Introduction
The Amazon Basin has the largest number of fish in the world, with approximately 6000 known species (Reis et al., Reference Reis, Albert, Di Dario, Mincarone, Petry and Rocha2016); however, information on the parasitic biodiversity of this region is still limited compared to its ichthyofauna (Ferreira et al., Reference Ferreira, Silva, Araújo, Hamoy, Matos and Videira2020).
Among the common fishes in the Neotropical region, Leporinus friderici (Bloch, 1794), popularly known as threespot leporinus, adopts a migratory strategy and has a high ecological importance in the ecosystem due to its herbivorous habits. Farming of this species also serves as a source of income for local fishermen in Brazil. Threespot leporinus can be found in rivers in Suriname, the Amazon, Paraná and Paraguay basins (Silva et al., Reference Silva, Martins, Prado, Utsunomia, Foresti and Porto-Foresti2020).
Representatives of the class, Myxozoa Grassé, 1970 (Kyger et al., Reference Kyger, Luzuriaga-Neira, Layman, Milkewitz Sandberg, Singh, Huchon, Peri, Atkinson, Bartholomew, Yi Soojin and Alvarez-Ponce2021), are considered obligate parasites (Okamura et al., Reference Okamura, Gruhl, Reft, Okamura, Gruhl and Bartholomew2015) and are morphologically characterized by spores formed via valves connected by a suture line. The sporoplasm of this class contains polar capsules and spiral polar filaments (Fiala et al., Reference Fiala, Bartošová-Sojková, Whipps, Okamura, Gruhl and Bartholomew2015; Naldoni et al., Reference Naldoni, Maia, Correa, Silva and Adriano2018). Within the subclass, Myxosporea Bütschli, 1881, and order Bivalvulida Shulman, 1959, Henneguya Thélohan, 1892 is the second largest genus in terms of the number of described species, with approximately 254 species worldwide (Vieira et al., Reference Vieira, Narciso, de Azevedo and da Silva2021; Rangel et al., Reference Rangel, Santos and Rocha2023). In Brazil, approximately 72 species of Henneguya sp. have been identified (Eiras, Reference Eiras2002; Eiras and Adriano, Reference Eiras and Adriano2012; Vidal et al., Reference Vidal, Iannacone, Whipps and Luque2017; Rangel et al., Reference Rangel, Santos and Rocha2023), with approximately 20 found in the Amazon region (Velasco et al., Reference Velasco, Videira, Nascimento, Matos, Gonçalves and Matos2016; Abrunhosa et al., Reference Abrunhosa, Sindeaux-Neto, Hamoy and Matos2018; Naldoni et al., Reference Naldoni, Maia, Correa, Silva and Adriano2018; Zatti et al., Reference Zatti, Atkinson, Maia, Bartholomew and Adriano2018; Ferreira et al., Reference Ferreira, Silva, Araújo, Hamoy, Matos and Videira2020).
Only 1 microparasite species in the class, Myxozoa, has been identified in L. friderici, Henneguya friderici (Casal et al., Reference Casal, Matos and Azevedo2003). Therefore, this study sought to reveal a new microparasite species affecting L. friderici using histological, molecular biology and phylogenetic analyses.
Materials and methods
Host collection
Specimens of L. friderici (n = 26); weight, 41.5 ± 3.5 (38–45) g; length, 11.8 ± 1.8 (10–13.6) cm, were collected from the Tartarugalzinho river (coordinates of the sampling point 1: N 01°30′32.5″ W 050°55′10.3″, point 2: N 01°32′12.5″ W 050°48′31.1″, point 3: N 01°30′32.2″ W 050°55′09.9″ and point 4: N 01°30′32.4″ W 050°55′10.9″), Tartarugalzinho municipality, Amapá state. The fishes were collected from December 2020 to November 2021 using fishing gear, such as a 20 mm gillnet between knots, and assistance from local fishermen. All analyses and collection procedures were approved by the Animal Use Ethics Committee of the Universidade Federal Rural da Amazônia (no. 8323110522) and registered in the Biodiversity Authorisation and Information System (SISBIO/ICMBIO; license number, 27119).
