Introduction
Myxozoans are the cnidarian parasites that have been reported from fish inhabiting freshwater, brackish water and marine environments (Lom and Dyková, Reference Lom and Dyková2006; Gürkanlı et al., Reference Gürkanlı, Okkay, Çiftci, Yurakhno and Özer2018; Okkay and Özer, Reference Okkay and Özer2020). Description of myxozoan parasites is mainly based on myxospore morphology and according to this traditional criteria, species of the genus Ortholinea Shulman, Reference Shulman and Bykhovskaya-Pavlovskaya1962 have spherical or subspherical myxospores that are lightly flattened or tapered posterior parallel to the sutural plane, containing 2 polar capsules, subspherical or pyriform, and some species have surface stripes (Lom and Dyková, Reference Lom and Dyková2006). However, taxonomic placement based solely on morphological criteria has been proven to be artificial and molecular data of the SSU rRNA gene provided more accurate allocations in the taxonomy of myxozoan parasites (Rangel et al., Reference Rangel, Rocha, Casal, Castro, Severino, Azevedo, Cavaleiro and Santos2017). Ortholinea is probably a genus known to have ancestors reviving marine habitats and it has recently been transferred from Ortholineidae to Myxobilatidae due to phylogenetic proximity (Karlsbakk et al., Reference Karlsbakk, Kristmundsson, Albano, Brown and Freeman2017). This genus is represented by 26 species worldwide including 4 species that have been reported from marine fishes in the Black Sea coasts of Türkiye (Lom and Dyková, Reference Lom and Dyková1992; Karlsbakk, Reference Karlsbakk, Costello, Emblow and White2001, Rangel et al., Reference Rangel, Rocha, Borkhanuddin, Cech, Castro, Casal, Azeveda, Severino, Szekely and Santos2014, Reference Rangel, Rocha, Castro, Severino, Casal, Azevedo, Cavaleiro and Santos2015, Reference Rangel, Rocha, Casal, Castro, Severino, Azevedo, Cavaleiro and Santos2017; Özer et al., Reference Özer, Özkan and Yurakhno2015a, Reference Özer, Özkan, Güneydağ and Yurakhno2015b; Gürkanlı et al., Reference Gürkanlı, Okkay, Çiftci, Yurakhno and Özer2018; Shin et al., Reference Shin, Jin, Sohn, Kim and Lee2023). Most of the Ortholinea species have been generally reported in the urinary bladder, but rarely in the kidney, gallbladder, and gill tissues of their host fishes (Rangel et al., Reference Rangel, Rocha, Borkhanuddin, Cech, Castro, Casal, Azeveda, Severino, Szekely and Santos2014, Reference Rangel, Rocha, Castro, Severino, Casal, Azevedo, Cavaleiro and Santos2015, Reference Rangel, Rocha, Casal, Castro, Severino, Azevedo, Cavaleiro and Santos2017; Gürkanlı et al., Reference Gürkanlı, Okkay, Çiftci, Yurakhno and Özer2018). In a recent study, Okkay and Özer (Reference Okkay and Özer2020), based on morphological criteria, reported Ortholinea orientalis from the urinary bladder of European anchovy, Engraulis encrasicolus (Linnaeus, 1758) and Pontic shad, Alosa immaculata Bennett, 1835, Ortholinea divergens from the kidney of grey wrasse, Symphodus cinereus (Bonnatterre, 1788) and Ortholinea sp. from the kidney of black goby, Gobius niger Linnaeus, 1758 collected from Sinop coasts of the Black Sea in Türkiye.
In the present study, we aimed to describe the phylogenetic peculiarities of above mentioned Ortholinea species and the description of possible new species among previously identified individuals based solely on myxospore morphology.
Materials and methods
Fish sampling and parasitological examination
In the present study, a total of 103 specimens of round goby Neogobius melanostomus (Pallas, 1814) were collected from a fisherman in the Sinop coast (42° 05′ 68″ N, 35° 10′ 55″ E) of the Black Sea, Türkiye, in the period September 2017–December 2019. Gills, fins, skin, urinary bladder, kidney, gall bladder, liver, intestine, smooth muscles and gonads of each fish species were investigated for the presence of Ortholinea parasites. Moreover, previously alcohol-preserved urinary bladder and kidney tissues of Gobius niger, Symphodus cinereus and Engraulis encrasicolus were re-investigated for Ortholinea myxospores by Okkay and Özer (Reference Okkay and Özer2020). Myxospores of Ortholinea were examined and photographed with an Olympus microscope (BX53) equipped with a digital camera (DP50), at 400 × and 1000 × magnifications and Nikon (H550S) with DIC attachment at the Faculty of Fisheries and Aquatic Sciences in Sinop, Türkiye. Measurements were based on 20 fresh myxospores from N. melanostomus and 20 alcohol-preserved myxospores from E. engrasicolus, and morphological terminology and definitions are explained by Lom and Dyková (Reference Lom and Dyková1992). All measurements are given with mean values ± standard deviation and min–max values in parentheses. The calculation of prevalence values (%) follows the definition by Bush et al. (Reference Bush, Lafferty, Lotz and Shostak1997), and the density values were semiquantitatively evaluated by applying a scale from ‘1 + ’ representing the lowest 1 + and ‘ + + + + + +’ representing the highest 6 + density, a methodology modified from 200 × magnification by Gürkanlı et al. (Reference Gürkanlı, Okkay, Çiftci, Yurakhno and Özer2018). The density of infection categorized according to the mean and range of myxosporean parasites in parentheses were determined as 1 + (1–9), 2 + (10–19), 3 + (20–29), 4 + (30–39), 5 + (40–49) and 6 + (>50).
