Introduction
Taenia solium is the aetiological agent of neurocysticercosis in humans, one of a number of neglected tropical diseases recognized by the World Health Organization (2015). The parasite is transmitted in a cycle between humans, who harbour the adult tapeworm in the small intestine (taeniasis), and pigs where the larval stage (cysticercus) develops in the muscles and brain after ingesting feces or other items contaminated with T. solium eggs. Humans may also develop cysticercosis by ingesting eggs from the feces of a person harbouring the T. solium tapeworm. Infection in the brain and other nervous tissue of humans by T. solium cysts (neurocysticercosis) is a serious cause of morbidity in areas having poor sanitation and free-roaming pigs (Garcia et al., Reference Garcia, Gonzalez and Gilman2020).
Efforts to prevent the transmission of T. solium and thereby reduce the incidence of neurocysticercosis rely on the treatment of patients with taeniasis, vaccination and medication of pigs, and improvements in sanitation and pig-rearing practices (Lightowlers, Reference Lightowlers2013).
Diagnostic tests for taeniasis are undertaken to determine the risk for transmission of cysticercosis in humans, identify endemic areas and to determine the outcomes of control programmes. Taenia solium taeniasis can be diagnosed by detection in the feces of eggs, tapeworm segments, parasite antigens or T. solium DNA in the feces (Praet et al., Reference Praet, Verweij, Mwape, Phiri, Muma, Zulu, van Lieshout, Rodriguez-Hidalgo, Benitez-Ortiz, Dorny and Gabriel2013), or by serology with recombinant antigens (Levine et al., Reference Levine, Lewis, Rodriquez, Jimenez, Khan, Lin, Garcia, Gonzales, Gilman and Tsang2007).
Human taeniasis is caused by 3 Taenia spp., including T. solium, T. saginata and T. asiatica. However, only T. solium causes neurocysticercosis and warrants a public health intervention. Taenia saginata, in particular, is widely distributed in areas where T. solium is prevalent; hence, diagnostic tests for T. solium taeniasis must differentiate T. solium from infection with other Taenia species. Egg morphology does not allow differentiation among Taenia species. Similarly, the coproantigen tests that have been described and well validated are unable to differentiate between Taenia spp. Recombinant antigens required to undertake species-specific serology (Levine et al., Reference Levine, Lewis, Rodriquez, Jimenez, Khan, Lin, Garcia, Gonzales, Gilman and Tsang2007) are not readily available. Serological methods are also limited because they are unable to differentiate a current infection from past infection. At least 20 different DNA-based methods have been described for species-specific differentiation of T. solium, including at least 15 different polymerase chain reaction (PCR)-based methods; however, few have been validated carefully with parasitologically proven fecal samples from patients with T. solium taeniasis (Lightowlers et al., Reference Lightowlers, Garcia, Gauci, Donadeu and Abela-Ridder2016).
Here we compare 4 tests for the diagnosis of taeniasis and evaluate coproantigen and 2 PCR-based tests for species-specific diagnosis of T. solium taeniasis.
Methods
Human fecal samples
Human fecal samples were collected from people in a contiguous area of Betafo and Mandoto provinces of Madagascar as part of baseline evaluations for a T. solium control programme. The region was known to be endemic for T. solium. Two fecal samplings were undertaken. Initially, 960 samples were collected from randomly selected individuals. Samples were examined freshly for the presence of eggs and aliquots of approximately 2 g were suspended in a 10× volume of 90% ethanol (for DNA analyses) and stored at room temperature. Subsequently, a further 960 fecal samples were selected by purposive sampling, selecting random individuals proportional to the number of pigs present in the area, instead of proportional to the number of people as it was done in the first sampling. These samples were examined and stored in ethanol as above, and in addition, a 2 g sample was placed into a 10× volume of 10% formalin (for coproantigen testing) and stored at room temperature. A further 9 egg-positive fecal samples stored in ethanol were available from a previous study undertaken in Madagascar (Ramiandrasoa et al., Reference Ramiandrasoa, Ravoniarimbinina, Solofoniaina, Andrianjafy Rakotomanga, Andrianarisoa, Molia, Labouche, Fahrion, Donadeu, Abela-Ridder and Rajaonatahina2020).
Egg detection
All fecal samples were examined using the Kato–Katz technique to identify taeniid eggs (Katz et al., Reference Katz, Chaves and Pellegrino1972). Most of the samples were evaluated using 1 slide for the Kato–Katz; however, during the second sampling, 78 samples were evaluated by using 2 slides from the same sample.