Live specimens were packaged and transported to the Laboratory of Morphophysiology and Animal Health at Universidade do Estado do Amapá (LABMORSA/UEAP) under artificial aeration supplied by electric pumps. The specimens were allowed to acclimate to the environment and were stored in aquariums comprising electric pumps and filters until morphological analysis and parasite collection.
Morphological analysis and parasite collection
The specimens were anaesthetized with tricaine methosulfonate (MS222 SIGMA) and subjected to neural myelotomy before measuring their weight (g) and length (cm). Briefly, the entire body surface was examined using a binocular stereoscopic microscope to detect lesions, cysts/xenomas and epidermal loss as part of the parasitological analyses. After external verification, the entire coelomic cavity was analysed by removing small fragments from each organ or tissue for visualization under a light microscope under a magnification of 400×. After detection of foci of infection, the material was collected and fixed in Davidson's solution (95% alcohol, formaldehyde, acetic acid and distilled water) for 24 h. Thereafter, the Ziehl–Neelsen technique (Luna, Reference Luna1968; Ferreira et al., Reference Ferreira, Silva, Araújo, Hamoy, Matos and Videira2020) was employed for histological analysis. Another section of the material containing a focus of infection was fixed in 80% alcohol for DNA extraction and molecular analysis. The methodology proposed by Bush et al. (Reference Bush, Lafferty, Lotz and Shostak1997) was employed to analyse the parasitic prevalence.
The recommendations of Lom and Arthur (Reference Lom and Arthur1989) were followed for the morphometric analyses of fresh myxospores (n = 35). The spore dimensions are expressed as arithmetic mean in μm, followed by standard deviation (s.d.). Images were obtained using MOTICAM 2300 3.0M Pixel coupled to a binocular microscope.
Molecular and phylogenetic analyses
Total DNA was extracted from each sample using the Wizard® Genomic DNA extraction kit (PROMEGA, Madison, USA), and quantified using a spectrophotometer (Biodrop DUO).
The molecular analyses were based on the 18S rDNA sequences (Table 1), which were amplified using the ERIB1(F) and ERIB10 (R) primers, followed by the MC3 (F) and MC5 (R) primers (NESTED PCR). The final polymerase chain reaction (PCR) volume of 25 μL consisted of 0.2 μL of Taq DNA polymerase (INVITROGEN, MA, USA), 10 μ m of each primer, 3.0 μL of the DNA sample and 16.8 μL of Master Mix (INVITROGEN).
Table 1. SSU rDNA primers used in this study, sequence and reference

Amplification was performed in a MyGene™ Series Peltier thermal cycler (Model MG96G) with the following cycling conditions for the ERIB1 and ERIB10 primers: initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, annealing at 56°C for 30 s, 72°C for 1 min and final extension at 72°C for 10 min. For the second amplification with the MC3 and MC5 primers, the following cycling program was employed: initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, annealing at 55°C for 50 s, 72°C for 1 min and final extension at 72°C for 10 min. The PCR results were analysed via electrophoresis on a 1.5% agarose gel in Tris-borate-EDTA buffer. The PCR product was purified using the Illustra™ GFX™ PCR DNA and Gel Band Purification kit, according to the manufacturer's recommendations.
The amplification products were sequenced on an ABI 3730 automated DNA analyser using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Partial sequences were assembled in the Codon Code Aligner software (CodonCode Corporation, Dedham, MA), and the generated product was compared with sequences deposited in GenBank (Table 2) using the Basic Local Alignment Search Tool (BLASTn) of the National Center for Biotechnology Information (NCBI). The SSU 18S rDNA nucleotide sequences were edited and aligned using the BioEdit program (Hall, Reference Hall1999), in which regions that were ambiguously aligned and unsigned regions in the SSU 18S rDNA datasets were removed (Holzer et al., Reference Holzer, Sommerville and Wootten2004; Gunter et al., Reference Gunter, Whipps and Adlard2009).
