitive to temperature changes, and many aspects of plant growth and development respond to temperature, for example, hypocotyl elongation or flowering (Wigge, Reference Wigge2013). However, both processes are also sensitive to light, and therefore the temperature and the light signals need to be taken into account together.
In principle, temperature and light could be sensed by two separate networks, and the integration could be achieved through further downstream elements. Nevertheless, it has become clear that in Arabidopsis thaliana, temperature and light sensing and signal integration are done by the same network (Franklin, Reference Franklin2009; Jung et al., Reference Jung, Domijan, Klose, Biswas, Ezer, Gao, Khattak, Box, Charoensawan, Cortijo, Kumar, Grant, JCW, Schäfer, Jaeger and Wigge2016; Legris et al., Reference Legris, Klose, Burgie, Rojas, Neme, Hiltbrunner, Wigge, Schäfer, Vierstra and Casal2016; Seaton et al., Reference Seaton, Toledo-Ortiz, Ganpudi, Kubota, Imaizumi and Halliday2018). Again, without mathematical analysis of the data and knowledge integration into dynamic mathematical models, progress in our conceptual understanding of how plants analyse ambient temperature and light conditions in a coordinated manner would be very difficult. The mathematical models allow for studying the effect of the different parts of the network, their interplay and role in the signal processing.
While dynamical systems approaches have been useful for understanding the interplay between temperature and light signal integration, many of the environmental signal integration networks in plants have been primarily interrogated with ‘Big Data’-based approaches that also provide insight into the structure and behaviours of signal integration networks. One research aim has been to infer the structure of large biological networks used in signal integration using transcriptomics data and either experimental or computational predictions of transcription factor binding (Brooks et al., Reference Brooks, Cirrone, Pasquino, Alvarez, Swift, Mittal, Juang, Varala, Gutiérrez, Krouk, Shasha and Coruzzi2019; Ezer et al., Reference Ezer, Shepherd, Brestovitsky, Dickinson, Cortijo, Charoensawan, Box, Biswas, Jaeger and Wigge2017; Gamboa-Tuz et al., Reference Gamboa-Tuz, Pereira-Santana, Zamora-Briseño, Castano, Espadas-Gil, Ayala-Sumuano, Keb-Llanes, Sanchez-Teyer and Rodríguez-Zapata2018; Greenham et al., Reference Greenham, Guadagno, Gehan, Mockler, Weinig, Ewers and McClung2017; Walker et al., Reference Walker, Boddington, Jenkins, Wang, Grønlund, Hulsmans, Kumar, Patel, Moore, Carter, Samavedam, Bonomo, Hersh, Coruzzi, Burroughs and Gifford2017). The resulting networks can be too complicated for us to easily comprehend how plants integrate environmental signals, and so there is an ongoing effort to identify submodules that perform specific environmental sensing and integration roles (Polanski et al., Reference Polanski, Rhodes, Hill, Zhang, Jenkins, Kiddle, Jironkin, Beynon, Buchanan-Wollaston, Ott and Denby2014).
When environmental signal integration networks are too complicated to model with dynamical systems, an alternative approach to understanding their behaviours is to directly predict phenotypic traits of interest from environmental input variables. Here, the aim is not to learn conceptually or model the precise network that plants use intrinsically to integrate signals, but rather to find a model that makes accurate predictions. These kinds of phenotypic forecasts have been especially useful in agricultural applications, where the primary aim of the researcher is to predict agriculturally relevant traits, given a certain set of abiotic environmental parameters. Innovations in this area come from deep learning techniques that integrate networks of sensor and satellite data (Aruul Mozhi Varman et al., Reference Aruul Mozhi Varman, Baskaran, Aravindh and Prabhu2017; Wang et al., Reference Wang, Tran, Desai, Lobell and Ermon2018; Wolanin et al., Reference Wolanin, Mateo-García, Camps-Valls, Gómez-Chova, Meroni, Duveiller, Liangzhi and Guanter2020) and statistical advances that integrate more time-series environmental data into these models, taking into account that a plants’ response to an environmental stimulus is time-of-day and season-dependent (Brestovitsky & Ezer, Reference Brestovitsky and Ezer2019; Kocian et al., Reference Kocian, Carmassi, Cela, Incrocci, Milazzo and Chessa2020; Newlands et al., Reference Newlands, Zamar, Kouadio, Zhang, Chipanshi, Potgieter, Toure and Hill2014).
