INTRODUCTION
The classification of Leishmania was initially based on the clinical symptoms of the disease that they generated. Parasites causing cutaneous leishmaniasis throughout the world were considered as being L. tropica and those causing visceral leishmaniasis as being L. donovani. The first move away from this position was by Nicolle (Reference Nicolle1908) who considered that Mediterranean visceral leishmaniasis was clinically and epidemiologically different from that of India, naming it L. infantum. Three years later Vianna (Reference Vianna1911) gave the name L. braziliensis to the parasite responsible for a case of disseminated leishmaniasis from Brazil. Following this, Yakimoff and Schokhor (Reference Yakimoff and Schokhor1914) considered that the parasites causing urban and rural cutaneous leishmaniasis in Asia were different varieties of L. tropica, denominating the urban form as var. minor and the rural form as var. major.
Over the years more parasites from different parts of the world were examined and in the 1980s Lainson and Shaw (Reference Lainson, Shaw, Lumsden and Evans1979, Reference Lainson, Shaw, Peters and Killick-Kendrick1987) concluded that there were three very different groups of parasites that warranted sub-generic status: L. (Leishmania), L. (Viannia) and L. (Sauroleishmania). However, the taxonomic position of some Leishmania-like parasites remained uncertain such as L. colombiensis and L. martiniquensis isolated from patients and L. enriettii, L. equatorensis, L. herreri and L. hertigi isolated from wild animals. Another dixenous parasite that like the Leishmania also produces promastigotes in its vector and in culture is Endotrypanum. This genus was created to accommodate endoerythrocytic sloth trypanosomatids described in French Guyana (Mesnil and Brimont, Reference Mesnil and Brimont1908).
Molecular phylogeny has helped to clarify the taxonomy of the trypanosomatids, including many pathogens. In 2012, a group formed by dixenous and monoxenous trypanosomatids originally called ‘slow evolving’, due to the high conservation of SSU rRNA sequences, was given subfamily status under the name Leishmaniinae (Jirkú et al. Reference Jirkú, Yurchenko, Lukeš and Maslov2012). The dixenous members of this subfamily included parasites of wild animals that may accidentally infect man, causing diseases generically known as leishmaniasis. The depth of the taxonomic complexity of the parasites originally described as Leishmania is presently being disclosed by molecular studies. The question is: should they all be classified as belonging to the same genus? The mere suggestion that organisms other than Leishmania species cause leishmaniasis is immediately boycotted or met with scepticism. But is this view acceptable in the light of our present day knowledge of their genetic diversity and phylogenetic relationships?
Renewed interests on trypanosomatids from wild animals have unveiled a diversified genetic repertoire within the genera Trypanosoma and Leishmania. Recent studies have provided relevant insights into the broader genetic diversity and new reservoirs of human- and non-human infective species of Leishmania (Cupolillo et al. Reference Cupolillo, Pereira, Fernandes, Catanho, Pereira, Medina-Acosta and Grimaldi1998; Asato et al. Reference Asato, Oshiro, Myint, Yamamoto, Kato, Marco, Mimori, Gomez, Hashiguchi and Uezato2009; Cassia-Pires et al. Reference Cassia-Pires, Boite, D'Andrea, Herrera, Cupolillo, Jansen and Roque2014; Pothirat et al. Reference Pothirat, Tantiworawit, Chaiwarith, Jariyapan, Wannasan, Siriyasatien, Supparatpinyo, Bates, Kwakye-Nuako and Bates2014). Consequently, there is an increasing number of ‘leishmanias’ from non-human hosts that cannot be classified in any of the existing subgenera of Leishmania, and are herein refereed as enigmatic or Leishmania-like trypanosomatids. The focus of this paper is to molecularly characterize a large number of new isolates from Amazonian wild mammals (sloth and porcupine) and sand flies from Central and South America, and to compare the data obtained with data available from other enigmatic leishmanias and reference species of all accepted subgenera of Leishmania, and their closest related monoxenous species. For this, we inferred taxon-rich phylogenetic trees based on gGAPDH and HSP70 genes, and used the data to critically discuss the generic status and nomenclature of the dixenous parasites that produce promastigotes in their vectors and cultures, some of which are the aetiological agents of the cohort of diseases known as leishmaniasis.
MATERIAL AND METHODS
Organisms and cultures
The organisms characterized in this study are routinely grown at 23–25 °C in TC100 medium (=Grace medium) supplemented with 10% FBS, and deposited in the Trypanosomatid Culture Collection of the University of São Paulo (TCC-USP). Routinely, upon inclusion in the TCC-USP, the predominant morphotype of each new isolate is recorded, while the isolate itself is barcoded by V7V8 SSU rRNA sequencing (Teixeira et al. Reference Teixeira, Borghesan, Ferreira, Santos, Takata, Campaner, Nunes, Milder, de Souza and Camargo2011). This procedure serves only as preliminary information for future phylogenetic analyses of selected samples based on gGAPDH sequences. The samples selected for this study comprised cultures of trypanosomatids isolated from phlebotomines, sloths and porcupines from different areas of Central and South America (Table 1). For comparison, the analyses included DNA sequences determined in this study or recovered from GenBank and genome data banks of all Leishmania-like and Endotrypanum species, and reference-species of all subgenera of Leishmania.
Table 1. Details of trypanosomatids used in the V7V8 SSU rRNA and HSP70/gGAPDH phylogenetic analyses
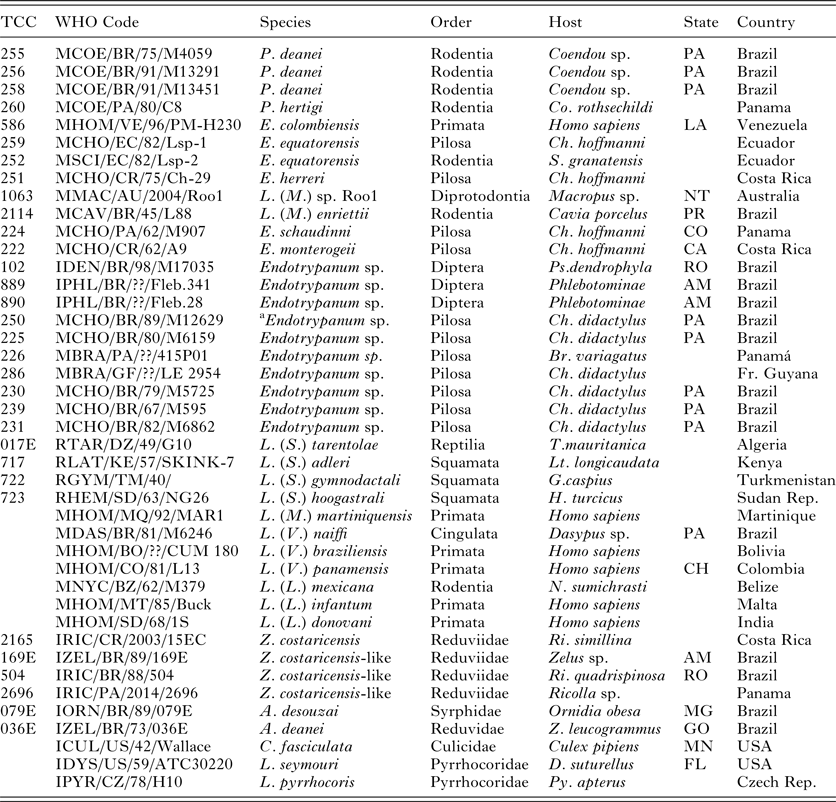
TCC, Trypanosomatid Culture Collection of the University of São Paulo;
P., Porcisia; E., Endotrypanum; L. (M.), L (Mundinia); Z., Zelonia; A., Angomonas; C., Crihtidia; Lp., Leptomonas. Br., Bradypus; Ch., Choloepus; Co., Coendou; D., Dysdercus; G., Gymnodactylus; H., Hemidactylus; Lt., Latastia. N., Nyctomys; S., Sciurus; T., Tarentola; Z., Zelus; Lu., Lutzomyia; Ps., Psathyromia; Py., Pyrrhocoris; Ri., Ricolla. State Abbreviations: Australia: NT, Northern Territory; Brazil: AM, Amazonas, GO, Goiás, MG, Minas Gerais, PA, Pará, PR, Paraná, RO, Rondônia; Colombia: CH, Choco Department; Costa Rica: CA, Cartago Province, HE, Heredia Province; Panama: CO, Colón Province; USA: MN, Minnesota; Venezuela: LA, Lara.