DNA isolation
Fecal DNA was isolated from all egg-positive fecal samples (n = 25) as well as from 200 randomly selected fecal samples which were found to be egg-negative. Approximately, 250 mg feces were placed in a 1.5 mL microtube and centrifuged at 1000 g for 1 min. The supernatant was discarded and the pellet was resuspended in 1 mL of distilled water by vortexing. Following re-centrifugation, the pelleted feces were processed for DNA isolation using the Qiagen QIAamp® PowerFecal® Pro DNA kit and Qiagen TissueLyser II homogenizer, according to the manufacturer's instructions with elution of fecal DNA in a volume of 50 μL. Purified DNA quantities were determined using the NanoDrop One instrument (ThermoFisher Scientific, Wilmington, DE, USA).
rrnS PCR
All egg-positive and 200 egg-negative fecal DNA samples were assessed by rrnS PCR. A 267 bp fragment of the small subunit of the mitochondrial ribosomal RNA was amplified, corresponding to positions 12 208–12 475 on the complete mitochondrial genome of T. solium (GenBank AB086256.1). The PCR was based on the generic Taenia spp. PCR described by Trachsel et al. (Reference Trachsel, Deplazes and Mathis2007) for the investigation of Taenia spp. infecting canines and also used by Ash et al. (Reference Ash, Okello, Khamlome, Inthavong, Allen and Thompson2017) for human taeniasis. Modified primers (hCest3 5ʹ TGA TTC TTT TTA GGG GAA GGT GTR GTG 3ʹ, hCest5 5ʹ GCG GTG TGT ACA TGA GYT AAA C 3ʹ) were designed more specifically to suit amplification of the sequences from the 3 Taenia sp. infecting humans. Magnesium ion concentration and annealing temperature in PCR were optimized using purified T. solium genomic DNA (Gauci et al., Reference Gauci, Ayebazibwe, Nsadha, Rutebarika, Poudel, Sah, Singh, Stent, Colston, Donadeu and Lightowlers2019). Fifty microlitre reaction volumes were prepared containing 3 mm MgCl2, 50 μm deoxynucleotide triphosphate (dNTP) (Promega), 0.5 μm hCest3 and hCest5 primers, GoTaq Green Reaction Buffer (Promega), 1.25 U GoTaq Flexi DNA polymerase (Promega) and, unless otherwise noted, 2 μL isolated fecal DNA. Controls included 40 pg purified genomic DNA from T. solium, T. saginata and/or T. asiatica (Gauci et al., Reference Gauci, Ayebazibwe, Nsadha, Rutebarika, Poudel, Sah, Singh, Stent, Colston, Donadeu and Lightowlers2019), and fecal DNA isolated from a volunteer known never to have been infected with Taenia sp. PCR conditions were 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 30 s, followed by the final extension at 72°C for 5 min.
Agarose electrophoresis and DNA sequencing
PCR products were electrophoresed in 1.5% agarose, 0.5× Tris-borate-ethylenediaminetetraacetic acid (EDTA) buffer (0.05 m Tris, 0.05 m boric acid, 0.01 m EDTA), 1:10 000 Gel Red (Biotium). PCRs in which a 267 bp rrnS product could be detected in agarose electrophoresis were processed for DNA sequencing. Briefly, 10 μL of PCR amplicon were treated with 20 U exonuclease 1 (ThermoFisher) and 2 U shrimp alkaline phosphatase (ThermoFisher) at 37°C for 30 min and 85°C for 15 min. The DNA sequences of PCR amplicons were determined using the hCEST5 primer by Macrogen, North Korea, using Sanger sequencing. NCBI BLAST comparisons of Taenia spp. DNA sequences amplified from fecal DNA with the appropriate segment of T. solium (GenBank AB086256.1), T. saginata (GenBank AY684274.1) or T. asiatica (GenBank AP017670.1) mitochondrial DNA sequences readily allowed species differentiation.
Tso31-nested PCR
Egg-positive (n = 25) and 200 egg-negative fecal DNA samples were assessed by nested PCR amplifying the single copy of the T. solium genomic gene Tso31 as previously described by Mayta et al. (Reference Mayta, Gilman, Prendergast, Castillo, Tinoco, Garcia, Gonzalez and Sterling2008). Initially, precisely the same commercial reagent suppliers and conditions described by Mayta et al. (Reference Mayta, Gilman, Prendergast, Castillo, Tinoco, Garcia, Gonzalez and Sterling2008) were used. The same nested PCR was undertaken using reagent sources as indicated above for the rrnS PCR and no improvement was found through the use of the particular reagent sources used by Mayta et al. (Reference Mayta, Gilman, Prendergast, Castillo, Tinoco, Garcia, Gonzalez and Sterling2008). For that reason, the following reagents and conditions were adopted for use in 50 μL Tso31-nested PCR reactions. Outer PCR: 3 mm MgCl2, 50 μm dNTP (Promega), 0.5 μm F1 primers (5ʹ ATG ACG GCG GTG CGG AAT TCT G 3ʹ) and R1 primer (5ʹ TCG TGT ATT TGT CGT GCG GGT CTA C 3ʹ) and 4 μL fecal DNA. Incubations were 95°C for 3 min followed by 25 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 1 min. The inner PCR used similar procedures with 2 μL of the outer PCR reaction, except for an MgCl2 concentration of 2.5 mm and 40 cycles, using primers F589 (5ʹ GGT GTC CAA CTC ATT ATA CGC TGT G 3ʹ) and R294 (5ʹ GCA CTA ATG CTA GGC GTC CAG AG 3ʹ). PCR amplicons were analysed on an agarose gel as described above.