There are a number of open challenges related to the study of how abiotic factors are integrated by plants, which merit quantitative investigation. For instance, there is a limit to how many combinations of environmental parameters can be perturbed in an experiment, so there is a need for new tools to help scientists iteratively design experiments (Ezer & Keir, Reference Ezer and Keir2019). These would help them choose growth conditions that would help them learn as much as possible about how plants integrate environmental signals, while adhering to time and budget constraints. A second major challenge is to efficiently reverse-engineer specific responses to abiotic stimuli, so we can efficiently engineer crops that are sustainable in the light of climate change.
11. Ecosystem complexity: interactions with pathogens and microbiome
Plant phenotype goes beyond their individual body: to understand their biology, one needs to account for the myriad of living organisms with which they interact. This represents a large field of research, notably because of the threat posed by pathogens on plant development. Needless to say, quantitative thinking is also at the heart of such biotic interactions.
The research field of plant immunity is deeply rooted in crop sciences with phytopathology and breeding for disease resistance traits. The gene-for-gene hypothesis by Flor established the field under the concept of qualitative disease resistance: the presence or absence of a matching pair of host resistance gene and pathogenic avirulent factor determines the qualitative traits, resistance or susceptibility (Flor, Reference Flor1942). This simple assumption has greatly contributed to the development of the plant immunity research field, resulting in a growing list of important immune receptors both in model and crop plant species and the deployment of such robust resistance traits in the crop fields.
Despite its initial contributions, the simple qualitative concept has been challenged with quantitative counterarguments. A resistance trait based on the one-on-one relationship would be predicted to break easily if fast-evolving pathogens come up with a strategy to overcome the resistance. However, most natural populations withstand stochastic pathogenic loads, reflecting their complex immune systems that would consist of resistance genes serving to recognise more than one pathogenic avirulent factor.
As the research field matures, evidence for the quantitative nature of disease resistance and plant immunity is accumulating. An obvious numbers game comes from a genomic view of plant immunity. Numerous genome sequencing datasets point out that the list of immune receptors on our hands, mostly discovered under the gene-for-gene concept, is only a tiny fraction of what a given plant species could carry. For example, out of approximately 160 genes belonging to NLR-type (formerly known as NBS-LRR) resistance genes found in Arabidopsis thaliana, only less than two dozen have been functionally assigned to resistance. The fraction of knowns in other species is far less than those found in the most well-studied model species (Kourelis et al., Reference Kourelis, Sakai, Adachi and Kamoun2020). What are the functions of the rest of these unannotated immune genes present in the plant genome? Are they ‘reservoir’ immune genes that would ensure the plant populations to be ready for stochastic pathogenic pressures? In this sense, does the immune complexity revealed from the functional genomics of plant immunity research reflect the phenotypic plasticity wired in their immune network? How much is the contribution of cryptic genetic variation accumulated in the system to overall plant fitness? Can the complexity be utilised to develop durable resistance in the field?
The current research focus is shifting towards a systematic and quantitative approach to understand the robustness of the plant immune system. Recent molecular findings point to cooperative modules of immune receptors and a diffuse relationship between receptor and ligands in the plant immune system (Cesari et al., Reference Cesari, Kanzaki, Fujiwara, Bernoux, Chalvon, Kawano, Shimamoto, Dodds, Terauchi and Kroj2014; Karasov et al., Reference Karasov, Kniskern, Gao, DeYoung, Ding, Dubiella, Lastra, Nallu, Roux and Innes2014; Wang et al., Reference Wang, Roux, Feng, Guy, Li, Li, Zhang, Lautier, Jardinaud and Chabannes2015; Williams et al., Reference Williams, Sohn, Wan, Bernoux, Sarris, Segonzac, Ve, Ma, Saucet and Ericsson2014). These findings suggest that we should adopt a network view on plant immunity to better understand innate complexity in the plant immune system (Adachi et al., Reference Adachi, Derevnina and Kamoun2019).
Copious omics-driven large-scale research has revealed that despite an obvious expansion in the peripheral nodes of the network which accommodates diverse recognition modes, plants have evolved a core machinery that activates rather conserved immune responses (Hillmer et al., Reference Hillmer, Tsuda, Rallapalli, Asai, Truman, Papke, Sakakibara, Jones, Myers and Katagiri2017; Mukhtar et al., Reference Mukhtar, Carvunis, Dreze, Epple, Steinbrenner, Moore, Tasan, Galli, Hao and Nishimura2011; Tsuda & Katagiri, Reference Tsuda and Katagiri2010; Wessling et al., Reference Wessling, Epple, Altmann, He, Yang, Henz, McDonald, Wiley, Bader and Glasser2014). Characteristic plant immune responses often culminate in immunological cell death events that would dispose of the infected local areas, while the activation of immune responses in return sacrifice growth in general. The two topics, the canalisation of diverse recognition events to canonical immune responses and the trade-off between immunity and growth, are the major areas that could benefit from the quantitative approaches already implemented in other areas of biology, particularly in plant development and signalling.