a This strain was considered initially to be Endotrypanum but was identified as L. deanei.
PCR amplification, sequencing and phylogenetic analysis
Total DNA was extracted from cultured flagellates using the traditional phenol–chloroform method. The V7V8 SSU rRNA and gGAPDH gene were PCR-amplified as previously described (Hamilton et al. Reference Hamilton, Stevens, Gaunt, Gidley and Gibson2004; Teixeira et al. Reference Teixeira, Borghesan, Ferreira, Santos, Takata, Campaner, Nunes, Milder, de Souza and Camargo2011). The amplification of HSP70 gene was done under the same reaction conditions adopted for gGAPDH and using the following primers: Hspf 5′-TGC GCA TCA TCA ACG AGC C-3′ and Hspr 5′-ATC TTG GTC ATGA TCG GGT TGC-3′.
PCR-amplified sequences were cloned using the TA Cloning Kit (Carlsbad, CA, USA), and from one to five clones were sequenced for each gene of each trypanosomatid isolate. Sequences were aligned using CLUSTALX (Thompson et al. Reference Thompson, Gibson, Plewniak, Jeanmougin and Higgins1997), and manually refined to obtain the following alignments: 1, V7V8 SSU rRNA sequences (~850 bp = barcodes); 2, gGAPDH sequences (768 bp); 3, HSP70 sequences (611 bp); 4, consisting of concatenated gGAPDH and HSP70 sequences (1·378 characters). Sequences determined in this study were all deposited in GenBank (Table S1). Sequences from Leptomonas costaricensis (Costa Rica), the isolate TCC169 (L. costaricensis-like from Brazil), Leptomonas seymouri, Leptomonas pyrrhocoris and Crithidia fasciculata were included in all analyses (Supplementary Table S1). Sequences from Angomonas deanei, Angomonas desouzai and species of other genera of trypanosomatids were used as outgroups of Leishmaniinae (Jirkú et al. Reference Jirkú, Yurchenko, Lukeš and Maslov2012). The dendrogram inferred using V7V8 SSU rRNA sequences of the Leishmaniinae subfamily and closely related trypanosomatids was done using the method of Maximum Parsimony (MP). The alignments of gGAPDH and HSP70 sequences were employed for phylogenetic inferences based on MP, Maximum Likelihood (ML) and Bayesian inference (BI) analyses. The MP and bootstrap analyses used the PAUP* version 4.0b10 software (Swofford, Reference Swofford2002), with 500 random sequence addition replicates followed by branch swapping (RAS-TBR), and the ML analyses used RAxML v.2.2.3 (Stamatakis, Reference Stamatakis2006). The general time reversible (GTR) model of nucleotide substitution with proportion of invariable sites and gamma distribution was selected for the datasets. We used the GTR model in individual analyses of each gene as well as in the combined analyses, which was run for 1 000 000 generations with trees sampled every 100 generations using four chains, and 25% of the early sample trees were discarded as ‘burn-in’.
RESULTS
Barcoding of Endotrypanum and Leishmania-like isolates and comparison with other species of trypanosomatids
In this study, we barcoded through V7V8 SSU rRNA sequences the reference isolates of Endotrypanum schaudinni and Endotrypanum monterogeii, and isolates previously classified in this genus by isoenzyme patterns: six from sloths (Choloepus hoffmanni, C. didactylus and Bradypus infuscatus) and three from sand flies (Lutzomyia gomezi and Psathyromyia dendrophyla). In addition, we also barcoded all isolates of Leishmania-like that are not positioned within the recognized subgenera of Leishmania: L. equatorensis, L. colombiensis, L. herreri, L. deanei and L. hertigi. All these organisms were found to be closely related to Endotrypanum species, sharing highly conserved V7V8 SSU rRNA sequences (average of 98·7%) with members of this genus. However, despite the high conservation, inferred dendrogram supported a branch formed by two main groups of isolates respectively headed by E. schaudinni and L. hertigi (Fig. 1) and separated by ~2·7% of sequence divergence. In addition, the analysis of V7V8 SSU rRNA sequences allowed the clustering of most species of Leishmania spp. in the three currently recognized subgenera, and uncovered two additional clusters headed by L. deanei and L. enriettii, respectively. A new group was formed by monoxenous flagellates of hemipterans, including L. costaricensis (Yurchenko et al. Reference Yurchenko, Lukes, Jirku, Zeledon and Maslov2006) from Costa Rica and three new isolates characterized in the present study: TCC169E and TCC504 from Brazilian Amazonia and TCC2696 from Panama, all sharing highly similar V7V8 SSU rRNA barcodes and placed into the subfamily Leishmaniinae (Fig. 1, Table S1).

Fig. 1. A dendrogram inferred by MP analysis using 33 sequences from V7V8 SSU rRNA (alignment A1, 820 characters) from references species of Heteroxenous trypanosomatids and new isolates from sand flies, sloth and porcupine. The numbers at nodes correspond to percentage of bootstrap values derived from 100 replicates (– support value <50%). The accession numbers of sequences in GenBank are in Supplementary Table S1. All new isolates grouped with Endotrypanum and Leishmania-like species.
Although the comparison of V7V8 SSU rRNA sequences is valuable to place new isolates in clusters representing genus/subgenera, more polymorphic sequences are required to distinguish the species within genera/subgenera, and to infer well-resolved phylogenetic relationships within and among them.
Phylogenetic relationships among isolates of Endotrypanum and Leishmania-like
In order to resolve the relationship between Endotrypanum and Leishmania-like isolates we inferred their phylogenetic relationships based on gGAPDH and HSP70 gene sequences. The analyses also included sequences from species of the complex L. enriettii, and the subgenera L. (Viannia), L. (Leishmania) and L. (Sauroleishmania). In the inferred phylogenies, all Leishmania spp. and Endotrypanum spp. formed a strongly supported monophyletic group (100% support) (Fig. 2). In addition, all analyses (P, ML and BI) using a single gene or concatenated gene sequences underscored (100% bootstrap support) three major monophyletic lineages (clades): L. hertigi, L. herreri and Endotrypanum spp.