Molecular analyses
To extract total genomic DNA from Ortholinea-infected host tissues of Gobius niger, Symphodus cinereus and E. engrasicolus of Okkay and Özer (Reference Okkay and Özer2020) and Neogobius melanostomus of the present study, an Invitrogen PureLink® Genomic DNA Mini Kit (USA) was employed. Extractions were performed according to the manufacturer's instructions and the DNA was hidden at −20 °C before use. To construct phylogenies, SSU rDNA was used as molecular marker. Amplification of the gene was carried out using primers of both MyxospecF (Fiala, Reference Fiala2006) and 18r (Whipps et al., Reference Whipps, Adlard, Bryant, Lester, Findlay and Kent2003). PCR amplifications were made using a Techne (TC-Plus) thermal cycler with the following procedure; 3 min of initial denaturation at 95 °C, followed by 40 cycles of denaturation at 94 °C for 1 min, annealing at 51 °C (−0.1 °C per cycle) for 1 min, and extension for 1.5 min at 72 °C. The final extension was facilitated at 72 °C for 10 min. For all PCR amplifications, a 50 μl reaction mixture was prepared with GoTaq® Colorless Master Mix 2× (Promega, Madison, U.S.A.), 0.5 pmol (final concentration) of each primer (Oligomer), genomic DNA<1 μg and sterile ddH2O (up to 50 μl). For electrophoresis (to check both genomic DNA and PCR products), 1% agarose gel prepared in 1× TBE buffer was used and visualizations of the gels were performed with the photo print imaging system (Vilber Lourmat, France). Nucleotide sequencings were performed commercially by Macrogen-Europe from both strands with the same primers used for PCR amplifications. Verification and assemblage of nucleotide sequencings were made with Software BioEdit (Hall, Reference Hall1999). For phylogenetic constructions, a data set was prepared in the light of available literature and also according to the results of BLAST (Basic Local Alignment Search Tool, https://blast.ncbi.nlm.nih.gov/Blast.cgi) search. Multiple nucleotide sequence alignment of the data set was performed with ClustalX (Thompson et al., Reference Thompson, Gibson, Plewniak, Jeanmougin and Higgins1997). Phylogenetic constructions were made using GTR + I + G (I: 0.312; G: 0.581) and TPM2 + I + G (I: 0.285; G: 0.533) evolutionary models that have been suggested by Akaike information criterion (Akaike, Reference Akaike1974) and Bayesian information criterion tests, respectively. These tests were performed using jModelTest v. 0.1 package program (Guindon and Gascuel, Reference Guindon and Gascuel2003; Posada, Reference Posada2008). To construct phylogenies, maximum-likelihood (ML), neighbour-joining (NJ) (Saitou and Nei, Reference Saitou and Nei1987) and maximum-parsimony (MP) (Eck and Dayhoff, Reference Eck and Dayhoff1966; Fitch, Reference Fitch1977) methods were applied. Software program PAUP* v. 4.0b10 was implemented using both NJ and MP analyses (Swofford, Reference Swofford1998). A heuristic search approach with a TBR swapping algorithm (10 incidental repetitions) was applied for MP analysis. The software program PhyML 3.0 (Guindon and Gascuel, Reference Guindon and Gascuel2003) was employed for ML analysis. Bootstrap tests were performed with 10 000 replicates for NJ and 1000 replicates for MP and ML analyses (Efron, Reference Efron1982; Felsenstein, Reference Felsenstein1985). BioEdit was used to resolute binary nucleotide sequence similarities. Genetic distances among genotypes, corrected in accordance with the previously mentioned evolutionary models, were computed using PAUP.
Our new 18S rDNA genotypes have been deposited in GenBank under accession numbers OR884251-OR884254 (Table 1).
