1. Introduction
The degree of genetic changes that occur in insect populations reared under laboratory conditions is an intriguing subject of study from both theoretical and applied points of view. Most control programmes of harmful insects entail the rearing of an insect under artificial conditions. The conditions that prevail in such colonies are often quite different from those experienced in natural populations of the species. The effect of these differences on the insect can be quite dramatic. Laboratory conditions may alter the developmental, physiological or behavioural responses of the insect in such a way that artificially reared colonies may soon become unsuitable for the purposes they were established in the first place.
In the last few years, we have become interested in the extent to which these genetic changes occur in the colonies of the olive fruitfly Bactrocera oleae (Gmelin) under conditions of artificial rearing, an issue first addressed by Bush & Kitto (Reference Bush and Kitto1979). B. oleae, the most important olive orchard pest in Greece, is a monophagous insect totally dependent on the olive fruit. In a preliminary study of the olive fruitfly, Bush & Kitto (Reference Bush and Kitto1979) examined one laboratory and two wild populations for 23 enzyme loci and found striking allele frequency differences for two: alcohol dehydrogenase (ADH) and 6-phosphogluconate dehydrogenase (6-PGD).
The ADH locus became subsequently the subject of more detailed studies, which revealed that this locus is under intensive selection pressure in the laboratory and that the factor responsible for this is the artificial larval diet (Zouros et al., Reference Zouros, Loukas, Economopoulos and Mazomenos1982, Reference Zouros, Loukas, Economopoulos, Vergini and Economopoulos1987; Loukas et al., Reference Loukas, Economopoulos, Zouros and Vergini1985; Economopoulos & Loukas, Reference Economopoulos and Loukas1986; Cosmides et al., Reference Cosmides, Loukas and Zouros1997; Cosmidis et al., Reference Cosmidis, Loukas and Zouros1999). At the same time, the profound changes in allele frequencies of the enzyme locus 6-PGD have left little doubt that this polymorphism is also under the influence of strong selection pressure.
Three active and one silent electrophoretically detectable alleles exist at 6-PGD locus in the Greek populations of B. oleae that have been studied so far. The active alleles are designated F (fast), I (intermediate) and S (slow) in order to signify their relative electrophoretic mobilities. In all the surveys for 6-PGD conducted so far in laboratory populations of B. oleae, the most common allele (allele I) declined from almost 0·64 to 0·25, the second most common allele (allele F) increased from 0·19 to 0·75, the third allele (allele S) declined from 0·15 to almost zero and the silent allele frequency either declined from almost 0·020 to zero or remained unchanged (Loukas et al., Reference Loukas, Economopoulos, Zouros and Vergini1985).
The enzyme 6-PGD (EC 1.1.1.44) is the third enzyme in the pentose phosphate pathway. It catalyses the oxidative decarboxylation of 6-phosphogluconate to ribulose 5-phosphate with the release of CO2 and the reduction of NADP. It is dimeric, NADP- and not metal-ion-dependent for almost all species (Rosemeyer, Reference Rosemeyer1987). It is also considered as one of the key enzymes of the pentose phosphate pathway. So far, it has been generally recognized that the two main functions of the pentose phosphate pathway are to provide NADPH for fat synthesis and ribose for nucleic acid synthesis (Wood, Reference Wood1985). The amino acid sequence has been reported in more than 40 different 6-PGDs, including sheep (Carne & Walker, Reference Carne and Walker1983), pig (Harbitz et al., Reference Harbitz, Chowdhary, Kran, Frengen, Gustavsson and Davies1990), human (Tsui et al., Reference Tsui, Chan, Waye, Fung and Lee1996) and Escherichia coli (Nasoff et al., Reference Nasoff, Baker and Wolfe1984). In Drosophila melanogaster, the X-linked 6-PGD gene is expressed in various tissues and its activity is particularly high in fat cells (Gutierrez et al., Reference Gutierrez, Christensen, Manning and Lucchesi1989).
