Since the time when Homo genus appeared on earth (about two to seven million years), only minor alterations have occurred in the human genome. Indeed, only a difference of about 1·6 % was found between modern humans and the most developed primates. Despite this overall stability of the human genome, very numerous minor alterations in gene structures have been found. In contrast, human ways of life, including dietary habits, have markedly evolved since the origin, especially during the last century.
Concepts in nutrigenetics
Basically, the concept is that some minor alterations in gene sequences can modulate, to some extent, specific metabolic pathways which make the corresponding subjects more or less prone to response to dietary intakes.
For instance, ethnic differences for various traits have long been observed. This has been reported for the risk for CVD or type 2 diabetes in African or white American children(Reference Lindquist, Gower and Goran1).
At individual level, polymorphisms of common genes are widespread, with allele frequencies ranging from 1/1 (very frequent) to 1/1000 (rare). For instance, in the field of lipid and lipoprotein metabolism, more than 250 single nucleotide polymorphisms (SNP) have been identified in about thirty genes encoding key proteins involved(Reference Ye and Kwiterowitch2, Reference Corella and Ordovas3). Regarding the metabolic syndrome, we have identified about 140 key genes potentially involved, including more than 800 relevant SNP (see http://www.ucd.lipgene).
Among various common types of alteration in DNA sequence, SNP are the most usually studied. A SNP is the single change of a base (A, T, G or C) at a given locus in the DNA sequence of a gene. The amplitude of biological impact will first depend on heterozygosity or homozygosity. The functional impacts on the encoded protein could be variable. If the mutation induces a modification of one important amino acid in the encoded protein sequence, this can alter the protein function/activity and thus the metabolic pathway involved. In contrast, a modification in the gene promoter sequence can alter the promoter activity and thus the level of gene transcription, resulting in altered encoded protein level and linked activity. In other cases, no effect on protein level or activity can be observed.
Most dietary intervention studies have clearly highlighted the very large inter-individual variability in metabolic responses. Generally, during intervention studies, most subjects show a change for a given parameter in one main direction with more or less amplitude, but a minority of subjects do not exhibit a marked change and few subjects can even show a change in the opposite direction. This clearly indicates that under the same environmental (dietary) pressure, different subjects can display both general traits and some particular susceptibility. This is the basis of the concept of gene–diet interaction in health and disease.
The high throughput tools recently developed allow easy, fast and cheap determination of lots of gene polymorphisms from a single blood sample, especially SNP (about 0·1€/point). This opens a new era where studies dedicated to interactions between diets, metabolic parameters, disease risk factors and gene polymorphisms can be carried out on small or large groups of healthy subjects or patients.
SNP–diet interactions in the Medi-RIVAGE study
Epidemiological studies(Reference Keys, Blackburn, Menotti, Fidanza and Aravanis4–Reference Martínez-González, Fernández-Jarne, Serrano-Martínez, Marti, Martínez and Martín-Moreno6) have raised and supported the concept that traditional Mediterranean diet is associated with reduced cardiovascular mortality. The Lyon Heart(Reference de Lorgeril, Renaud, Mamelle, Salen, Martin, Monjaud, Guidollet, Touboul and Delaye7) and the Indo-Mediterranean(Reference Singh, Dubnov, Niaz, Ghosh, Singh, Rastogi, Manor, Pella and Berry8) studies were the first trials based on a Mediterranean-type dietary intervention and showed a beneficial effect on the survival rate after myocardial infarction (secondary prevention). In the field of cardiovascular primary prevention, the first two intervention studies have recently shown the protective effects of a Mediterranean-type diet on cardiovascular risk factors in subjects with the metabolic syndrome in Italy(Reference Esposito, Marfella, Ciotola, Di Palo, Giugliano, Giugliano, D’Armiento, D’Andrea and Giugliano9), or at moderate cardiovascular risk in France(Reference Vincent-Baudry, Defoort and Gerber10).
One of the goals of the Medi-RIVAGE study(Reference Vincent-Baudry, Defoort and Gerber10, Reference Vincent, Gerber and Bernard11) was to investigate the interactions between gene polymorphisms in the subjects included and the intervention diets (Mediterranean-type diet or low-fat, low-cholesterol diet) on the resulting changes in cardiovascular risk factors after a 3- and 12-month follow-up. Extensive biological investigations were performed with a special focus on fasting and postprandial lipid and lipoproteins parameters.
The Medi-RIVAGE intervention study: design and methods
The design and methods of the Medi-RIVAGE study have been reported(Reference Vincent, Gerber and Bernard11) and the results in terms of reduction of cardiovascular risks after a 3-month follow-up have been published(Reference Vincent-Baudry, Defoort and Gerber10). Briefly, volunteers with a habitual western-type diet and moderate cardiovascular risks (hypercholesterolaemia, hypertriglyceridaemia, elevated BMI or hypertension) were provided with detailed nutritional recommendations. They were advised to have either a low-fat diet or a Mediterranean type-diet for 3 months but the two groups experienced, in fact, Mediterranean type-diets(Reference Vincent-Baudry, Defoort and Gerber10). At entry (n 212) and at the end of the 3-month period (n 169), biochemical analyses were performed as previously described(Reference Vincent-Baudry, Defoort and Gerber10, Reference Vincent, Gerber and Bernard11).
For polymorphism detection, genomic DNA were prepared from white blood cells isolated from whole blood samples by a standard proteinase K–phenol method as reported(Reference Gastaldi, Dizière, Defoort, Portugal, Lairon, Darmon and Planells12, Reference Borel, Moussa and Reboul13). The polymorphisms were genotyped by a PCR-RFLP (polymerase chain reaction-restriction fragment length polymorphism) assay. Fifteen SNP in eleven different genes have been determined, such as genes coding for: apo A-IV (Ser347Thr, A/T, rs675), apo B (−516, C/T, rs934197), apo C-III (SstI S1/S2, C/G), apo E (e2/e3/e4, rs7412; −219, G/T), MTP (−493, G/T), I-FABP (Ala54Thr, A/G, rs1799883), SR-BI (exon1, G/A, rs4238001; exon8, C/T, rs5888; intron5, G/A), CETP (TaqIB, C/T, rs708272), lipoprotein lipase (−93, G/T, rs1800590), hepatic lipase (−480, C/T, rs1800588) and ACE (insertion/deletion).
Gene polymorphisms and diet interactions
As already published(Reference Vincent-Baudry, Defoort and Gerber10), we observed after a 3-month intervention reductions in BMI, glycemia, insulinaemia, HOMA (homeostatic model assessment) score, total and LDL-cholesterol, triglycerides and Framingham risk score.
In a first global approach we have looked at the possible association of gene polymorphisms with all biological variables by projection on latent structure (PLS) analysis using SIMCA P-11 software. It is possible to evaluate how the various genotypes can influence the phenotypic outcomes (blood parameters) upon a dietary intervention using a PLS multivariate approach. Briefly, this statistical method allows reducing both the overall gene variants (x variables) and the phenotypical outcomes (biochemical variables; y variables) of each individual into few score values. Hence, each individual will be characterised by one point in a two-dimensional space, defined by its genetic score and its phenotypic score as coordinates. The relationship between the genetic scores and the phenotypic scores of a population can thus be displayed as a scatter-gram, and the index of determination, R 2, calculated (Fig. 1). These data clearly indicate the existence of a relationship between genotypes and phenotypes in those subjects.