If one can visualise characteristic immune responses, for example by defining and utilising immune responsive elements, detailed image analysis in a spatiotemporal manner would certainly uncover the hidden rules of immune signalling and cell death execution. Such details in immune responses in plants are expected to provide a way to modulate the responses to quantitatively pinpoint how much either acute or residual immune responses affect plant performance during the course of development. As local cell death could disturb the developmental system drastically, rerouting of developmental signalling in a quantitative matter would strengthen our understanding of phenotypic plasticity of plants under external stress.
Another important research area of plant immunity lies in its connection to field sciences. Microbial ecology is an inseparable research area from that of plant immunity. With recent advances in microbiome research platforms, the relative importance of the host-microbe interaction starts to be unveiled in its contribution to plant performance (Chen et al., Reference Chen, Nomura, Wang, Sohrabi, Xu, Yao, Paasch, Ma, Kremer and Chen g2020; Hacquard et al., Reference Hacquard, Spaepen, Garrido-Oter and Schulze-Lefert2017). Such efforts widen the past focus on plant–pathogen interactions to embrace different kinds of plant–microbe interactions including symbiosis. Definitely, the expansion of the plant immunity field to embrace the multitude of interactions between plants and microbes as well as between plant immune components will require sophisticated quantitative tools for analysis to build up a comprehensive view of the plant immune system.
12. Back to society: plants and humans
Engineering technology and approaches have always been a weighty facilitator of research and innovation in life sciences, including plant sciences. In turn, many areas of plant sciences are identified as the next frontiers of bioengineering, particularly in light of working towards mitigating climate disasters and realising a sustainable future (Wintle et al., Reference Wintle, Boehm, Rhodes, Molloy, Millett, Adam, Breitling, Carlson, Casagrande, Dando, Doubleday, Drexler, Edwards, Ellis, Evans, Hammond, Haseloff, Kahl, Kuiken and Sutherland2017). The interface between biology and engineering can be classified into three types: engineering for biology, with biology and inspired by biology.
First, engineering can help biology with quantitative tools. It could be a totally new method, or more often a new improved version of an available technology. For example, how fast and cost-effectively we can sequence DNA has kept transforming the size of the datasets we can afford to produce, and hence the depth and breadth of information we can extract from them. Plant genomes tend towards the large end of the spectrum compared to ones from the other kingdoms (e.g., Nystedt et al., Reference Nystedt, Street, Wetterbom, Zuccolo, Lin, Scofield, Vezzi, Delhomme, Giacomello, Alexeyenko, Vicedomini, Sahlin, Sherwood, Elfstrand, Gramzow, Holmberg, Hällman, Keech, Klasson and Jansson2013). The phenotypic variation is high even within a species, which has prompted genomic sequencing of a collection of accessions (e.g., Alonso-Blanco et al., Reference Alonso-Blanco, Andrade, Becker, Bemm, Bergelson, Borgwardt, Cao, Chae, Dezwaan, Ding, Ecker, Exposito-Alonso, Farlow, Fitz, Gan, Grimm, Hancock, Henz, Holm and Zhou2016). New bioimaging technology has revealed more and more about plant structures, ever since Robert Hooke saw ‘cells’ in oak cork. In the past 20years, plant science has been leading the emerging field of morphodynamics. Community efforts to capture growth and development in 4D have kept improving the capacity in data acquisition, processing, extraction and analysis (e.g., Barbier de Reuille et al., Reference Barbier de Reuille, Routier-Kierzkowska, Kierzkowski, Bassel, Schüpbach, Tauriello, Bajpai, Strauss, Weber, Kiss, Burian, Hofhuis, Sapala, Lipowczan, Heimlicher, Robinson, Bayer, Basler, Koumoutsakos and Smith2015; Wolny et al., Reference Wolny, Cerrone, Vijayan, Tofanelli, Barro, Louveaux, Wenzl, Strauss, Wilson-Sánchez, Lymbouridou, Steigleder, Pape, Bailoni, Duran-Nebreda, Bassel, Lohmann, Tsiantis, Hamprecht, Schneitz and Maizel2020). Phenotypic platforms have been developed, driving innovative solutions to capture morphodynamics in situ, generating a wealth of quantitative data. Even root development that occurs underground and is usually optically inaccessible has been visualised, with such technologies as transparent soil and CT scan protocols (Bao et al., Reference Bao, Aggarwal, Robbins, Sturrock, Thompson, Tan, Tham, Duan, Rodriguez, Vernoux, Mooney, Bennett and Dinneny2014; Rellán-Álvarez et al., Reference Rellán-Álvarez, Lobet, Lindner, Pradier, Sebastian, Yee, Geng, Trontin, LaRue, Schrager-Lavelle, Haney, Nieu, Maloof, Vogel and Dinneny2015). Satellite data, together with machine learning techniques, are increasingly used to monitor ecosystem evolution at continent scale (Newman & Furbank, Reference Newman and Furbank2021).