Table 1. Source information of Ortholinea isolates obtained in this study and Myxozoan species obtained from NCBI (given with references) for phylogenetic analyses

Results
In the present study, only the kidney of the round goby, N. melanostomus was found to be infected by a species of the genus Ortholinea Shulman, Reference Shulman and Bykhovskaya-Pavlovskaya1962 (Myxozoa: Ortholineidae) based on the following distinguishing characteristics of the genus: (1) myxospore morphology, (2) dimensions of myxospore length and width, (3) morphology of polar capsules and dimensions of their length and width, (4) molecular level peculiarities. Myxospore morphology and morphometry of the examined parasites corresponded well with O. gobiusi that was presented in its previous reports from the same fish host. On the other hand, the infected tissue samples previously used by Okkay and Özer (Reference Okkay and Özer2020) were revisited for the Ortholinea species reported from E. engrasicolus, and a more detailed investigation of 20 previously alcohol-preserved myxospores from infected urinary bladder together with molecular evaluation revealed a new Ortholinea species namely O. hamsiensis n. sp. The details of taxonomic summary, morphology and infection indices of both species are provided below;
Taxonomic summary of Ortholinea gobiusi Naidenova, Reference Naidenova1968
Phylum: Cnidaria Hatschek, 1888
Subphylum: Endocnidozoa Schuchert, 1996
Class: Myxozoa Grasse, 1970
Subclass: Myxosporea Bütschli, 1881
Order: Bivalvulidae Shulman, 1959
Suborder: Variisporina Lom and Noble, 1984
Family: Ortholineidae Lom & Noble, 1984
Genus: Ortholinea Shulman, Reference Shulman and Bykhovskaya-Pavlovskaya1962
Name: Ortholinea gobiusi Naidenova, Reference Naidenova1968 (Fig. 1A,B)
Host: Neogobius melanostomus (Pallas, 1814) round goby
Locality: Sinop coasts of the Black Sea, Türkiye (42° 05′ 68″ N, 35° 10′ 55″ E)
Prevalence of infection: 4.8% (6 females out of 103)
Density of infection: 1–5 individuals in the field of view (1 + ) (200 × magnification)
Description of myxospores: The characteristic feature is a round
or mostly ovoid myxospores, and the myxospore surfaces have external striations (Fig. 1C). Two polar capsules of the parasite are rounded and positioned almost in opposite directions. Parasite individuals were detected in the presporogonic and sporogonic stages (Fig. 1D). All morphometric data of fresh myxospores are provided in Table 2.
Taxonomic summary of Ortholinea hamsiensis n. sp. (Fig. 1E–H)
Phylum: Cnidaria Hatschek, 1888
Subphylum: Endocnidozoa Schuchert, 1996
Class: Myxozoa Grasse, 1970
Subclass: Myxosporea Bütschli, 1881
Order: Bivalvulidae Shulman, 1959
Suborder: Variisporina Lom and Noble, 1984
Family: Ortholineidae Lom & Noble, 1984
Genus: Ortholinea Shulman, Reference Shulman and Bykhovskaya-Pavlovskaya1962
Type host: Engraulis encrasicolus (Linnaeus, 1758) European anchovy
Type locality: Sinop coasts of the Black Sea, Türkiye (42° 02’ 68” N, 35° 10’ 55”E)
Prevalence of infection: 1.4% (2 females out of 72)
Density of infection: 1–5 individuals in the field of view (1 + ) (200 × magnification)
Type material: One holotype (MyxoOH 2023.1) and 1 paratype (MyxoOH 2023.2) were hidden at the Faculty of Fisheries and Aquatic Sciences Parasitological Collection of the Sinop University, Sinop, Türkiye
Etymology: Parasite species is derived from the local fishery name in Türkiye ‘hamsi’ of the host, E. engrasicolus
Description
Myxospores of Ortholinea hamsiensis n. sp.
Immature and developing myxospores are oviform and slightly tapering down to the tip in the frontal view. Mature myxospores are subspherical with slight tapering down to the less pronounced tip in the frontal view and subspherical in the sutural view (Fig. 1E,F,G,H) with measurements of 9.1 ± 0.25 (8.8–9.9) μm in length, 9.2 ± 0.11 (8.9–9.4) μm in thickness and 8.4 ± 0.33 (8.2–9.1) μm in width. Two polar capsules equal in size, located nearly at the higher 1/3 level of the myxospores, measuring 3.1 ± 0.11 (3.0–3.3) μm in length and 2.7 ± 0.11 (2.6–2.9) μm in width. The polar tubule had 3–4 coils.

Figure 1. A fresh spore of Ortholinea gobiusi, (A) frontal view, (B) sutural view, (C) surface ridges indicating the presence of striations, (D) developmental sporogonic stage with developing myxospores; A myxospore of O. hamsiensis n. sp. observed by DIC objective, E. frontal view, F. sutural view; hand drawing of O. hamsiensis n. sp. G. frontal view, H. sutural view.
Table 2. Site of infection, hosts, geographical localities and dimensions (μm, ± SD) of species of the genus Ortholinea found in marine fish

PTC, number of polar tubule coils. –: no data.
Differential diagnosis of Ortholinea hamsiensis n. sp.
A comparison of myxospore characteristics of presently reported new species with those of the original description of O. gobiusi from grass goby Zosterisessor ophiocephalus by Naidenova (Reference Naidenova1968) shows that myxospores of the new species in the present study are more subspherical and slightly tapering down to the less pronounced tip in frontal view, while myxospores of O. gobiusi are oviform and sharply tapering down to a pronounced tip with smaller myxospore dimensions. The same situation occurs when compared with O. gobiusi from the same fish host N. melanostomus inhabiting the same sampling locality (Özer et al., Reference Özer, Özkan, Güneydağ and Yurakhno2015b). Myxospores of Ortholinea divergens from Parablennius sanguinolentus (Özer et al., Reference Özer, Özkan, Güneydağ and Yurakhno2015b) are more rounded than those observed in the new species. The present species also differs from O. divergens in having smaller polar capsule dimensions. The shapes of the polar capsules of the new species and O. mullusi have different appearances, oval in the previous and pyriform in the latter species.