Apart from Drosophila, our knowledge of 6-PGD in insects is limited and is mostly concentrated on two Tephritidae species, the medfly Ceratitis capitata and the olive fruitfly B. oleae. In the Mediterranean fruitfly C. capitata, 6-PGD has been mapped to chromosome 5 (Zacharopoulou, Reference Zacharopoulou1990). Stinner and House (Reference Stinner and House1990) reported that the enzyme in C. capitata is dimeric, encoded by a single locus and represented by two alleles but, later on, Gasperi et al. (Reference Gasperi, Guglielmino, Malacrida and Milani1991), who analysed populations of C. capitata collected from the African region and the Mediterranean basin, revealed a third 6-PGD allele. Moreover, the genomic sequences of 6-PGD from both species has already been determined (Goulielmos et al., Reference Goulielmos, Cosmidis, Eliopoulos, Loukas and Zouros2006).
In both larvae and adults of several insects, up to 40% of glucose catabolism is channelled through the pentose pathway. Diapausing insects such as B. oleae are able to withstand relatively low temperatures for extended periods (Manikas, Reference Manikas1974). This cold hardiness is achieved principally through the production of polyols which involves polyol dehydrogenase, an NADPH-requiring enzyme (Holden & Storey, Reference Holden and Storey1994; Joanisse & Storey, Reference Joanisse and Storey1994). Hence, an alteration in NADPH levels that might result from differences in the properties of the 6-PGD alleles could be of adaptive significance for B. oleae during diapause period.
The experiments described here were designed to answer two obvious questions: (1) Are the 6-PGD allozymes the direct target of selection in B. oleae laboratory population or do they serve as markers of selection acting at closely linked but undetected sites of the genome? (2) What is the molecular basis of the 6-PGD allozyme polymorphism concerning alleles F and I?
2. Materials and methods
In order to answer the first question, we established 11 new populations (which we refer to as the perturbation populations) using 20 lines, homozygous for alleles F and I. These lines were derived from a composite population which was founded after mixing equal number (500) of individuals from three large laboratory populations kept separately under artificial rearing for more than 270 generations with a minimum disturbance in their genetic background. The aforementioned laboratory populations originated from three wild populations collected from different localities in southern Greece. The composite population consisted of 1500 adults and did not contain the S allele. The two alleles F and I, in this colony, were in a frequency of 0·74 and 0·26, respectively, and were at approximate equilibrium for about 20 generations. In this composite population, non-random associations between 6-PGD alleles and undetected variants in the background genotype were expected to have ceased to exist or to be greatly reduced.
Ten homozygous isofemale lines for each allele (F and I) were isolated from the composite population and combined into two new populations designated FF and II. After 20 generations, pupae from populations FF and II were taken and used to start perturbation populations. These populations were established by mixing homozygous individuals for alleles F and I in different ratios. The initial frequencies of alleles F and I for 6-PGD were chosen, with the exception of populations 8 and 10, far from the equilibrium frequencies observed in previous experiments.
All populations were started with 1500 adults and maintained in this size for all consecutive generations. The technique for rearing the olive fruitfly in the laboratory has been described by Tsitsipis (Reference Tsitsipis1982). The frequency of 6-PGD alleles was monitored in cage populations for 12 generations. In all populations, equilibrium had been reached up to the 8th generation. We continued the experiments up to the 12th generation to ensure that the equilibrium was in fact maintained. In all populations, a random sample of 300 adults was removed and scored for 6-PGD in every generation to provide an estimate of the actual gene frequency. The 6-PGD alleles were separated on horizontal 10% starch gel. Electrode buffer, gel buffer, staining recipe and zymogram pattern can be found in Loukas and Krimbas (Reference Loukas, Economopoulos, Zouros and Vergini1980).
(i) RNA preparation, cDNA synthesis and sequencing of 6-PGD I allele
The B. oleae individuals used for RNA preparation were derived from lines homozygous for allele I. Total RNA was extracted twice, from 30 pupae each time, which had been frozen in liquid nitrogen and stored at −80°C. The two samples consisted of pupae carrying the 6-PGD I allele. Total RNA was extracted by using the RNeasy mini kit (Qiagen Extraction kits; Qiagen, Valencia, CA, USA).