Fig. 2. Phylogenetic tree of heteroxenous trypanosomatid genera of the subfamily Leishmaninae inferred by MP, ML and Bayesian analysis, based on concatenated heat shock protein 70 and glycosomal glyceraldehyde-3-phosphate dehydrogenase (gGAPDH) genes sequences (alignment A2, 1354 characters). The accession numbers of HSP70 and gGAPDH sequences of these trypanosomatids in GenBank are in Table X. Numbers at the major nodes correspond respectively to MP, ML and BI support values (– support value <50%). The four heteroxenous genera Endotrypanum, Porcisia, Zelonia and Leishmania are strongly supported and the new subgenus L. (Mundina), is the most basal taxonomic group of the genus Leishmania.
The monophyletic group containing L. herreri, L. equatorensis, L. colombiensis and Endotrypanum species comprises two clades (100% support), headed by Endotrypanun spp. and L. herreri, separated by 2·0 and 4·2% of HSP70 and gGAPDH sequence divergences, respectively. The clade grouping the reference species E. monterogeii (Costa Rica, C. hoffmanni) and E. schaudinni (Panama, C. hoffmanni) also harboured L. colombiensis (isolated from a patient from Venezuela), TCC 226 (from the sloth, B. infuscatus of Panama), TCC 102 (from the sand fly Psanthyromya dendrophyla, from the state of Rondônia, Brazilian Amazonia) with strongly support values of 100% for P, ML and BI. The clade harbouring L. herreri (Costa Rica) clustered six new isolates: TCC889 and TCC890 from sand flies from the State of Amazonas, Brazil, TCC225 from Choloepus didactylus, and TCC230, TCC231 and TCC239 from C. hoffmanni from the state of Pará, Brazil. The trypanosomatids of L. herreri group share similarity of ~96 and 99% of gGAPDH and HSP70 gene sequences, respectively. The similarity between the sequences of L. equatorensis from Sciurus granatensis (squirrel) and C. hoffmanni (sloth) from Ecuador was 100% for both HSP70 and gGAPDH genes.
Basal to the major clade comprising Endotrypanum and L. herreri subclades was the clade composed of porcupine parasites: L. hertigi from Panama and L. deanei from Brazil, with support of 100% in all analyses. The isolates forming the complex hertigi-deanei share great similarity in the sequences of gGAPDH and HSP70 (99 and 100% respectively). However, this is a very distant clade, separated by ~12 and 5·0% of divergences in gGAPDH and HSP70 sequences respectively from Endotrypanum and other Leishmania-like.
Leishmania enriettii complex
In the inferred phylogenies based on gGAPDH and HSP70 sequences, L. enriettii, the Australian Leishmania sp. from the red kangaroo and L. martiniquensis compose a clade positioned basal (support values of 75, 80 and 96% for P, ML and BI, respectively) to the major clade comprising L. (Viannia), L. (Leishmania) and L. (Sauroleishmania) species. The L. enriettii clade was well-supported (96, 100 and 100% for P, ML and BI, respectively) as the more distant clade using concatenated genes, whereas the analyses based on single genes showed lower support values. This clade comprises species separated by large genetic distances, sharing 5·0 and 4·0% of gGAPDH and HSP70 sequence divergences, respectively. Even though, in all analyses this was the most basal clade of the genus Leishmania. The results further demonstrate that parasites belonging to the L. enriettii complex shares the greatest similarity of gGAPDH and HSP70 sequences with the species of the subgenus L. (Leishmania) (92 and 96%, respectively) than with the species of the subgenus L. (Viannia) (87 and 94%, respectively).
DISCUSSION
The creation of the subgenus L. (Viannia) and the validation of the subgenera L. (Leishmania) and L. (Sauroleishmania) (Lainson and Shaw, Reference Lainson, Shaw, Peters and Killick-Kendrick1987) was a major step forward to organizing the taxonomy of the leishmaniasis parasites. However, in the 1990s biochemical and molecular data began to show that some flagellates classified as Leishmania did not fall comfortably within any of the three subgenera (Croan et al. Reference Croan, Morrison and Ellis1997). The parasite that spearheaded this group, but was not immediately linked with leishmaniasis, was Endotrypanum, an enigmatic endoerythrocytic sloth parasite described in French Guyana (Mesnil and Brimont, Reference Mesnil and Brimont1908) that develops as promastigotes in culture and sand flies (Shaw, Reference Shaw1963, Reference Shaw1969, Reference Shaw and Canning1981).
Phylogenetic analysis (Croan et al. Reference Croan, Morrison and Ellis1997) based on the genes encoding the DNA polymerase alpha catalytic polypeptide (POLA) and the RNA polymerase II largest subunit (RPOIILS) showed that L. hertigi and Endotrypanum formed two groups that were distinct from the Leishmania subgenera L. (Leishmania), L. (Viannia) and L. (Sauroleishmania). Multilocus enzyme electrophoresis (MLEE), sialidase activity, ITS rRNA restriction profiles and minicircle kDNA sequences (Cupolillo et al. Reference Cupolillo, Pereira, Fernandes, Catanho, Pereira, Medina-Acosta and Grimaldi1998) showed that two leishmania, L. colombiensis and L. equatorensis grouped within the Endotrypanum clade. Following these results a revised classification was proposed (Cupolillo et al. Reference Cupolillo, Medina-Acosta, Noyes, Momen and Grimaldi2000) that created two informal groups. One, the Paraleishmania, contained the L. hertigi and Endotrypanum clades and the other, the Euleishmania, contained the subgenera L. (Leishmania) and L. (Viannia).
Taxonomy: amendments to the subfamily Leishmaniinae and the genus Endotrypanum and the creation of the new genera Porcisia, Zelonia and the new subgenus Leishmania (Mundinia)
Phylum Euglenozoa (Cavalier-Smith, 1981); Class Kinetoplastea Honigberg 1963; Order Trypanosomatida (Kent 1880; Hollande 1982), Family Trypanosomatidae (Doflein 1901), Leishmaniinae (Maslov and Lükes, 2012 emend Espinosa et al. 2016)
Subfamily Leishmaniinae Maslov and Lukeš (Jirkú et al. Reference Jirkú, Yurchenko, Lukeš and Maslov2012) emend Shaw, Camargo and Teixeira 2016.