In the previous study by Okkay and Özer (Reference Okkay and Özer2020), an Ortholinea species was found in the urinary bladder of E. encrasicolus, and based on the comparisons of myxospore morphology and morphometry with the previous wide range of host and geographical locality reports in the literature, they identified it as O. orientalis. However, a more detailed examination of these previously alcohol-preserved infected tissue myxospores of Ortholinea species, namely O. hamsiensis n. sp. in the present study, revealed that there were some differences when compared with the previous reports of O. orientalis from other host species inhabiting a wide range of geographical localities. The myxospores of the presently reported new species are subspherical with slight tapering down to the less pronounced tip in frontal view and O. orientalis has subspherical to triangular myxospores, with the broadest anterior and pointed posterior end together with a conspicuous triangular intercapsular process occurs at the anterior end of the myxospore (Karlsbakk and Køie, Reference Karlsbakk and Køie2011). Myxospore dimensions of the new species are smaller than those of O. orientalis from navaga Eleginus gracilis (Tilesius, 1810) and Eleginus nawaga (Walbaum, 1792) but larger than those of O. orientalis from Atlantic herring Clupea harengus Linnaeus, 1758, Pacific herring, Clupea pallasi Valenciennes, 1847, Alaska Pollock, Gadus chalcogrammus Pallas, 1814, red mullet, Mullus barbatus ponticus Essipov, 1927, Black Sea shad, Alosa tanaica (Grimm, 1901) (Shulman and Shulman-Albova, Reference Shulman and Shulman-Albova1953; Aseeva, Reference Aseeva2000; Karlsbakk and Køie, Reference Karlsbakk and Køie2011; Özer et al., Reference Özer, Özkan and Yurakhno2015a). The polar capsules of the new species are larger than those of O. orientalis from C. pallasii and E. nawaga, O. labracis from the European seabass, Dicentrarchus labrax (Linnaeus, 1758), O. scatophagi from the spotted scat, Scatophagus argus (Linnaeus, 1766) but, smaller than those of O. mullusi from M. barbatus ponticus, O. auratae from the gilthead seabream, Sparus aurata (Linnaeus, 1758), O. alata from the northern butterflyfish, Chaetodon rainfordi McCulloch, 1923, and O. striateculus from silver fish, Leptatherina presbyteroides (Richardson, 1843) (Shulman and Shulman-Albova, Reference Shulman and Shulman-Albova1953; Kent and Moser, Reference Kent and Moser1990; Su and White, Reference Su and White1994; Özer et al., Reference Özer, Özkan and Yurakhno2015a; Rangel et al., Reference Rangel, Rocha, Casal, Castro, Severino, Azevedo, Cavaleiro and Santos2017; Gürkanlı et al., Reference Gürkanlı, Okkay, Çiftci, Yurakhno and Özer2018; Chandran et al., Reference Chandran, Zacharia and Sanil2020). Ortholinea saudii from marbled spinefoot Siganus rivulatus (Abdel-Baki et al., Reference Abdel-Baki, Soliman, Saleh, Al-Quraishy and El-Matbouli2015) own too large polar capsules and myxospores compared to the new species in the present study.
Molecular analyses
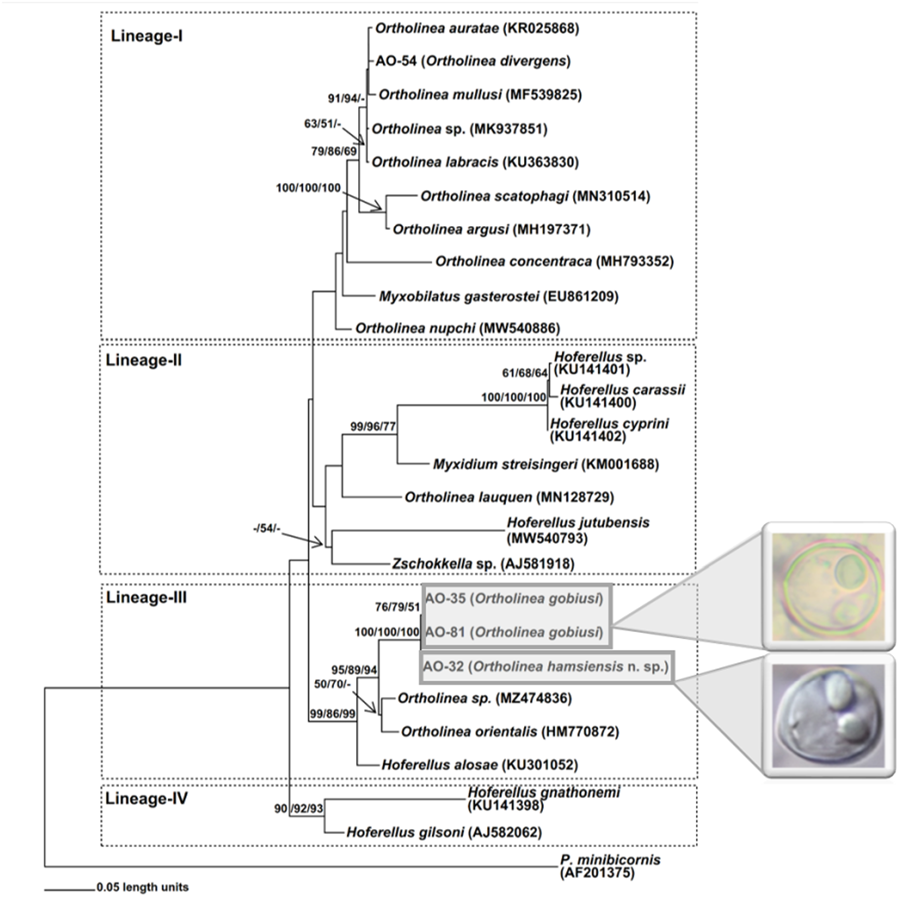
A total of 4 isolates of Ortholinea including AO-81 from N. melanostomus, AO-35 from G. niger, AO-54 from S. cinereus and AO-32 from E. encrasicolus were studied for molecular analysis. As a result of nucleotide sequencings, approximately 1700 bp of SSU rDNA were obtained from myxosporean specimens observed in infected host fish tissues. The codes for myxosporean genotypes obtained from different host fishes are given in Table 1. Concordant with the initial microscopic observations, BLAST searches associated all 4 genotypes obtained in this study with the genus Ortholinea, and thus, a data set was constituted with SSU rDNA sequences of available Ortholinea species together with some allied myxosporean species which are readily available in GenBank (Table 1). Of the 26 binomial species within the genus Ortholinea, only 11 of them had