Table 2. Species, hosts and GenBank accession number for SSU rDNA sequences from Henneguya spp., Myxobolus spp. and Kudoa spp. (outgroup) used for phylogenetic analysis (except the sequence in this study)

The phylogenetic relationships obtained using maximum parsimony and Bayesian inference (BI) were initially employed in the PAUP 4.0 b10 program (Swofford and Sullivan, Reference Swofford, Sullivan, Salemi and Vandamme2003) and then in the MrBayes 3.1.2 program (Ronquist and Huelsenbeck, Reference Ronquist and Huelsenbeck2003). The nucleotide substitution models were selected using the Bayesian criterion (BIC) implemented in jModelTest 2.1.10 (Darriba et al., Reference Darriba, Ta boada, Doallo and Posada2012), based on phylogenetic analyses of the evolutionary model of general time reversible (GTR + R), which was selected as the best model for use in nucleotide replacement for the SSU 18S rDNA datasets.
Maximum parsimony analysis was performed with a heuristic search digit, which was assigned equal weight to transcription and transversion. Insertions and deletions (indels) were treated as missing data. The confidence level for the most parsimonious nodes in the tree was determined using 1000 bootstrap replicates (Felsenstein, Reference Felsenstein2004). Bayesian analysis was performed using the Monte Carlo Chain Markov algorithm (MCMC), implemented in BEAST v.1.8.4 (Drummond et al., Reference Drummond, Suchard, Xie and Rambaut2012) with 10 000 000 generations sampled every 10 000 steps (Brooks et al., Reference Brooks, Gelman, Jones and Meng2011). The BI products were used to build a phylogenetic tree from a set of myxozoan sequences.
Results
Species: taxonomic summary
Kingdom Metazoa Linnaeus, 1758
Phylum Cnidaria Hatscheck, 1888
Class Myxozoa Grassé, 1970 (Kyger et al., Reference Kyger, Luzuriaga-Neira, Layman, Milkewitz Sandberg, Singh, Huchon, Peri, Atkinson, Bartholomew, Yi Soojin and Alvarez-Ponce2021)
Subclass Myxosporea Bütschli, 1881
Order Bivalvulida Shulman, 1959
Family Myxobolidae Thélohan 1892
Genus Henneguya Thélohan, 1892
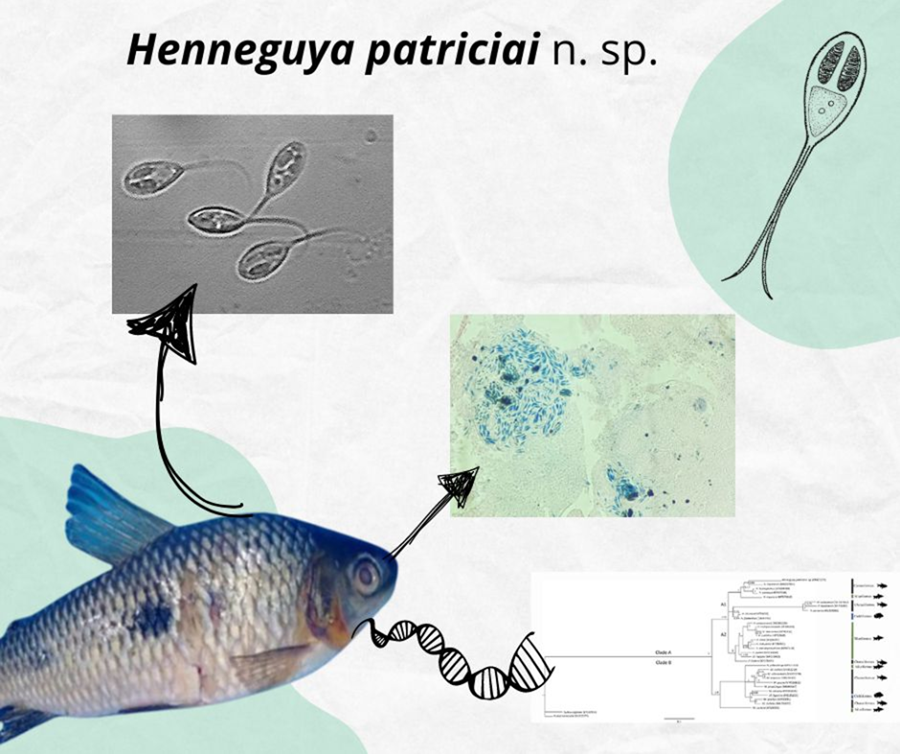
Species Henneguya patriciai n. sp. (Figs 1 and 2A)
Host: Leporinus friderici

Figure 1. Schematic drawing of the spore frontal view of Henneguya patriciai n. sp. (1) Polar capsules; (2) polar filaments; (3) sporoplasm; (4) nucleus; (5) valve elongation. Scale bar 20 μm.