The subfamily Leishmaniinae (Jirkú et al. Reference Jirkú, Yurchenko, Lukeš and Maslov2012) was erected for a group of monoxenous and dixenous trypanosomatid parasites based on a phylogenetic analysis of a concatenated dataset of SSU rRNA and gGAPDH gene sequences. This previous study included more monoxenous than dixenous trypanosomatids and did not compare sequences from the Leishmania-like herein taxonomically revised. Our phylogenetic analyses (MP, ML and BI) based on combined gGAPDH and HSP70 sequences and using trypanosomatids not included in the subfamily Leishmaniinae as outgroup (Jirkú et al. Reference Jirkú, Yurchenko, Lukeš and Maslov2012) strongly support two major clades, one comprising Leishmania–Endotrypanum and the other containing of monoxenous trypanosomatids herein represented by C. fasciculata, L. seymouri and L. pyrrrochoris (Fig. 2). Phylogenetic analyses based on concatenated V7V8 SSU rRNA and gGAPDH support congruent branching patterns (data not shown). Our findings are consistent with published phylogenetic trees inferred using SSU rRNA and gGAPDH sequences supporting the subdivision of the subfamily Leishmaniinae into two clades (Jirkú et al. Reference Jirkú, Yurchenko, Lukeš and Maslov2012; Kostygov et al. Reference Kostygov, Dobakova, Grybchuk-Ieremenko, Vahala, Maslov, Votypka, Lukeš and Yurchenko2016). In face of the data gathered in the present study on well-resolved and taxon-rich phylogenies, including Leishmania and Leishmania-like, Leptomonas and Crithidia species, we suggest that the subfamily Leishmaniinae is limited to the strongly supported clade that includes the dixenous trypanosomatids and, in addition, also harbours some trypanosomatids that are thought to be monoxenous positioned into the new genus Novymonas (Kostygov et al. Reference Kostygov, Dobakova, Grybchuk-Ieremenko, Vahala, Maslov, Votypka, Lukeš and Yurchenko2016) and into the clade headed by L. costaricensis (Figs 1 and 2), which was erected to a new genus in the present study (section below). New subfamilies may eventually be further created to accommodate the monoxenous trypanosomatids that nested into the other major clade (Crithidia, Lotmaria and Leptomonas) of the originally proposed subfamily Leishmaniinae when their phylogenetic relationships are more clearly defined.
The distinction between monoxenous and heteroxenous trypanosomatids as a taxonomic criterion must be viewed with caution. Presently, molecular characters support this general division, but it is conceivable that as more trypanosomatids are analysed mono- and heteroxenous trypanosomatids may cluster together in several trypanosomatid taxa. The fact cannot be ignored that the apparently monoxenous trypanosomatids L. costaricensis (Yurchenko et al. Reference Yurchenko, Lukes, Jirku, Zeledon and Maslov2006) and Novymonas sp. (Kostygov et al. Reference Kostygov, Dobakova, Grybchuk-Ieremenko, Vahala, Maslov, Votypka, Lukeš and Yurchenko2016) are more closely related to the heteroxenous Leishmania and Endotrypanum spp. than to the monoxenous Crithidia and Leptomonas. Another trypanosomatid (G755) isolated from Guatemala (Noyes et al. Reference Noyes, Arana, Chance and Maingon1997) is also a member of the L. costaricensis clade. Knowing so little about the life history of these parasites it is difficult to say whether they are in fact truly monoxenous. A parasite isolated from diffuse lesions of a HIV patient from Martinique was considered to be a monoxenous trypanosomatid (Dedet and Pratlong, Reference Dedet and Pratlong2000). Further analysis showed that it was a leishmanine parasite, but it did not belong to the subgenera previously associated with human leishmaniasis (Noyes et al. Reference Noyes, Pratlong, Chance, Ellis, Lanotte and Dedet2002). Other parasites also considered to be monoxenous parasites have been isolated from HIV patients from Spain (Jimenez et al. Reference Jimenez, Lopez-Velez, Molina, Canavate and Alvar1996) and Brazil (Pacheco et al. Reference Pacheco, Marzochi, Pires, Brito, Madeira and Barbosa-Santos1998). Their phylogenetic position was not determined. The 18S rRNA sequence of a parasite isolated from the blood of a HIV patient living in France (Morio et al. Reference Morio, Reynes, Dollet, Pratlong, Dedet and Ravel2008) showed a 99·8% similarity to Herpetomonas samuelpessoai. Of 33 strains from Indian cases of visceral leishmaniasis, the ITS1 rDNA of 21 matched that of L. seymouri (Ghosh et al. Reference Ghosh, Banerjee, Sarkar, Datta and Chatterjee2012). These monoxenous organisms are from HIV patients or patients with infections, such as L. (L.) donovani that also depress the immune system. These observations show that the line between monoxenous and heteroxenous is a very fine one. It also suggests that infections of monoxenous organisms may occur in immunological competent individuals, but since they are resolved quickly they are not detected.
Endotrypanum and some enigmatic Leishmania, parasites of Neotropical animals, have previously been grouped together under the name paraleishmania (Cupolillo et al. Reference Cupolillo, Medina-Acosta, Noyes, Momen and Grimaldi2000; Pothirat et al. Reference Pothirat, Tantiworawit, Chaiwarith, Jariyapan, Wannasan, Siriyasatien, Supparatpinyo, Bates, Kwakye-Nuako and Bates2014). Actually, they form two natural clades. One is composed of flagellates previously classified as either Leishmania or Endotrypanum, and the other of parasites from American porcupines.
Porcisia n. gen. Shaw, Camargo and Teixeira
Type species: Porcisia hertigi (Herrer, Reference Herrer1971) (Synonym: Leishmania hertigi).
Diagnosis: Our phylogenetic analyses of five isolates of the complex L. hertigi/L. deanei confirm their uniqueness and clustering in a clade strongly supported in all phylogenetic analyses and separated from its sister clade formed by Leishmania-like and Endotrypanum by relevant genetic distance. With the above points in mind we consider that the L. hertigi complex warrants generic status and propose the name Porcisia for the new genus.
Etymology: The genus name is based on first four letters of porcupine; this animal is the principal host.
Historical comments: Leishmania hertigi was discovered by Aristides Herrer (Herrer, Reference Herrer1971) in porcupines, Coendou rothschildi, from Central Panama and was later found in Costa Rica (Zeledon et al. Reference Zeledon, Ponce and de Ponce1977). Similar parasites were found in porcupines in the Brazilian states of Piaui and Pará (Deane et al. Reference Deane, da Silva and de Figueiredo1974; Lainson and Shaw, Reference Lainson and Shaw1977). The Brazilian parasites were biochemically and morphologically distinct from P. hertigi and were named L. hertigi deanei (Lainson and Shaw, Reference Lainson and Shaw1977). In 1987, this subspecies was given specific status as L. deanei (Lainson and Shaw, Reference Lainson, Shaw, Peters and Killick-Kendrick1987). Later, a parasite was identified by partial 18S rDNA sequence as L. hertigi in a porcupine from the city of Brasília, which is located in centre-west region of Brazil (Silva et al. Reference Silva, Madeira Mde, Barbosa Filho, Schubach, Barros and Figueiredo2013). This was most probably P. deanei and not P. hertigi, but more detailed studies of the Brasilia parasite are needed to confirm this. The 18S rDNA sequences are too conserved and consequently may not distinguish between these closely related species. These two parasites occur in different species of porcupines, distinct geographical regions and in our study P. hertigi and P. deanei are clearly separated. The porcupine parasites are biochemically, molecularly and morphologically different from the other species of Leishmaniinae. Their vectors are unknown.
Molecularly validated species: Porcisia hertigi and Porcisia deanei SSU rRNA, gGAPDH and HSP70 gene sequences were deposited in GenBank under the accession numbers listed in Table S1.
So far all parasites of the genus Porcisia have been found in porcupines. In the present study, one isolate (TCC 250), whose preliminary identification as Endotrypanum was based on it being isolated from the blood of a two-toed sloth, was molecularly identified as P. deanei. Porcupines and sloths are arboreal animals and it is possible that this represents a rare infection of a P. deanei in a sloth. However, we could not discard the possibility of misleading cultures.