genetic records (SSU rDNA genotypes) in GenBank, thus, we were able to perform a genetic comparison with only this limited number of species. In addition, we also had to ignore 2 of the genetically available species, O. saudii and O. amazonica, due to their short SSU rDNA sequences in GenBank which caused a serious loss of genetic information in the aligned data set. As Ortholinea represents a paraphyletic lineage, we have also included several related species from the genera Myxobilatus, Hoferellus and Myxidium, in our data set. Phylogenetic analyses were performed over 981 (excluding gaps) aligned nucleotides with 392 segregated characters (509 substitution mutations). The ML trees created using GTR + I + G (I: 0.312; G: 0.581) and TPM2 + I + G (I: 0.285; G: 0.533) models were topologically similar, however, the ML tree with the initial model have suggested higher bootstrap values, thus considered in this study. The same situation was also observed in the NJ analysis. The Parsimony analysis that was conducted with 202 synapomorphic characters produced 48 single most parsimonious trees with 801 steps (CI: 0.635456; RI: 0.682609 and HI: 0.364544). In this study, the ML tree created using GTR + I + G model is given, additionally, bootstrap values obtained from NJ (with GTR + I + G model) and MP analyses have also been stated on each related node (Fig. 2).

Figure 2. ML phylogram based on 18S rDNA nucleotide sequences of Ortholinea isolates obtained in this study (AO-32, AO-35, AO-54 and AO-81) and some closely related Myxozoa species downloaded from NCBI (given with GenBank accession numbers). On each related node bootstrap values (⩾50%) obtained from ML, NJ and MP analyses are stated with the given order. The tree is rooted with Parvicapsula minibicornis (Kent et al., Reference Kent, Khattra, Hedrick and Devlin2000).
On the phylogenetic trees 4 main lineages (-I, -II, -III and -IV) appeared (Fig. 2). However, the positioning of certain species, such as Ortholinea nupchi, Myxobilatus gasterostei, Ortholinea concentraca, Hoferellus jutubensis and Zschokkella sp. (AJ581918), displayed discrepancies among the trees generated using the ML, NJ and MP algorithms. Therefore, the placement of these species within a particular lineage lacks robust support, signified by the absence of bootstrap values in the phylogenetic trees. Conversely, the new genotypes examined in this study consistently demonstrated stable phylogenetic relationships within the respective lineages they were positioned.
Notably, the first 3 lineages proved to be paraphyletic, encompassing Ortholinea species along with those from other myxozoan genera such as Myxobilatus (Lineage-I), Myxidium, and Zschokkella (Lineage-II), as well as Hoferellus (Lineages-II and -III). On the other hand, Lineage-IV presented as monophyletic, comprising 2 Hoferellus species.
The genotype AO-54, originating from S. cinereus and initially identified as Ortholinea divergens based on morphological criteria in Okkay and Özer (Reference Okkay and Özer2020), has placed within Lineage-I as sister to Ortholinea auratae (KR025868). The nucleotide sequence similarity and genetic distance between AO-54 and O. auratae were determined as 97.6% and 0.0337, respectively (see Supplementary Table). In all phylogenetic trees, Ortholinea mullusi consistently emerged as a sister to the lineage mentioned earlier, exhibiting 94.5% nucleotide sequence similarity and 0.05911 genetic distance with AO-54. On the other hand, bootstrap analyses unveiled a polytomy and suggested a single bootstrap value for the lineage encompassing AO-54, O. auratae and O. mullusi. For this very reason no bootstrap values were assigned to support the intralineage relationships within this group. Additionally, O. labracis, O. scatophagi, O. argusi, O. concentraca, O. nupchi, Ortholinea sp. (MK937851) and Myxobilatus gasterostei have also appeared within Lineage-I.