Figure 2. (A) Light microscopy of spore frontal view of Henneguya patriciai n. sp. Scale bar: 40 μm. (1) Spore body; (2) valve elongation; (3) polar filament; (4) sporoplasm; (5) nucleus. Insert: sutural view of the spore of Henneguya patriciai n. sp. highlighting the suture line (arrow). Scale bar: 5 μm. (B) Histological section of the caudal kidney with Henneguya patriciai n. sp. cysts. Stained in Ziehl–Neelsen. Scale bar: 100 μm.
Prevalence: 100% (16 specimens)
Site of infection: Henneguya cysts and spores in the abdominal cavity and caudal kidney
Collection site: Tartarugalzinho river, Tartarugalzinho county, Amapá state.
Species deposit: Glass slide with Ziehl–Neelsen stained spores was deposited in the collection of the Amazon Research Institute (INPA), Manaus, Amazonas state, Brazil (accession number: INPA 93)
Etymology: specific epithet in honour of Patricia Matos (in memoriam), great researcher in the field of describing new species of microparasites in the Amazon region.
Vegetative phase
According to Molnár (Reference Molnár2002), Henneguya is a cyst-forming parasite. Based on microscopic analysis, irregular cysts with different sizes were found in the tissues containing the parasites (Fig. 2B). In this case, it was not possible to measure them, due to irregularities in sizes and shapes.
Morphological description of the spores
Fresh spores of H. patriciai n. sp. were measured (n = 35). The total length of the spore was 38.4 ± 2.5 (35.9–40.9) μm, the average spore body was 14.4 ± 1.1 (13.3–15.5) μm, and the width was 7.3 ± 0.6 (6.7–7.9) μm. The 2 polar capsules had a length of 5.1 ± 0.4 (4.7–5.5) μm and a width of 2.0 ± 0.1 (1.9–2.1) μm in the same pear shape. Each of these polar capsules contained 9–11 turns of coiled filaments (Table 3).
Table 3. Comparative table of measurements (μm) with standard deviation of Henneguya patriciai n. sp. and other Henneguya spp. described in the Amazon basin of the northern Brazilian region and Leporinus spp. in Brazil

IS, infection site; TL, total length; T, tail length; TAS, tail appendages size (=, tail appendages of equal size; ≠, or of different size); BL, body length; BW, body width; PL, polar capsule length; PW, polar capsule width; PCS, polar capsule size (=polar capsules of equal size; ≠, or of different size); PF, number of coils in the polar filaments.
Phylogenetic analyses
The partial SSU rDNA sequence of H. patriciai n. sp. obtained in the present study had 978 base pairs (GenBank accession number: OR421275), which comprised G + C (A = 0.2495, C = 0.1969, G = 0.2864, T = 0.2672). Assuming a GTR + G model of nucleotide substitution, the estimated nucleotide substitution rates were A–C = 1.0659, A–G = 2.9412, A–T = 1.4484, C–G = 0.7866, C–T = 4.5161 and G–T = 1.0000, with a gamma distribution (G) of 0.3700.
To construct the phylogenetic tree, 30 sequences of species from the Myxobolidae family available in GenBank were used. A BLAST search revealed that H. patriciai n. sp. did not match any other sequences deposited in GenBank, but Henneguya piaractus Martins et al., 1997 was the closest sequence found in GenBank, with just 84.79% similar to H. patriciai n. sp.
The phylogenetic tree revealed 2 main clades, A and B, with strong nodal support (Fig. 3). Both clades (A and B) were formed by hosts belonging to the orders, Characiformes, Siluriformes and Cichliformes. In clade B, only H. polarislonga Jorge et al., 2022 was found; the other parasites were identified to belong to the genus, Myxobolus.