Zelonia n. gen Shaw, Camargo and Teixeira
Type species: Zelonia costaricensis (Yurchenko et al. Reference Yurchenko, Lukes, Jirku, Zeledon and Maslov2006) syn: L. costaricensis
Type host: Ricolla simillima (Heteroptera, Reduviidae).
Type locality: El Ceibo (10 × 20 kN, 84 × 05 kW), La Virgen, Province Heredia, Costa Rica
Etymology: The name is based on the surname of the famous Costarican protozoologist, Professor Rodrigo Zeledon, whose pioneering studies contributed enormously to trypanosomatid research in Central America and especially in Costa Rica.
The type species of the new genus was isolated from a predator hemipteran from Costa Rica and originally described as L. costarricensis by Yurchenko et al. (Reference Yurchenko, Lukes, Jirku, Zeledon and Maslov2006). Previous phylogenetic analyses (Jirkú et al. Reference Jirkú, Yurchenko, Lukeš and Maslov2012; Kostygov et al. Reference Kostygov, Dobakova, Grybchuk-Ieremenko, Vahala, Maslov, Votypka, Lukeš and Yurchenko2016) also have shown that it is distant from L. pyrrhocoris and L. seymouri. It is therefore unreasonable to continue to consider it to be a species of Leptomonas and so we have created the genus Zelonia to accommodate the clade headed by this parasite. The barcoding of our trypanosomatid collection revealed three additional isolates sharing highly similar V7V8 SSU rRNA sequences with Z. costaricensis. Interestingly, all isolates of the genus Zelonia were obtained from predatory hemipterans (Reduviidae) in Equatorial regions of Costa Rica, Brazil (Amazonia) and Panamá (Fig. 1, Table 1). The isolate TCC169 exhibits promastigote and opistomatigote-like forms in log- and stationary axenic cultures, and produced a few intracellular amastigotes in macrophage cultures; however, these forms disappeared in 3–4 days (unpublished results). The trypanosomatid from the digestive tract of a hemipteran (Rhopalidae) from Ecuador and recently described under the new genus Novymonas was the most closely phylogenetically related to Zelonia costarricensis in a tree based on 18S SSU rRNA, 28S LSU rRNA and Hsp83. The authors (Kostygov et al. Reference Kostygov, Dobakova, Grybchuk-Ieremenko, Vahala, Maslov, Votypka, Lukeš and Yurchenko2016) considered that ‘It does not cluster within either the Leishmania clade or the Leptomonas–Lotmaria–Crithidia group’. In agreement with these previous studies, in our phylogenetic analyses the new genus Zelonia is one basal group of the re-defined Leishmaniinae subfamily, therefore, more phylogenetically related to Endotrypanum–Leishmania than to Leptomonas–Lotmaria–Crithidia. Jirkú et al. (Reference Jirkú, Yurchenko, Lukeš and Maslov2012) clearly show that Z. costaricensis and Leptomonas barvae belong to a separate group they designated as Le, which coincides with our redefinition of the subfamily Leishmaniinae. Further analyses are still required regarding the apparently most basal species of this subfamily, L. barvae, Leptomonas moramango and Blastocrithidia miridarum (Kostygov et al. Reference Kostygov, Grybchuk-Ieremenko, Malysheva, Frolov and Yurchenko2014; Maslov et al. Reference Maslov, Yurchenko, Jirku and Lukes2010), all unquestionably requiring a taxonomic reappraisal at subfamily and genus levels.
Amendment to the genus Endotrypanum (Mesnil and Brimont, Reference Mesnil and Brimont1908) emend Shaw, Camargo and Teixeira 2016.
Type species: Endotrypanum schaudinni Mesnil and Brimont, Reference Mesnil and Brimont1908. Type material not available.
Host Choloepus didactylus;
Type Locality: French Guyana.
Diagnosis: Phylogenetic positioning within the family Trypanosomatidae was based on gGAPDH and HSP70 gene sequences deposited in GenBank (Supplementary Table S1).
Genus composition and historical comments: There are two strongly supported groups within the genus. One contains a species previously placed within the genus Leishmania – E. herreri, and the second group contains two other species previously considered to be Leishmania – E. colombiensis and E. equatorensis. These two groups also contain parasites of sloths from Central and South America identified as Endotrypanum species.
Our analysis and past ones (Croan et al. Reference Croan, Morrison and Ellis1997; Cupolillo et al. Reference Cupolillo, Medina-Acosta, Noyes, Momen and Grimaldi2000; Noyes et al. Reference Noyes, Pratlong, Chance, Ellis, Lanotte and Dedet2002; Asato et al. Reference Asato, Oshiro, Myint, Yamamoto, Kato, Marco, Mimori, Gomez, Hashiguchi and Uezato2009; Pothirat et al. Reference Pothirat, Tantiworawit, Chaiwarith, Jariyapan, Wannasan, Siriyasatien, Supparatpinyo, Bates, Kwakye-Nuako and Bates2014) using a variety of different markers consistently show a strongly supported monophyletic group that includes the following parasites: E. schaudinni, E. monterogeii, L. colombiensis, L. equatorensis and L. herreri. Based on RNA Pol II sequences (Pothirat et al. Reference Pothirat, Tantiworawit, Chaiwarith, Jariyapan, Wannasan, Siriyasatien, Supparatpinyo, Bates, Kwakye-Nuako and Bates2014) Endotrypanum and L. colombiensis group together and are separate from both L. equatorensis and L. herreri. Our results follow this pattern (Fig. 2). Previous analyses of this group were hampered by the fact that Endotrypanum was not compared with isolates considered to be Leishmania. Endotrypanum was not included in the differential diagnosis in the original descriptions of L. colombiensis (Kreutzer et al. Reference Kreutzer, Corredor, Grimaldi, Grogl, Rowton, Young, Morales, McMahon-Pratt, Guzman and Tesh1991) and L. equatorensis (Grimaldi Junior et al. Reference Grimaldi Junior, Kreutzer, Hashiguchi, Gomez, Mimory and Tesh1992). It was also absent in subsequent publications in which four visceral isolates of L. colombiensis from Venezuelan patients differed from four cutaneous isolates and from a standard of this species that was isolated from L. gomezi (Rodriguez-Bonfante et al. Reference Rodriguez-Bonfante, Bonfante-Garrido, Grimaldi, Momen and Cupolillo2003).
Karyotype analyses of 17 isolates labelled as Endotrypanum from Brazilian, Panamanian and Colombian sloths and Brazilian sand flies unveiled four groups (Lopes et al. Reference Lopes, Iovannisci, Petrillo-Peixoto, McMahon-Pratt and Beverley1990). The first and second groups were closely related, the first being composed of isolates from Colombia, Brazil and Panama and the second of strains from Panama. Isolates in the third and fourth groups were all from Brazil. Parasites of the first two groups have different levels of virus-like-particles that are absent in the last two groups. The fourth group, consisting of two strains, was so different that the authors suggested that it was in fact not Endotrypanum, echoing the opinion of the group who isolated and first characterized them (Arias et al. Reference Arias, Miles, Naiff, Povoa, de Freitas, Biancardi and Castellon1985). Both strains failed to react with Endotrypanum-specific monoclonals (Lopes and McMahon-Pratt, Reference Lopes and McMahon-Pratt1989) confirming that they were not Endotrypanum. Our results reflect a similar situation. The strains in the group headed by TC889 are all from the Brazilian Amazon, while the second group contains E. colombiensis (TCC583, TCC586) from Venezuela, Endotrypanum isolates from Panama (TCC 226), French Guyana (TCC286), Rondônia, Brazil (TCC102), E. schaudinni (TCC224) from Panama and E. monterogeii (TCC222) from Costa Rica.