The other 3 novel genotypes obtained in this study, AO-32, AO-35 and AO-81, along with genotypes, O. orientalis (HM770872), Ortholinea sp. (MZ474836), and Hoferellus alosae (HM770872) collectively formed Lineage-III. The intraspecific relationships within this lineage appeared as consistent, as evidenced by significant bootstrap values observed at all nodes. Notably, genotypes AO-81 and AO-35, originating from N. melanostomus and G. niger, respectively, both belonging to the Gobiidae family, exhibited the highest nucleotide sequence similarity at 99.9%. Additionally, they displayed the lowest genetic distance, recorded at 0.00062, positioning them as closely related sisters. Genotype AO-32, which was previously designated as O. orientalis depending on morphological criteria (Okkay and Özer, Reference Okkay and Özer2020), appeared as closely related to the group mentioned earlier rather than O. orientalis genotype HM770872. In accordance with this, the nucleotide sequence similarity and genetic distance between AO-32 and AO-81 were 98.7% and 0.01120, additionally were 98.8% and 0.01057 between AO-32 and AO-35. However, contrasting figures emerged with AO-32 and the O. orientalis genotype HM770872 where these values were 90.2% and 0.09983 (Fig. 2, Supp. Table).
Discussion
Ortholinea (Shulman, Reference Shulman and Bykhovskaya-Pavlovskaya1962), the target myxozoan genus in this study, is composed of coelozoic 26 binominal parasite species that infect mainly the urinary bladder of mostly marine and rarely freshwater fishes (Lom and Dyková, Reference Lom and Dyková2006; Shin et al., Reference Shin, Jin, Sohn, Kim and Lee2023). Despite its limited biological diversity when compared with some other myxosporean genera such as Myxobolus, Myxidium, etc., reported species from fishes of this genus reveal a worldwide distribution (Lom and Dyková, Reference Lom and Dyková1992; Rangel et al., Reference Rangel, Rocha, Borkhanuddin, Cech, Castro, Casal, Azeveda, Severino, Szekely and Santos2014, Reference Rangel, Rocha, Castro, Severino, Casal, Azevedo, Cavaleiro and Santos2015, Reference Rangel, Rocha, Casal, Castro, Severino, Azevedo, Cavaleiro and Santos2017; Gürkanlı et al., Reference Gürkanlı, Okkay, Çiftci, Yurakhno and Özer2018; Shin et al., Reference Shin, Jin, Sohn, Kim and Lee2023). Concordant with this data, 4 Ortholinea species (O. divergens, O. gobiusi, O. orientalis, O. mullusi) have been reported from the Black Sea coasts of Türkiye thus far (Özer et al., Reference Özer, Özkan and Yurakhno2015a, Reference Özer, Özkan, Güneydağ and Yurakhno2015b; Gürkanlı et al., Reference Gürkanlı, Okkay, Çiftci, Yurakhno and Özer2018).
Until the end of the 20th century, species identification processes within the genus Ortholinea have been solely based on morphological and morphometric features of myxospores just like in other myxosporean genera. However, only these morphological characters are limited in numbers and inadequate in variations, thus they are mostly insufficient to reveal the true genealogy of myxozoa (Fiala et al., Reference Fiala, Bartošová-Sojková, Whipps, Okamura, Gruhl and Bartholomew2015). Moreover, molecular phylogenetic studies depending on nucleotide sequences of SSU rDNA gene that were published in the last 2 decades clearly revealed the incongruences between molecular phylogeny and myxospore morphology-based classification systems in most myxosporean genera such as Myxobolus, Henneguya, Sphaerospora, Myxidium, Zschokkaella and Chloromyxum. All these genera appeared as polyphyletic or paraphyletic taxa in the phylogenetic trees (Kent et al., Reference Kent, Andree, Bartholomew, El-Matbouli, Desser, Devlin, Feist, Hedrick, Hoffmann, Khattra, Hallet, Lester, Longshaw, Palenzuala, Siddall and Xiao2001; Fiala, Reference Fiala2006). Additionally, in a comprehensive study including the genera Myxobolus, Kudoa, Henneguya, Chloromyxum, Sphaerospora, Sphaeromyxa and Myxidium, it has been concluded that just restricted morphological characters are concordant with phylogeny obtained from SSU rDNA data because of the plasticity in myxospore morphology (Fiala and Bartošová, Reference Fiala and Bartošová2010). Likewise, the genus Ortholinea appeared as another paraphyletic myxosporean genus in phylogenetic studies since some species of Acauda, Myxobilatus, and Hoferellus genera appeared within the same lineage together with Ortholinea species (Rangel et al., Reference Rangel, Rocha, Borkhanuddin, Cech, Castro, Casal, Azeveda, Severino, Szekely and Santos2014, Reference Rangel, Rocha, Casal, Castro, Severino, Azevedo, Cavaleiro and Santos2017; Alama-Bermejo and Hernandez-Orts, Reference Alama-Bermejo and Hernandez-Orts2018; Alama-Bermejo et al., Reference Alama-Bermejo, Viozzi, Waicheim, Flores and Atkinson2019; Chandran et al., Reference Chandran, Zacharia and Sanil2020). For this very reason, in today's systematic concept, molecular data are indispensable for the diagnosis of myxozoan specimens and the identification of new species. However, despite its necessity, the identification of the most valid Ortholinea species is still solely dependent on the morphological features and only 11 nominal species have molecular data (SSU rDNA nucleotide sequence) in GenBank. In this context, this study aims to obtain and phylogenetically analyse the SSU rDNA genotypes of some Black Sea-originated Ortholinea specimens reported in a previous study (Okkay and Özer, Reference Okkay and Özer2020) in addition to some original Ortholinea specimens obtained from Neogobius melanostomus.