Figure 3. Phylogenetic tree generated by Bayesian inference (IB) through partial alignment of Henneguya patriciai n. sp. with SSU r DNA gene sequences of select Henneguya and Myxobolus species. Node numbers are indicated for posterior probabilities values calculated by IB.
Henneguya patriciai n. sp. belongs to clade A1 in a subclade formed by parasites whose hosts belong to the orders, Characiformes [Piaractus brachypomus: H. brachypomus (Capodifoglio et al., Reference Capodifoglio, Adriano, Naldoni, Meira, Silva and Maia2020); Piaractus mesopotamicus: H. piaractus (Martins and Souza, Reference Martins and Souza1997); P. brachypomus: H. tapariensis (Capodifoglio et al., Reference Capodifoglio, Adriano, Naldoni, Meira, Silva and Maia2020)]; and Siluriformes [Eugerres brasilianus: H. lagunensis (Azevedo et al., Reference Azevedo, Negrelli, de Oliveira, Abdallah, Camara, Matos and Vieira2021)]. Unlike clade B, clade A did not include any species of the genus, Myxobolus. The p-distance analysis revealed large genetic divergence among other species of Henneguya spp. (Table 4).
Table 4. The uncorrected P-distances recorded between pairs of Henneguya spp. that comprise the clade of registered Henneguya spp. in Brazilian Amazon

Discussion
For L. friderici, the presence of some myxozoans has already been reported, such as Myxobolus sp., Ceratomyxa sp. and Henneguya sp. (Carvalho et al., Reference Carvalho, Júnior, Silva Ferreira, Araújo, Matos and Videira2021). Henneguya friderici, a species of the genus, Henneguya (i.e. H. patriciai n. sp.) is the second species of this genus described for this host and the third identified in the state of Amapá. The morphology and morphometry of H. patriciai n. sp. corroborate the description of the genus, Henneguya, provided by Lom and Dyková (Reference Lom and Dyková2006). These authors said that the genus Henneguya has ellipsoid spores with biconvex in sutural view, where each valve continues as a caudal projection. As a rule, this genus has 2 polar capsules very elongated and binucleate sporoplasm.
The morphometry, host and organ/tissue data, and DNA sequences were compared with those of other Henneguya spp. identified in the northern region of the Brazilian Amazon and Henneguya spp. described in Leporinus spp.
Analysis of the spore morphology of H. patriciai n. sp. and comparison with H. friderici revealed that H. patriciai n. sp. had larger spore bodies and valve elongation, and a wider spore body than H. friderici spores. These findings were confirmed by morphometric analyses of both species, and were supported by the results of phylogenetic analyses, which verified that they are different species.
Based on the measures obtained from the spores of H. patriciai n. sp., this new parasite had similar measurements to H. quelen (Abrunhosa et al., Reference Abrunhosa, Sindeaux-Neto, Hamoy and Matos2018), H. friderici and H. tapajoensis (Zatti et al., Reference Zatti, Atkinson, Maia, Bartholomew and Adriano2018). Despite the approximation of these cited species, H. patriciai n. sp. was smaller than all Henneguya spp. listed in this study. Based on spore body length, H. patriciai n. sp. was like H. tucunarei (Zatti et al., Reference Zatti, Atkinson, Maia, Bartholomew and Adriano2018) and H. testicularis (Azevedo et al., Reference Azevedo, Corral and Matos1997). In terms of the number of coils, H. patriciai n. sp. was similar to H. rhamdia (Matos et al., Reference Matos, Tajdari and Azevedo2005) but differed from the other species.
All other species of Henneguya spp. from the northern region of the Brazilian Amazon differed from H. patriciai n. sp. and had different hosts, except H. friderici. According to a BLAST search, H. patriciai n. sp. also differed from all SSU rDNA sequences of other myxozoans in the database; this finding confirms that H. patriciai n. sp. is a new species.