Fig. 1 Partial least-square (PLS) plot displaying the relationship among the genotypes score values and the phenotype score values during the dietary transition (0–3 months) (█, baseline; ▵, 3 months)
A more detailed investigation of interactions between some SNP and the anti-oxidant status of the subjects at baseline clearly evidenced that β-carotene concentrations were significantly different in subjects bearing different SNP in apo B and SR-BI, while lycopene concentrations were different in subjects bearing different SNP in apo A-IV and apo B(Reference Borel, Moussa and Reboul13).
From data presented in Fig. 1, we can calculate that the overall SNP analysed could predict up to 23 % of the change in blood biochemical pattern, during the dietary intervention (0 to 3 months of the nutritional challenge). As summarised in Table 1, reductions in most cardiovascular risk markers were significantly associated in univariate analysis with the presence of some of the polymorphisms (SNP) determined. The most important ones seem to be in genes coding for apo B (the main apoprotein constitutive of LDL and very low-density lipoproteins (VLDL), involved in tissue secretion and uptake), apo E (a key apoprotein in LDL, VLDL and chylomicrons, involved in tissue secretion and uptake), apo A-IV (an apoprotein specifically secreted by the small intestine), MTP (microsomial transfer protein, a key intra-cellular protein involved in VLDL and chylomicrons assembly in the liver and small intestine), CETP (cholesteryl ester transfer protein, a plasma protein that facilitates cholesterol esters and triglycerides reciprocal exchange between lipoproteins) and SR-BI (a key membrane transporter controlling absorption and exchanges of lipids and cholesterol).
Table 1 Reductions in cardiovascular risk factors after a 3-month dietary intervention significantly (P < 0·05) associated with gene polymorphisms of included subjects (n 169)
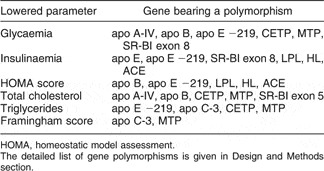
HOMA, homeostatic model assessment.
The detailed list of gene polymorphisms is given in Design and Methods section.
The detailed data obtained for the interaction between the reduction in LDL-cholesterol, after a 3-month intervention, and SNP are illustrated in Fig. 2. The significant interactions between reduction in LDL-cholesterol and SNP in genes coding for apo E and MTP are highlighted.