Ortholinea gobiusi is one of the valid species that is lacking molecular data in the genus Ortholinea. This species was first identified by Naidenova (Reference Naidenova1968) from the urinary bladder of Gobius ophiocephalus in the northern Black Sea and for the next nearly 50 years no record was given for this species until 2015 when Özer et al., reported O. gobiusi from the urinary bladder of Neogobius melanostomus (Pallas, 1814) collected from the Sinop coast of Türkiye (southern Black Sea). According to the morphological features and morphometric data of myxospores, researchers identified and reported this species with 4.1% prevalence out of 76 fish samples (Özer et al., Reference Özer, Özkan, Güneydağ and Yurakhno2015b). Five years later, Okkay and Özer (Reference Okkay and Özer2020) reported Ortholinea specimens similar to O. gobiusi from the kidney of another gobiid Gobius niger. However, they did not designate these specimens as O. gobiusi but named them as Ortholinea sp. particularly because of the differences in the polar capsule dimensions. As mentioned, none of these studies included molecular data. In this study, however, we identified some Ortholinea specimens from N. melanostomus using both morphological and molecular techniques (AO-81). Additionally, we also analysed O. gobiusi specimens (AO-35), previously reported as Ortholinea sp. from G. niger in Okkay and Özer (Reference Okkay and Özer2020) from a molecular phylogenetic perspective. The morphological and morphometric data (Table 2) of the new Ortholinea specimens obtained from N. melanostomus were consistent with the O. gobiusi features reported by Özer et al. (Reference Özer, Özkan, Güneydağ and Yurakhno2015b) and Naidenova (Reference Naidenova1968). Although the polar capsules of the Ortholinea specimens from G. niger were relatively smaller as mentioned earlier, other morphological and morphometric features were fitting well with O. gobiusi descriptive features (Naidenova, Reference Naidenova1968; Okkay and Özer, Reference Okkay and Özer2020). As a result of molecular analyses, these 2 Ortholinea specimens showed SSU rDNA genotypes with 99.9% nucleotide sequence similarity and 0.00062 genetic distance. This much identity and low genetic distance between SSU rDNA genotypes of specimens, AO-35 and AO-81, clearly indicates that they belong to the same species (O. gobiusi). Morphological data of AO-35 (Okkay and Özer, Reference Okkay and Özer2020) and AO-81 (obtained in this study) also supported this inference (Table 2). As a result, depending on molecular and morphological data we designated AO-81 as O. gobiusi. In this study, we have provided the first molecular data, SSU rDNA sequences, of O. gobiusi (genotypes AO-81 and AO-35) and thus completing the deficiency in the description of this species.
In the present study, O. gobiusi was found in the kidney of N. melanostomus and this new information about its site of infection makes a new contribution to our current knowledge about its tissue selection that is being solely reported from the urinary bladder of its gobiid fish hosts. The infection prevalence in this study was determined as 4.8% and this value is very similar to that of its previous report 4.1% from the urinary bladder of the host fish from the same locality by Özer et al. (Reference Özer, Özkan, Güneydağ and Yurakhno2015b).