Henneguya patriciai n. sp. was found to parasitize the caudal kidney and pyloric caeca of all specimens of L. friderici. To our knowledge, this is the first report of a species of Henneguya spp. from the Brazilian Amazon that parasitizes the pyloric ceca of fish. Of note, H. friderici was found in the gills, intestine and liver of L. friderici, with a prevalence of 30% (Casal et al., Reference Casal, Matos and Azevedo2003).
According to Molnár and Eszterbauer (Reference Molnár, Eszterbauer, Okamura, Gruhl and Bartholomew2015), histozoic myxozoans, such as Henneguya, tend to have tissue/organ specificity for their host; however, some species of Henneguya spp. parasitize more than 1 tissue/organ, such as H. longisporoplasma (Zatti et al., Reference Zatti, Marinho, Adriano and Maia2022), which parasitizes the gills, fins and kidneys of Plagioscion squamosissimus (Heckel 1840). Henneguya torpedo (Azevedo et al., Reference Azevedo, Casal, Matos, Alves and Matos2011) was found in the brain and spinal cord of Brachyhypopomus pinnicaudatus (Hopkins 1991), similar to H. patriciai n. sp., which was found in more than 1 organ or tissue.
Phylogenetic analysis revealed that H. patriciai n. sp. grouped into a subclade (A1) composed of hosts of different orders of L. friderici, which opposes studies that defended the affinity for the order or family of the host as an important signal for phylogeny within the Myxobolidae family (Videira et al., Reference Videira, Velasco, Azevedo, Silva, Gonçalves, Matos and Matos2015; Velasco et al., Reference Velasco, Videira, Nascimento, Matos, Gonçalves and Matos2016; Abrunhosa et al., Reference Abrunhosa, Sindeaux-Neto, Hamoy and Matos2018; Zatti et al., Reference Zatti, Atkinson, Maia, Bartholomew and Adriano2018). Henneguya friderici was grouped in the subclade (A2) that contained species of Henneguya spp. and Myxobolus spp., unlike H. patriciai n. sp., which was present in its subclade and was the only species of the genus, Henneguya.
When the P-distance between the species of Henneguya spp. from the Amazon region was analysed, the dissimilarity between H. patriciai n. sp. and both H. paraensis and H. tapariensis was 16%, which was the minimum, and between H. friderici, another species of Henneguya described in L. friderici, was 20%. This result indicates that H. patriciai n. sp. differs from other species of the same genus.
In conclusion, based on phylogenetic, molecular and morphological/morphometric analyses, Henneguya sp. found in L. friderici from Tartarugalzinho river is a new species of the genus, H. patriciai n. sp. Overall, H. patriciai n. sp. is the second species of the genus Henneguya, to be identified in L. friderici in the Amazon region.
Data availability statement
The DNA sequences are deposited in GenBank (OR421275). All data generated or analysed during this study are included in this published article.
Acknowledgements
The authors would like to thank the fishermen, Nelson, of the Tartarugalzinho river, for their local knowledge, support and availability in this study and to all the members of research grupe ‘Sanidade de Organismos Aquáticos da Amazônia’ (SOAA) for all the support in this study. The authors also thank Coordination for the Improvement of Higher Education Personnel (CAPES) and National Council for Scientific and Technological Development (CNPq).
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Abthyllane Amaral de Carvalho, Lilia Suzane de Oliveira Nascimento, Saturo Cardoso Morais, Roger Leomar da Silva Ferreira and Luize Cristine Pantoja dos Reis. The manuscript was written by Abthyllane Amaral de Carvalho, Edilson Rodrigues Matos and Marcela Nunes Videira, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Financial support
Partial financial support was received from the Coordination for the Improvement of Higher Education Personnel (CAPES), through a doctoral scholarship granted to Abthyllane Amaral de Carvalho; from the National Council for Scientific and Technological Development (CNPq) through Dr. Edilson Matos; and from the State University of Amapá for resolution 175/2017 of subsidies to projects of the research group Sanity of Aquatic Organisms of the Amazon, led by Marcela Nunes Videira.
Competing interests
None.
Ethical standards
This study was performed in line with the principles of the Animal Use Ethics Committee of the Federal Rural University of the Amazon: no 8323110522; and Biodiversity Authorization and Information System: licence 50376-1.