In order to generate cDNA, the gene-specific 5′-ATGTCAGCTAA AGCGGATATTGCACTG-3′ upstream and 5′-GCCTGGTATGTA CTTGCGAGACATTAC-3′ downstream primers were used, derived from the nucleotide sequence of the genomic region of the B. oleae 6-PGD F allele (NCBI GenBank® accession number AJ517226). cDNA was synthesized by using the Qiagen OneStep RT-PCR kit (Qiagen) from 2 mg of total RNA, using the reverse transcriptase mix provided with the kit, as described by the manufacturer. The mixture was incubated at 50°C for 30 min, reverse transcriptase was heat-inactivated and, subsequently, the HotStart Taq DNA polymerase (Qiagen) was activated by applying an initial heating step at 95°C for 15 min. Therefore, cDNA was amplified by the PCR (40 cycles of 94°C for 30 s, 52°C for 45 s, 72°C for 1 min and, finally, 72°C for 10 min). Aliquots of PCR products were resolved by 1% agarose gel electrophoresis. Sequencing of the double-stranded plasmid was carried out using a LI-COR 4200 analyser. Analysis of the sequenced fragments was performed by using the DNA GeneImagIR program. The resulting PCR product of 1443 bp, from three independent PCR amplifications, was sequenced to verify that it represented the successfully generated cDNA fragment and, therefore, to determine the coding sequence of the 6-PGD gene of B. oleae ‘II’ strain.
3. Results and discussion
Table 1 gives the F allele frequency of 6-PGD surveyed in the first 12 generations of 11 laboratory populations. In order to interpret the results shown, the standard error (S.E.) of the F allele frequency is used. It is obtained from the binomial distribution as S.E.=[p(1−p)/2N]1/2, where p stands for allele frequency and N for the sample size. Approximately 95% confidence intervals can be obtained by taking two S.E. values around the frequency estimate. When the confidence intervals do not overlap, the null hypothesis that F allele frequencies are not different can be rejected at a=0·05. Thus, the S.E. test can be used to see whether there is a significant difference in the frequency of allele F between any two generations.
Table 1. Frequencies of allele F at 6-PGD locus in 11 laboratory populations of the olive fruitfly B. oleae
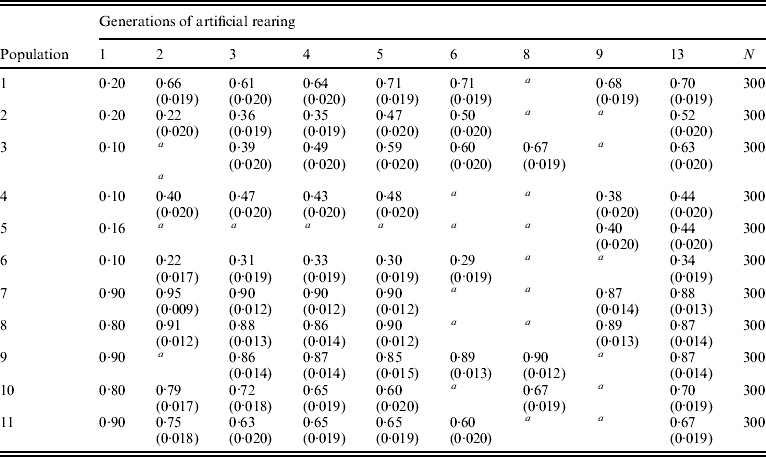
The founder population corresponds to generation 1. Frequencies of the I allele are not given since they are complementary to those of allele F. S.E. values are given in parentheses.
N is the number of individuals sampled in every generation and population. Populations were not sampled for 6-PGD in generations 7, 10, 11 and 12.
a Not sampled in this generation. More explanations are given in the text.
The frequency of allele F in populations 1, 3, 10 and 11 rebounded and returned to a level close enough to the equilibrium frequency of allele F displayed by the composite population (0·76). The response of the populations towards the new equilibrium was as quick as in colonies established afresh from individuals collected from the wild (Bush & Kitto, Reference Bush and Kitto1979; Loukas et al., Reference Loukas, Economopoulos, Zouros and Vergini1985). Besides, F allele frequency increased considerably in populations 2, 4, 5 and 6, whereas in populations 7, 8 and 9, allele F sustained high frequencies with minor deviations from the values in the founder populations. It would appear from these observations that no genetic changes have occurred in laboratory populations of B. oleae that would neutralize the selection forces acting on the 6-PGD locus.
L'Hertier & Tessier (Reference L'Hertier and Tessier1934) were the first geneticists to use population cages to maintain large laboratory populations of Drosophila and to follow changes in gene frequencies due to selection. Since then, this technique has been used to study selection on a variety of morphological markers, chromosomal polymorphisms and biochemical polymorphisms.