Similar to O. gobiusi, another valid Ortholinea species that lacks molecular data is O. divergens. This species was initially identified and named as Sphaerospora divergens by Thélohan (Reference Thélohan1895) from the English Channel and subsequently transferred to the genus Ortholinea as the type species of the genus by Shulman (Reference Shulman and Bykhovskaya-Pavlovskaya1962). As can be expected from a relatively old species, there is no type sample available for comparison. And over the years O. divergens reported from diverse geographical locations and hosts including; Reinhardtius hippoglossoides off the Labrador and Barents Sea, the North Atlantic Ocean, and the Bering Sea, the North Pacific Ocean (Wierzbicka, Reference Wierzbicka1990a, Reference Wierzbicka1990b, Reference Wierzbicka1992), Reinhardtius platessoides and Hippoglossoides platessides in North Atlantic (Zubchenko, Reference Zubchenko1980, Reference Zubchenko1985), Aidablennius sphynx, Diplodus annularis, Lipophrys pavo (Syn. Salaria pavo), Liza aurata (Syn. Chelon auratus), Parablennius sanguinolentus, P. tentacularis, Symphodus roissali, S. ocellatus, S. cinereus and Salaria pavo in the northern Black Sea (Ukrainian coasts) (Yurakhno, Reference Yurakhno2009). This species has also been reported in the southern Black Sea (Turkish coasts) from P. sanguinolentus (Özer et al., Reference Özer, Özkan, Güneydağ and Yurakhno2015b) and S. cinereus (Okkay and Özer, Reference Okkay and Özer2020). In the present study, we phylogenetically analysed the nucleotide sequence of the SSU rDNA gene of O. divergens specimens (AO-54) from Okkay and Özer (Reference Okkay and Özer2020). As a result of phylogenetic analyses, O. divergens turned out as a sister to O. auratae on 97.6% nucleotide sequence similarity and 0.0337 genetic distance. These 2 species also revealed significant morphological differences such as O. divergens possessing round or ovoid myxospores and pyriform polar capsules while O. auratae myxospores were ellipsoidal and spherical. Additionally, only 2–3 developing spores were observed in the plasmodium of O. divergens, whereas the size of the glycocalyx-like sheet-covered plasmodium was quite large inhabiting numerous developing myxospores of O. auratae in Rangel et al. (Reference Rangel, Rocha, Borkhanuddin, Cech, Castro, Casal, Azeveda, Severino, Szekely and Santos2014). The glycocalyx-like sheet covering the plasmodia is a rather evident characteristic differing from the other species. As a result, here in the present study, we have provided the first molecular data of O. divergens for international databases. These data are particularly important for future molecular-based systematic studies concerning the genetic boundaries, diversity and systematic conflicts of the genus Ortholinea since this species is the type species of the genus.
The most commonly reported member of the genus Ortholinea is Ortholinea orientalis which was initially identified by Shulman and Shulman-Albova (Reference Shulman and Shulman-Albova1953) from Clupea harengus and Eleginus navaga obtained from the White Sea. Subsequently, it has been reported from several fish species classified within the families Clupeidae, Gadidae, Alosidae and Mullidae which were collected from diverse localities including Denmark, Japan Sea, Fars East Sea and the Black Sea (Aseeva, Reference Aseeva2000, Reference Aseeva2002; Karlsbakk and Køie, Reference Karlsbakk and Køie2011; Özer et al., Reference Özer, Özkan and Yurakhno2015a). As can be expected from a myxozoan species reported from such different hosts and localities, morphometric data given in these studies were quite diverse and this situation makes species boundaries of O. orientalis quite wide and thus questionable. To test whether this prediction is valid, in the present study, we phylogenetically analysed O. orientalis specimens (AO-32) obtained from E. encrasicolus, a member of another family within Clupeiformes, collected previously off the Sinop coast of the Black Sea by Okkay and Özer (Reference Okkay and Özer2020). Our results revealed that this prediction is valid, and AO-32 appeared as distantly related to O. orientalis genotype obtained from GenBank but instead was sister to O. gobiusi (Fig. 2). The nucleotide sequence similarity and genetic distance between AO-32 and O. orientalis specimen (HM770872) was only 90.2% and 0.09983 which were not enough to consider these specimens as a single species. Moreover, the nucleotide sequence similarities (98.7–98.8%) and genetic distances (0.01057–0.0112) between AO-32 and its sister species, O. gobiusi (AO-35 and AO-81), were also not sufficient to consider this specimen as O. gobiusi. The reason for this inference is; the intraspecific sequence similarities of valid Ortholinea species (O. orientalis: 99.6%; O. labracis: 100%; O. auratae: 99.8%; O. concentrica: 99.6%) are reported as higher than 99.5% (Gürkanlı et al., Reference Gürkanlı, Okkay, Çiftci, Yurakhno and Özer2018). Additionally, the morphometric differences in the myxospore lengths of AO-32 and O. gobiusi specimens also supported this inference (Okkay and Özer, Reference Okkay and Özer2020; Table 2). As a result, depending on both morphological and molecular phylogenetic evidences provided above, we suggest AO-32 as a new species namely Ortholinea hamsiensis n. sp.
In conclusion, significant results obtained in this study can be summarized as follows; (i) a novel myxosporean species, namely Ortholinea hamsiensis n. sp. have been identified from the urinary bladder of Engraulis engrasicolus, (ii) the first molecular records for Ortholinea divergens, the type species of this genus, and (iii) the first molecular records for O. gobiusi have been provided. With this new data, the missing molecular parts of the descriptions of these 2 species have been completed and phylogenetic relationships of these species with other Ortholinea species have been revealed.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182024000325.
Data availability statement
Sequence data is available on the NCBI GenBank database. All other necessary data are included in the article and its supplementary materials.
Author contributions
All authors designed and conducted laboratory work and all of them were involved in the manuscript and approved the final version.
Financial support
This study received no grant from a funding agency.
Competing interests
The authors declare that they have no conflict of interest.
Ethical standards
All applicable international, national and institutional guidelines for the care and use of animals were followed.