The nomenclature of the genus Endotrypanum remains extremely complex. The type species E. schaudinni does not exist, so a neotype will have to be designated for one parasite from French Guiana where it was originally described. If we choose the isolate TCC286 (MBRA/GF/??/LE 2954), reportedly isolated from French Guiana, as the neotype of E. schaudinni then it suggests that E. colombiensis is a synonym of E. schaudinni since the two are identical in our analysis. Of the two endotrypanums from Central America the one from Panama was named E. schaudinni (MCHO/PA/62/M907) and the one from Costa Rica E. monterogeii (MCHO/CR/62/A9). In our analysis, both are identical, but differ from the French Guiana isolate, then strain M907 should be called E. monterogeii and not E. schaudinni. From this it follows that the name that has priority for Central American Endotrypanum spp., is E. monterogeii. E. herreri from Costa Rica and E. equatorensis from Ecuador are distinct and appear to be valid species. It is possible that the TC889 group represents a new taxon within the genus Endotrypanum. We recommend that more data are gathered before proposing to the ICZN a neotype for the amended genus Endotrypanum and to define species within this genus.
The vectors of Endotrypanum species appear to be phlebotomine sand flies in which they have a peripylarian development (Shaw, Reference Shaw1963, Reference Shaw and Canning1981; Christensen and Herrer, Reference Christensen and Herrer1976; Franco et al. Reference Franco, Tesh, Guzman, Deane and Grimaldi Junior1997). Different Endotrypanum isolates have been obtained from wild sand flies in Brazil, Panama, Costa Rica, Ecuador and Venezuela. There is some evidence of vectorial specificity of endotrypanums. After three days flagellates were found from the cardia to the rectal ampulla in experimental infections of E. colombiensis in L. trapidoi but in P. papatasi parasites were limited to the blood meal (Kreutzer et al. Reference Kreutzer, Corredor, Grimaldi, Grogl, Rowton, Young, Morales, McMahon-Pratt, Guzman and Tesh1991).
The E. colombiensis is the only species that has been associated with leishmaniasis. So far, none of the endotrypanums examined to date produce lesions or lasting infections in experimental animals, such as hamsters or mice. E. colombiensis produced intracellular amastigotes in cultures of J774 macrophages, but from 72 h onwards the parasites began to die and by 120 h they had disappeared (Kreutzer et al. Reference Kreutzer, Corredor, Grimaldi, Grogl, Rowton, Young, Morales, McMahon-Pratt, Guzman and Tesh1991). In their original description of E. herreri Zeledon et al. (Reference Zeledon, Ponce and de Ponce1977) reported that it produced amastigotes in tissue culture and hamster but they did not give details of the cells used nor if lesions were produced in hamsters.
Our phylogenetic analysis shows deep rooted and well-supported differences within the heteroxenous parasites confirming that Endotrypanum is a valid genus within the emended family Leishmaniinae. It follows that it should not be synonymized with the genus Leishmania.
Leishmania (Mundinia) n. subgenus Shaw, Camargo and Teixeira 2016
Type species: Leishmania (Mundinia) enriettii Muniz and Medina, Reference Muniz and Medina1948
Host: Domestic Guinea-pig (Cavia porcellus); Type Locality: Curitiba, Parana State, Brazil
Diagnosis: gGAPDH and HSP70 gene sequences deposited in GenBank (Table S1).
Molecularly validated species: L. (M.) enriettii, L. (M.) martiniquensis, a parasite of the red Kangaroo
Etymology: The name Mundinia is in honour of Muniz (Mun) and Medina (din) who discovered and named this parasite.
Comments on nomenclature: In 1995, a paper was published (Dedet et al. Reference Dedet, Roche, Pratlong, Cales-Quist, Jouannelle, Benichou and Huerre1995) reporting the presence of a very unusual trypanosomatid isolated from a case of diffuse cutaneous leishmaniasis in an HIV positive patient. Initially, it was thought to be a monoxenous trypanosomatid, but a phylogenetic analysis (Noyes et al. Reference Noyes, Pratlong, Chance, Ellis, Lanotte and Dedet2002) showed that it belonged to the L. enriettii clade. This parasite received the name Leishmania martiniquensis (Desbois et al. Reference Desbois, Pratlong, Quist and Dedet2014). Subsequently cytochrome b studies (Asato et al. Reference Asato, Oshiro, Myint, Yamamoto, Kato, Marco, Mimori, Gomez, Hashiguchi and Uezato2009) confirmed the uniqueness of the L. enriettii clade.
More enigmatic parasites associated with leishmaniasis in cows and horses were discovered in Europe and the United States (Müller et al. Reference Müller, Welle, Lobsiger, Stoffel, Boghenbor, Hilbe, Gottstein, Frey, Geyer and von Bomhard2009; Lobsiger et al. Reference Lobsiger, Muller, Schweizer, Frey, Wiederkehr, Zumkehr and Gottstein2010; Reuss et al. Reference Reuss, Dunbar, Calderwood Mays, Owen, Mallicote, Archer and Wellehan2012). Based on ITS1 rDNA analyses the aetiological agents of these infections were identified as L. siamensis but no parasites were isolated. Unfortunately, the specific name siamensis is a nomem nudum as it was not described in accordance with Article 13 of the International Code of Zoological Nomenclature. As such this name becomes unavailable. The authors (Sukmee et al. Reference Sukmee, Siripattanapipong, Mungthin, Worapong, Rangsin, Samung, Kongkaew, Bumrungsana, Chanachai, Apiwathanasorn, Rujirojindakul, Wattanasri, Ungchusak and Leelayoova2008) used the wording ‘suspected new species’ and did not include the name siamensis in their 2008 publication. The name first appears as Leishmania sp. siamensis in GenBank (entries EF200012 and EF200011), and the next in a publication a year later when material from horses was considered to be close to Leishmania sp. siamensis (Müller et al. Reference Müller, Welle, Lobsiger, Stoffel, Boghenbor, Hilbe, Gottstein, Frey, Geyer and von Bomhard2009). Since then the name has been extensively used in the literature and at the time of writing 18 citations were found in a PubMed search using Leishmania siamensis. In 2012, a parasite from the Trang region of Thailand was found to be phylogenetically closest to L. enriettii (Bualert et al. Reference Bualert, Charungkiattikul, Thongsuksai, Mungthin, Siripattanapipong, Khositnithikul, Naaglor, Ravel, El Baidouri and Leelayoova2012). A recent ITS1 analysis (Pothirat et al. Reference Pothirat, Tantiworawit, Chaiwarith, Jariyapan, Wannasan, Siriyasatien, Supparatpinyo, Bates, Kwakye-Nuako and Bates2014) showed that sequences previously identified as L. siamensis were identical to those of L. martiniquensis. So besides being a nomem nudum L. siamensis becomes a synonym of L. martiniquensis. This ITS1 rDNA analysis also showed that a yet unnamed kangaroo Leishmania (Rose et al. Reference Rose, Curtis, Baldwin, Mathis, Kumar, Sakthianandeswaren, Spurck, Low Choy and Handman2004) grouped with L. martiniquensis, and that an isolate previously identified as L. siamensis from Trang Thailand grouped with L. enriettii. These same workers (Pothirat et al. Reference Pothirat, Tantiworawit, Chaiwarith, Jariyapan, Wannasan, Siriyasatien, Supparatpinyo, Bates, Kwakye-Nuako and Bates2014) showed that these two groups formed a monophyletic L. enriettii complex based on RNA pol II sequences (Pothirat et al. Reference Pothirat, Tantiworawit, Chaiwarith, Jariyapan, Wannasan, Siriyasatien, Supparatpinyo, Bates, Kwakye-Nuako and Bates2014). The main difference between the ITS1 (the most polymorphic sequences) and RNA Pol II analyses was that in the latter both the kangaroo Leishmania and Trang isolate (MHOM/TH/2010/PCM2) grouped with L. enriettii. This indicates that at least two different parasites are responsible for leishmaniasis in Thailand. One is L. martiniquensis and the other remains to be named. Recently another species of this subgenus has been described from Ghana (Kwakye-Nuako et al. Reference Kwakye-Nuako, Mosore, Duplessis, Bates, Puplampu, Mensah-Attipoe, Desewu, Afegbe, Asmah, Jamjoom, Ayeh-Kumi, Boakye and Bates2015) that is phylogenetically closer to L. enriettii than to L. martiniquensis.