Fig. 2 Partial least-square (PLS) correlation coefficient calculated among various single nucleotide polymorphisms (SNP) and LDL-cholesterol. Positive values are associated with increased plasma total cholesterol, and negative values to a decrease. Statistical level of significance is calculated by the jack-knife method, using a 95 % CI, visualised as error bars. SNP with error bars not crossing the reference line are significantly associated to plasma total cholesterol (names in black boxes)
Sex-specific association of I-FABP and MTP gene variants with response to dietary intervention
A detailed investigation of some interactions provides a good example of the potential interest but the complexity of the nutrigenetics approach.
A common mutation has been described in the FABP2 gene encoding the I-FABP (intestinal fatty acid binding protein), a key protein involved in the intracellular lipid trafficking in the enterocyte(Reference Baier, Sacchettini, Knowler, Eads, Paolisso, Tataranni, Mochizuki, Bennett, Bogardus and Prochazka14, Reference Dworatzek, Hegele and Wolever15). This gene variant results in a Ala/Thr substitution at position 54 in the protein sequence, which alters its interactions with fatty acids. We found that men and women homozygous for Thr54 presented an opposite pattern for triglyceride-rich lipoprotein cholesterol at baseline with highest values observed in Thr/Thr men and lowest values in Thr/Thr women. Nevertheless, the FABP2 genotype did not interact noticeably with the amplitude of response to the dietary intervention(Reference Gastaldi, Dizière, Defoort, Portugal, Lairon, Darmon and Planells12).
The MTTP gene encodes the microsomial transfer protein (MTP), an intra-cellular protein involved in VLDL and chylomicrons assembly in the liver and small intestine, and shows a common mutation in the promoter sequence (−493 G/T polymorphism)(Reference Karpe, Lundahl, Ehrenborg, Eriksson and Hamsten16, Reference Ledmyr, McMahon, Ehrenborg, Nielsen, Neville, Lithell, MacFarlane, Packard and Karpe17). We found that men and women exhibit a different pattern for insulin values at baseline with highest values in T/T men and lowest values in T/T women(Reference Gastaldi, Dizière, Defoort, Portugal, Lairon, Darmon and Planells12). However, men and women homozygous for the T allele displayed a significantly more pronounced response after the 3-month dietary intervention for plasma cholesterol and triglycerides as illustrated in Figs. 3 and 4, while the lowering of the Framingham score for cardiovascular risk was more pronounced in T/T men than T/T women (Fig. 5).

Fig. 3 Plasma cholesterol response to dietary intervention according to MTP −493 G/T polymorphism. P = 0·030 for comparison between polymorphisms in their response to diet (tested with repeated measures general model), adjusted for menopausal status in women. From reference(Reference Gastaldi, Dizière, Defoort, Portugal, Lairon, Darmon and Planells12) (␣, at baseline; ![]() , after a 3-month diet)
, after a 3-month diet)

Fig. 4 Plasma triglycerides response to dietary intervention according to MTP −493 G/T polymorphism. P = 0·036 for comparison between polymorphism in their response to diet (tested with repeated measures general model), adjusted for menopausal status in women and BMI. From reference(Reference Gastaldi, Dizière, Defoort, Portugal, Lairon, Darmon and Planells12) (␣, at baseline; ![]() , after a 3-month diet)
, after a 3-month diet)

Fig. 5 Framigham risk score response to dietary intervention according to MTP −493 G/T polymorphism. P = 0·008 for comparison in men between polymorphism in their response to diet (tested with repeated measures general model), adjusted for BMI, smoking status and professional activity (␣, at baseline; ![]() , after a 3-month diet)
, after a 3-month diet)
Conclusions
The data reported thus provide the first evidence of the interaction between some gene polymorphisms (SNP) and metabolic responses to a dietary pattern of the Mediterranean type. The fact that the variations of very important parameters, which are acknowledged cardiovascular risk factors, are influenced by gene polymorphisms is indicative of the key importance of genetic susceptibility regarding responses to diets. Moreover, the fact that some gene polymorphisms (genes coding for apo B, apo A-IV, apo E, MTP, CETP) are concomitantly associated to variations of several parameters, which take part in various metabolic pathways, does indicate that these polymorphisms are of particular importance in modulating responses to diets and controlling nutritional status and metabolic homeostasis.
Further large-scale studies on gene–diet interactions are necessary to thoroughly evaluate the relevant ones, as recently reviewed(Reference Williams, Ordovas, Lairon, Hesketh, Lietz, Gibney and van Ommen18). This newly accumulated knowledge might constitute the basis of new kinds of dietary recommendations. Indeed, the alternative to the present recommendations to the general public or some broad groups (kids, teenagers, elderly people, pregnant women, etc.) can be more focussed recommendations based on the specific responses to particular dietary patterns or nutrients which are, at least in part, modulated by various gene polymorphisms and their combinations. This can raise and support the concept of ‘personalised nutrition’ aiming at providing targeted dietary advices to specific groups of people or even individuals. Such a strategy needs accumulation of new knowledge, should take into consideration ethical and psychological issues related to genetic screening and requests that sufficient means are devoted to efficient advising and follow-up of people. A long way ahead.