Bioengineering technology is not limited to hardware or data acquisition. Breakthroughs in in silico platforms for data processing, analysis and sharing underscore rapid progress in quantitative experimentation, lately including AI and machine learning. These are only few examples; engineering is constantly rewriting what is possible to find in the chemistry and physics of living organisms, and it has been driving biology towards quantitative data collection.
Second, engineering with biology involves a much deeper connection. Notably, through domestication, this corresponds to a form of co-evolution between plants and humans. Since the dawn of civilisation, people have indeed engineered food, drugs and other health remedies, materials and energy sources with living organisms, in the forms of agriculture, horticulture, forestry, fermentation and so on. Genetic engineering deepened the level of orientation towards engineering in that it is design-led.
In the past 20years, the new field of ‘synthetic biology’ has been expanding rapidly in microbial classes first but later in multicellular systems including plants. Synthetic biology is sometimes called ‘engineering biology’, for its commitment to the engineering principles in design-led problem solving, such as simplicity, universality, efficiency, consistency and predictability. Because of its streamlined and user-considered designs, synthetic biology enables complex genetic engineering; small and well-characterised standard parts of promoters, terminators, functional coding sequences and logic gates all promote efficient cloning of many-part, multigene constructs. Simpler and faster genetic construction calls for large-scale and efficient, likely automated, characterisation platforms. Tools are constantly expanded (while each of them simplified); new technology, should it be CRISPR or directed evolution, is adopted quickly and transformed into usable and sharable parts. Predictability and reproducibility are overarching goals in synthetic biology, and thus predictive modelling plays a crucial role. Synthetic biology builds upon systems biology, or systems biology prompted synthetic biology; synthetic biology could be viewed as an experimental platform for systems biology, and systems biology as a theoretical platform for synthetic biology.
The two most major industrial applications of plant sciences—metabolic engineering and molecular breeding—entail complex genetic engineering to tinker with biosynthetic or developmental pathways that are controlled by multiple genes that often cross-regulate among themselves. As such, synthetic biology technology has already been employed to the pioneering crop engineering projects such as the GM omega-3 project, C4 Rice Project and RIPE (Realizing Increased Photosynthesis Enhancement) Project (Napier et al., Reference Napier, Haslam, Beaudoin and Cahoon2014; Parry et al., Reference Parry, Andralojc, Scales, Salvucci, Carmo-Silva, Alonso and Whitney2013; Wang et al., Reference Wang, Vlad and Langdale2016). Step changes in biotechnology and agriculture are anticipated, as more countries approve gene-edited organisms in their policy and regulation.
Development of universal standards is a key objective of the synthetic biology field, and plant synthetic biologists, in particular, have been leading the efforts to enhance community sharing. The unified design overhangs for standard parts to make DNA assembly compatible named PhytoBricks, and the simple and open material transfer agreement called ‘Open MTA’ were both proposed and developed by plant scientists first and then adopted by wider circles of synthetic biologists (e.g., the student genetic engineering competition iGEM and the largest DNA repository Addgene; Patron et al., Reference Patron, Orzaez, Marillonnet, Warzecha, Matthewman, Youles, Raitskin, Leveau, Farré, Rogers, Smith, Hibberd, AAR, Locke, Schornack, Ajioka, Baulcombe, Zipfel, Kamoun and Haseloff2015; Kahl et al., Reference Kahl, Molloy, Patron, Matthewman, Haseloff, Grewal, Johnson and Endy2018).