An important assumption in the studies with single gene markers is that the changes observed are due to selection at the locus being observed, and not due to selection at a linked gene or block of genes. One way to partially avoid this complication is to introduce the marker into the laboratory populations on as many independently derived chromosomes as possible. This decreases the probability that the marker will be in linkage disequilibrium with other genes. Ohta & Kimura (Reference Ohta and Kimura1970) have shown that the variance in linkage disequilibrium expected due to random drift in experimental populations is equal to 1/(n−1) in the first generation, if it is produced by extracting n chromosomes from a population in linkage equilibrium. In the study presented in this paper, we used 20 homozygous lines for 6-PGD F and I alleles taken from a population in which the genetic milieu for 6-PGD locus was greatly randomized. Our experiments revealed that no reduction of selection pressure on the 6-PGD locus had occurred. In all perturbation populations, the F allele of 6-PGD increased from low frequencies (0·10, 0·16 and 0·20) to a level between 0·34 and 0·70. The unfailing repeatability of this result leads us to the conclusion that it is a real phenomenon that cannot be attributed to a random process. Under natural conditions, this polymorphism is apparently maintained at equilibrium by some form of balancing selection. Laboratory conditions alter the direction of the selection pressures and drive the colony to a new polymorphic equilibrium.
Nevertheless, it can also be seen from Fig. 1 that populations with low and high frequencies of F allele do not totally converge after 12 generations of laboratory rearing. A plausible explanation for this would be linkage disequilibrium caused by random genetic drift, which will arise in any population as long as it is finite. The approximate magnitude of linkage disequilibrium and associative overdominance were estimated in various situations in natural populations (Ohta, Reference Ohta1971). They mainly depend on N ec, the product of the effective population number (N e) and the recombination fraction (c). Although precautions were taken to homogenize the genetic background and to maintain our experimental populations with a large number of individuals (1500), we cannot exclude the possibility that selection acts on hidden genetic polymorphisms in linkage disequilibrium with the 6-PGD locus rather than on the locus per se.
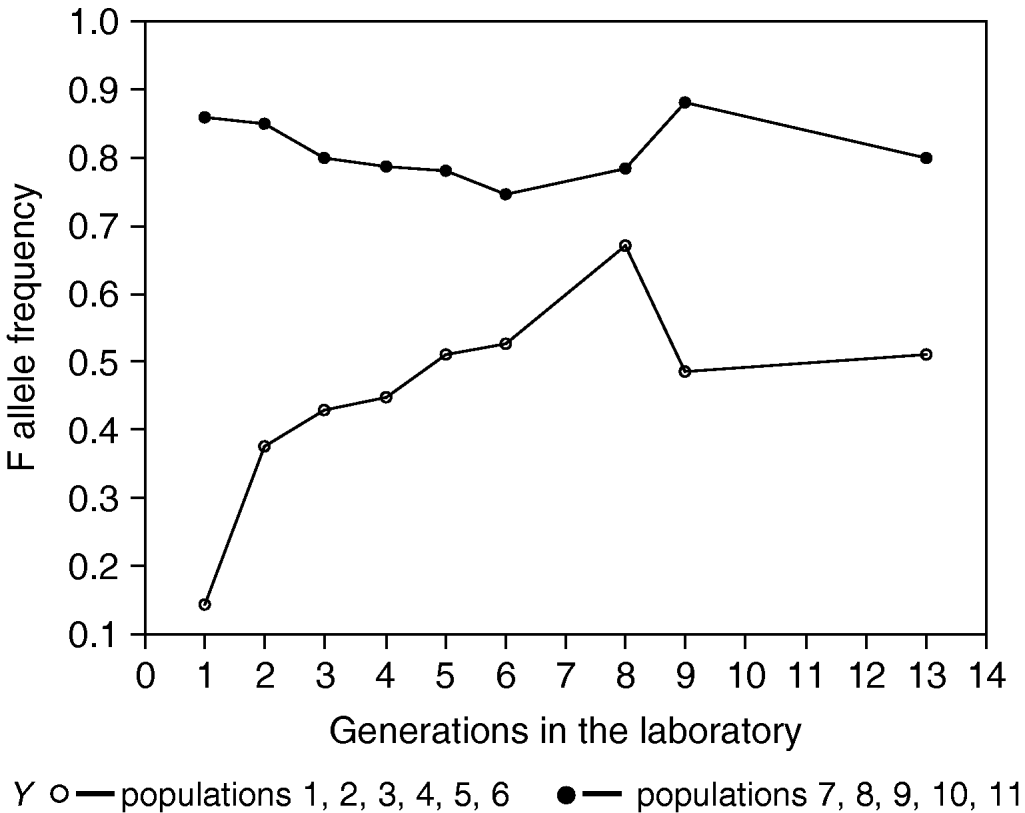
Fig. 1. Frequency of 6-PGD F allele in laboratory populations of B. oleae. Each value in the Y-axis represents the mean for all the corresponding populations.