Unfortunately, the specific name australiensis is not available for the Australian red kangaroo parasite. The name was used in a recent review paper (Akhoundi et al. Reference Akhoundi, Kuhls, Cannet, Votypka, Marty, Delaunay and Sereno2016) without any formal description. The name was written as ‘L. australiensis’. The use of inverted commas does not change the validity of a name. So L. australiensis is a nomem nudum.
The growing reluctance to formally name parasites in accordance with the International Code of Zoological Nomenclature that are obviously distinct or considered to be distinct by their authors is frustrating. The cases of L. siamensis and L. australiensis have different motives, but clearly the use of the specific name helps authors communicate. We have refrained from correctly naming these two parasites as clearly this should be done by the researchers involved in their discovery. But it is their apparent indisposition to name them that has led to the present nomenclature problems. A species name is essential in following a parasite in the literature and draws attention to the parasite in question. If later studies show that it is the same as another previously described parasite the name will become a subspecies or synonym. A classical case was the discussion as to whether L. (L.) chagasi should be called L. (L.) infantum. The general conclusion was the L. (L.) chagasi should no longer be considered as a species but as a subspecies of L. (L.) infantum. However, the work that led up to this generated data that was important in understanding the epidemiology of visceral leishmaniasis in the Americas.
Our Hsp70 and gGPDH analyses (Fig. 2) also show that L. martiniquensis, L. enriettii and the kangaroo Leishmania are distinct species that form a monophyletic group within the genus Leishmania. The above data strongly supports our proposal that this group previously called the L. enriettii complex should be given sub generic status within the genus Leishmania.
The type species of the subgenus is Leishmania (Mundinia) enriettii Muniz and Medina, Reference Muniz and Medina1948. Its vector is unknown but biting midges (Diptera, Ceratopogonidae) have been incriminated as vectors of the kangaroo parasite and L. (M.) enriettii (Dougall et al. Reference Dougall, Alexander, Holt, Harris, Sultan, Bates, Rose and Walton2011; Seblova et al. Reference Seblova, Sadlova, Vojtkova, Votypka, Carpenter, Bates and Volf2015). It is possible that biting midges may also prove to be vectors of the subgenus L. (Mundinia). A monoxenous trypanosomatid isolated from the midgut of naturally infected biting midges was classified as Sergeia podlipaevi, which is closely related to the Trypanosomatidae EVA from the sand fly Lutzomyia evansi from South America (Svobodová et al. 2007). In addition, the monoxenous trypanosomatids Herpetomonas trimorpha and H. zitplika were also isolated from biting midges (Podlipaev et al. Reference Podlipaev, Votypka, Jirku, Svobodova and Lukes2004; Zidkova et al. Reference Zidkova, Cepicka, Votypka and Svobodova2010). Both Sergeia and Herpetomonas exhibited promastigote in cultures and were placed in phylogenetic trees distant from Leishmaniinae. Novymonas esmeraldas was also detected in biting midges (Kostygov et al. Reference Kostygov, Dobakova, Grybchuk-Ieremenko, Vahala, Maslov, Votypka, Lukeš and Yurchenko2016). Finally, a trypanosomatid was reported in a fossil ceratopogonid from the early cretaceous (Poinar, Reference Poinar2008).
The lack of genetic variation within isolates identified by comparison of their ITS1 sequences as L. (M.) martiniquensis form very different geographical regions of the world is intriguing (Pothirat et al. Reference Pothirat, Tantiworawit, Chaiwarith, Jariyapan, Wannasan, Siriyasatien, Supparatpinyo, Bates, Kwakye-Nuako and Bates2014) and needs to be investigated in greater detail.
Taxonomic summary
The revised classification and nomenclature of Leishmaniinae parasites, some of which are associated with leishmaniasis in man*.
Phylum Euglenozoa Cavalier-Smith, 1981
Class Kinetoplastea Honigberg, 1963 emend., Vickerman, 1976
Subclass Metakinetoplastina Vickerman, 2004
Order Trypanosomatida Kent, 1880 stat. nov. Hollande, 1952
Family Trypanosomatidae Doflein, 1951
Subfamily Leishmaniinae, Maslov & Lükes 2012 emend Shaw, Teixeira and Camargo 2016
Type species of the genus Leishmania donovani (Laveran & Mesnil 1903)
Genus Leishmania Ross 1908
Subgenus L. (Leishmania) Safjanova 1982
Type species: Leishmania (Leishmania) donovani (Laveran Mesnil 1903)
L. (.L) donovani * (Laveran Mesnil 1903); L. (L.) infantum (Nicolle 1908); L. (L.) archibald * Castellani and Chalmers 1919; L. (L.) tropica * (Wright, 1903) Lühe, 1906; L. (L.) aethiopica *Bray, Ashford Bray, Reference Zidkova, Cepicka, Votypka and Svobodova1983; L. (L.) major * Yakimoff & Schkolor; L. (L.) arabica * Peters et al. 1986; L. (L.) turanica Strelkova et al. 1990; L. (L.) gerbilli Wang et al. 1964; L. (L.) mexicana Biagi, 1953 emend Garnham 1962; L. (L.) amazonensis * Lainson & Shaw 1982; L. (L.) aristidesi Lainson and Shaw, Reference Lainson, Shaw, Lumsden and Evans1979; L. (L.) venzuelensis * Bonfante-Garrido 1980; L. (L.) pifanoi * Medina & Romero 1959, emend Medina & Romero, 1962; L. (L.) waltoni * Shaw, Pratlong & Dedet 2015; specific status requires confirmation L. (L.) garnhami * Scroza et al. 1979; L. (L.) forattinii Yoshida et al. 1993.