Finally, biology can also inspire engineering. Among all the living organisms great and small that together exhibit a fantastic variety of functional and structural features, plants have been the primary source of inspiration for engineering. This may be due to their sessile nature. The plant mode of living is not as dependent on rapid and constant movement, and thus their structures are closer to engineered constructions. Many functional structures are made of no-longer living components and feasible to emulate via engineering. For example, the functional appendages that aid seed flight in winged or haired diaspores (such as of maples and dandelions) are made solely of the cell wall remnants of no-longer living cells. The intricate surface textures that confer differential functionalities, such as structural colour and hydrophobicity, are patterned with wax. In fact, the representative examples of biomimetic innovations, such as the burr-mimicking VELCRO and self-cleaning materials that resemble the lotus leaf surface, were inspired by plant structures (Koch et al., Reference Koch, Bhushan and Barthlott2009). This trend continues in robotics, and plant tropisms motivated a large interdisciplinary consortium effort to create self-growing robots, which also embody sensors and modify their growth in response to physical cues from the environment (Fiorello et al., Reference Fiorello, Del Dottore, Tramacere and Mazzolai2020).
Engineered mimics often highlight what we do not as yet know about the living structures that they emulate. Therefore, biomimetic engineering, as well as engineering with biology, can be used as learning via building. This is why in bioengineering, Richard Feynman’s famous words—‘What I cannot create, I do not understand.’—are so often quoted. Reconstruction and recapitulation are a highly effective process of redefining questions. The engineering investigative framework dictates the circular iteration of design–build–test stages. They are reminiscent of the modern scientific framework in which a question is defined as hypothesis that is tested via experimentation. The engineering cycle is indeed a variation of the general framework in research and project development of any kinds. However, there is a benefit to explicitly casting this concept onto applied and basic research in natural sciences, because of the emphasis on its interactive nature. Research typically does not end with the ‘test’ stage with a clear and definitive conclusion. Any project or scientific endeavour is not a linear process; rather, it is iterations of inquisition. There is no failure of a project; any sound scientific experimentation only brings us closer to new insights and findings about plants and how to apply such gained knowledge better.
13. Conclusion: a proposition
At the end, this overview of quantitative plant biology in the 21st century is an invitation to explore new paradigm changes in fields including high-resolution cell dynamics and computational reconstruction of gene networks, or plant and environment interactions through multiscale metamodels. This synthesis must also include yet another important player: the citizen. With the rise of participative science, notably through education and digital technologies, it appears that data acquisition, analysis and interpretation is more and more open to plant amateurs. This is facilitated by fair science and open access to data. By sharing lab and citizen data, new questions arise. How to deal with heterogeneous measurements, from people with different expertise? How to compare results obtained in controlled conditions but in small numbers and results obtained in the field in large numbers? Even more interestingly, situated knowledge, for instance, relating to agronomical practices, are now fuelling basic research in the labs. This notably includes the synergistic properties of varietal mixtures, an emerging field of study in genetics and ecophysiology. It seems therefore that quantitative plant biology is taking a turn from inter- to transdisciplinary research.
Building on this fertile field and these exciting developments, we proposed the founding of a new journal, Quantitative Plant Biology, that also includes a new transdisciplinary field of research, involving scientists and nonscientists. This will likely require new ways to coordinate between disciplines and community resources, for instance, when considering stock centres [special code repositories, coordination on file specifications (like SBML did for other areas of biology), and stock centres of synthetic biology parts, databases of images and shared software for image storage and analysis] or new standards in article format and reviewing (e.g., running scripts and models in published notebooks with the article, access to code and data). This will also require the formation of the next generation of inter- and transdisciplinarians. Several summer schools and workshops are supporting these efforts. We hope this open access journal, affiliated to both a public research institution like the John Innes Centre and a nonprofit publisher (Cambridge University Press), is the ideal forum for these stimulating endeavours.
Acknowledgement
We are thankful to our colleagues and the larger community for fruitful discussions on this topic.
Financial support
This work received no specific grant from any funding agency, commercial or not-for-profit sectors.
Conflict of interest
The authors declare that there is no conflict of interest.
Authorship contributions
All authors contributed to specific sections of the article, and all reviewed the entire manuscript.
Data availability statement
Data availability is not applicable to this article as no new data were created or analysed in this study.
Ethical standards
All the authors comply with the journal’s publishing ethics.





Comments
Dear Enrico,
Here is our opinion piece on the definition of Quantitative Plant Biology. Although most of the editorial board is involved, we still need to have external reviewers for this. Reviewers may simply give their opinion on areas we should have covered or important things we have missed or shortening suggestions (it’s quite long!).
Best wishes,
Olivier