These findings are coupled with nucleotide differences detected between the F and I alleles of 6-PGD. Nucleotide sequences for the 6-PGD F and I alleles have been deposited in the EMBL database with accession numbers AJ517226 and AM778416, respectively. Apart from a few synonymous nucleotide substitutions, two missense mutations at position 501 (AAC for allele F and AAG for allele I) and position 730 (GAG for allele F and AAG for allele I) resulted in obtaining different amino acids in ‘II’ strain.
The predicted amino acid sequence obtained from B. oleae 6-PGD ‘II’ strain results in a lysine residue at amino acid position 167 and also a lysine residue at position 244. In contrast, the sequence from the ‘FF’ strain had the residues asparagine and glutamic acid at the respective positions. The 6-PGD sequence of B. oleae ‘II’ strain was the same in both samples.
All the highly conserved regions of 6-PGD (which are important for the catalytic activity of the enzyme), such as those forming the substrate and coenzyme binding sites, and several amino acid residues responsible for substrate and coenzyme specificity have already been localized (Goulielmos et al., Reference Goulielmos, Eliopoulos, Loukas and Tsakas2004, Reference Goulielmos, Cosmidis, Eliopoulos, Loukas and Zouros2006). According to this model, two peripheral regions located at the distant parts of the two domains of the enzyme with respect to their interface are highly variable. These regions may accumulate several mutations, since they can be characterized as regions of non-functional importance and are predicted to be antigenic, thus reflecting possible regions for antibody recognition.
The differences in the amino acid sequence between strains ‘F’ and ‘I’ of B. oleae, defined in the present study, are not localized on any of the aforementioned highly conserved areas of the protein, but are located in the previously assumed as probable sites that accommodate variations appearing at the allozymic variants of B. oleae. Indeed, loops 164–168 and 247–250 had been defined as located in external, non-conserved regions of the protein chain and it was believed that they could be possible positions where the F, I and S variants of B. oleae might diversify, due to the introduction of any hydrophobic or differentially charged amino acid residues (Goulielmos et al., Reference Goulielmos, Eliopoulos, Loukas and Tsakas2004, Reference Goulielmos, Cosmidis, Eliopoulos, Loukas and Zouros2006). Although the structural location of the two substitutions away from the binding site may not affect its enzymatic mechanism, they may affect the dynamic properties of the enzyme, resulting in selection between alleles of 6-PGD locus.
This study complements previous studies on the genetic changes following the domestication of the olive fruitfly B. oleae. The experiments described here are a reinforcement of the hypothesis that selection acts on the 6-PGD locus itself. Because of the strong correlation between changes at 6-PGD locus and the colony's performance, these allozyme changes might be used as tools for quality assessment, even if the causal relationship between the two sets of characters remains unknown. Of course, this relies on the assumption that certain allozymes will always be associated with low performance. As long as this association holds, it is not necessary to know, for the sake of quality testing, what is the exact cause–effect relationship on which it rests. The very properties usually required from a successful mass-rearing system (low-cost production necessitating the maintenance of large numbers of individuals in limited space, rapid rate of reproduction and minimal fluctuations in the ambient environment) represent the opposite extreme of the conditions under which natural populations have evolved in many ways. By knowing which enzymes are more affected by artificial rearing, it could be possible to identify the factors that must be modified in order to improve the method. Thus, a long series of laboratory and field experiments is needed before the profound response of 6-PGD allozymes to the shift from natural to artificial rearing can be of use for the amelioration of B. oleae laboratory populations.