Subgenus L. (Viannia) Lainson and Shaw, Reference Lainson, Shaw, Peters and Killick-Kendrick1987
Type species: Leishmania (Viannia) braziliensis * Vianna, Reference Vianna1911, emend Matta 1916
L. (V.) braziliensis * Vianna, Reference Vianna1911; L. (V.) peruviana * Velez, 1913; L. (V.) guyanensis * Floch, 1954; L. (V.) panamensis * Lainson & Shaw, 1972; L. (V.) lainsoni * Silveira et al. 1987; L. (V.) shawi * Lainson et al. 1989; L. (V.) naiffi * Lainson & Shaw, 1989; L. (V.) lindenbergi * Silveira et al. 2002; L. (V.) utingensis Braga et al. 2003.
Subgenus L. (Sauroleishmania) Ranque 1973 emend. Safyanova 1982
Type species: L. (Sauroleishmania) L. (S.) tarentolae Wenyon 1921
L. (S.) adleri Heisch 1954; L. (S.) agamae David 1929; L. (S.) ceramodactyli Adler & Thoedor 1929; L. (S.) davidi Strong 1924; L. (S.) gulikae Ovezmuchammedov & Safjanova 1987; L. (S.) gymnodactyli Khodukin & Sofiev 1929; L. (S.) helioscopi Chodukin & Sofieff 1940; L. (S.) hemidactyli Mackie et al. 1923; L. (S.) hoogstraali McMillan 1965; L. (S.) nicollei Chodukin & Sofieff 1940; L. (S.) phrynocephali Chodukin & Sofieff 1940; L. (S.) platycephala Telford 2008; L. (S.) senegalensis Ranque 1973; L. (S.) sofieffi Markov et al. 1964; L. (S.) tarentolae Wenyon 1921; L. (S.) zmeevi Andruchko & Markov 1955; L. (S.) zuckermani Paperna et al. 2011; L. (S.) sp., I Telford 1979; L. (S.) sp., II Telford 1979.
L. (S.) henrici † Leger 1918; L. (S.) chamaeleonesis † Wenyon 1921: † Most probably intestinal flagellates as parasites were in the cloaca
Subgenus L. (Mundinia) Shaw, Camargo and Teixeira 2016
Type species: L. (Mundinia) enriettii (Muniz and Medina, Reference Muniz and Medina1948)
L. (M.) enriettii (Muniz and Medina, Reference Muniz and Medina1948) L. (M.) martiniquensis * (Desbois et al. Reference Desbois, Pratlong, Quist and Dedet2014; Syn. L. siamensis), L. (M.) spp. Kangaroo [MMAC/AU/2004/Roo1]; L. (M.) spp. * Ghana [MHOM/GH/2012/GH5 (LV757)]; L. (M.) spp. * Trang, Thailand [MHOM/TH/2010/PVM2].
Genus Porcisia Shaw, Camargo and Teixeira 2016
Type species: Porcisia hertigi (Herrer, Reference Herrer1971)
P. hertigi (Herrer, Reference Herrer1971); P. deanei (Lainson and Shaw, Reference Lainson and Shaw1977 emend., Lainson and Shaw, Reference Lainson, Shaw, Peters and Killick-Kendrick1987)
Genus Endotrypanum Mesnil and Brimont, Reference Mesnil and Brimont1908
Type species: Endotrypanum schaudinni Mesnil and Brimont, Reference Mesnil and Brimont1908
E. schaudinni Mesnil and Brimont, Reference Mesnil and Brimont1908; E. monterogeii Shaw, Reference Shaw1969; E. colombiensis * (Kreutzer et al.1991); E. equatorensis (Grimaldi Junior et al.Reference Grimaldi Junior, Kreutzer, Hashiguchi, Gomez, Mimory and Tesh1992); E. herreri (Zeledon, Ponce, Murillo,1979)
Genus Zelonia Shaw, Camargo and Teixeira 2016
Type species: Z. costaricensis (Yurchenko et al. Reference Yurchenko, Lukes, Jirku, Zeledon and Maslov2006)
Z. costaricensis, strain G755 (Noyes et al. Reference Noyes, Pratlong, Chance, Ellis, Lanotte and Dedet2002), TCC169E, 504 and 2696 (present study)
Genus Novymonas Kostygov and Yurchenko 2016
Type species: Novymonas esmeraldas Votýpka, Kostygov, Maslov and Lukeš 2016
N. esmeraldas
Concluding remarks
Up to now the phylogenetic robustness of the Leishmaniinae has been based on the use of a limited number of gene sequences. Recently Harkins et al. (Reference Harkins, Schwartz, Cartwright and Stone2016) used a bioinformatics’ method employing 200 000 informative sites across the genomes of 13 species of Leishmaniinae that included three L. (Leishmania) species, 5 L. (Viannia) species, L. (S.) adleri, L. (M.) enriettii, two Porcisia species and Endotrypanum. The validity of the genera Porcisia and Endotrypanum is supported by Harkins et al. (Reference Harkins, Schwartz, Cartwright and Stone2016) who showed that their origin predated that of the four Leishmania subgenera. These same authors showed that using a relaxed molecular clock that dixenous leishmanine radiation began around 80 mya. This agrees with a previous hypothesis (Shaw, Reference Shaw1997) that had suggested that a monoxenous insect flagellate established itself in mammals some 90 mya and that after this the now dixenous line radiated in the evolving mammalian orders. In essence, this means that the leishmanine trypanosomatids evolved in mammals from an insect parasite and not from a reptilian parasite, in agreement with phylogenetic inferences (Croan et al. Reference Croan, Morrison and Ellis1997; Noyes et al. Reference Noyes, Pratlong, Chance, Ellis, Lanotte and Dedet2002; Pothirat et al. Reference Pothirat, Tantiworawit, Chaiwarith, Jariyapan, Wannasan, Siriyasatien, Supparatpinyo, Bates, Kwakye-Nuako and Bates2014; Harkins et al. Reference Harkins, Schwartz, Cartwright and Stone2016).
For many years, leishmaniasis’ vectors worldwide were considered to be Phlebotomus sand flies. This position became untenable as the true genetic diversity of the subfamily Phlebotominae became evident. It is now accepted that, with perhaps the exception of L. (Mundinia) parasites, the leishmaniasis’ vectors are phlebotomine sand flies and not Phlebotomus sand flies. Similarly, our greater understanding of the genetic complexity of the parasites causing leishmaniasis means that it is more correct to say that the leishmaniases are caused by leishmanine parasites rather than Leishmania parasites.
SUPPLEMENTARY MATERIAL
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182016002092
ACKNOWLEDGEMENTS
We are grateful to many colleagues that contributed with trypanosomatid samples and to Marta Campaner for cultures of the organisms characterized in this study, and to Carmen S. A. Takata and Tânia E. Matsumoto for assistance in DNA sequencing.
FINANCIAL SUPPORT
This work was supported by grants from the following national and state Brazilian funding agencies CNPq (PROTAX and PROSUL) and CAPES (PNIPB) to M. M. G. T. and E. P. C., and FAPESP to M. G. S. and J. J. S; and O. A. E. was supported by a Panamanian SENACYT Ph.D. fellowship.