Coproantigen
The stool samples stored in 10% formalin were examined for coproantigens using the polyclonal antibody-based enzyme-linked immunosorbent assay as described by Allan et al. (Reference Allan, Avila, Garcia Noval, Flisser and Craig1990) with slight modifications (Mwape et al., Reference Mwape, Phiri, Praet, Muma, Zulu, Van den Bossche, de Deken, Speybroeck, Dorny and Gabriel2012). To determine the test result, the optical density of each stool sample was compared with the mean of a series of 8 reference Taenia-negative stool samples plus 3 standard deviations (cut-off).
Results
Egg detection
Four (0.4%) of the 960 fecal samples randomly selected from the study population were found to have Taenia eggs present using the Kato–Katz method. A further 12 fecal samples were found to have Taenia eggs present among the 960 samples obtained by purposive sampling (1.25%). All the egg-positive samples were identified as positive among the samples tested using a single slide, except for one, which was identified among the 78 samples tested by 2 slides, and was positive in both slides.
Species-specific diagnosis of T. solium taeniasis
A total of 25 fecal samples from different people, which were found to be positive for Taenia spp. eggs by Kato–Katz, were examined using rrnS and Tso31-nested PCRs, as well as 200 randomly selected egg-negative fecal samples. The concentration of DNA obtained from fecal samples using the PowerFecal® Pro DNA kit ranged from 0.1 to 120.1 ng μL−1, depending on the organic content of the fecal sample. Twenty of the 25 egg-positive samples (80%) were positive in rrnS PCR. Of those 20 samples, the DNA sequence of the PCR product was successfully obtained from 18 samples, all of which were confirmed to be T. solium (GenBank accession no. OR098460). Twelve of the 25 egg-positive samples (48%) were positive in the Tso31-nested PCR, all of which were also positive by rrnS PCR.
Analytical sensitivity and specificity of the Tso31 PCR
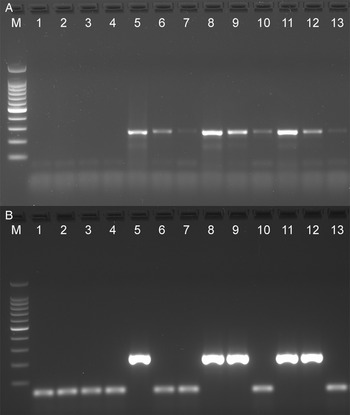
The analytical sensitivity of the Tso31-nested PCR was determined using purified T. solium genomic DNA (Fig. 1A) for comparison with the data published by Mayta et al. (1998). Positive reactions were seen using 500 fg DNA but not at 200 fg. Specificity of the assay is demonstrated in Fig. 1B. No reaction products were found with T. saginata genomic DNA; however, a weak band was evident with genomic DNA from T. asiatica. This band was sequenced using the F589 primer which revealed a sequence (GenBank OQ476203) having 93% identity with the corresponding segment of the Tso31-nested gene.

Figure 1. Agarose gel electrophoresis of PCR products showing analytical sensitivity and specificity of the Tso31-nested PCR. (A) Titration of T. solium DNA in PCR; 100 bp markers, lanes 1–6 PCRs containing 10 pg, 5 pg, 1 pg, 500 fg, 200 fg, 100 fg, DNA respectively. (B) PCR with different Taenia sp. DNA; 100 bp markers, lane 1 T. solium, lane 2 T. saginata, lane 3 T. asiatica.
Sensitivity comparison of PCRs with fecal DNA
The rrnS and Tso31-nested PCRs were compared using dilutions of DNA isolated from the feces of proven T. solium tapeworm carriers. Figure 2 shows the results of rrnS PCR (top panel) and Tso31-nested PCR (bottom panel) using the same DNA samples. DNA from egg-positive fecal samples remained detectably positive in rrnS PCR at 1:10 and 1:100 dilutions of the fecal samples; however, there was an absence of detectable product in some of the same dilution samples using the Tso31-nested PCR (Fig. 2).

Figure 2. Comparative sensitivity of rrnS PCR (A) and Tso31-nested PCR (B) using dilutions of DNA isolated from fecal samples. 100 bp markers, lane 1 distilled water, lanes 2–4 fecal DNA from a known taeniasis-negative individual undiluted (2 μL), 1:10 dilution and 1:100 dilution, respectively; lanes 5–7, 8–10, 11–13 dilutions of fecal DNA from 3 fecal samples with proven T. solium infection, undiluted (2 μL), 1:10 dilution and 1:100 dilution, respectively.
Diagnostic sensitivity comparison of egg detection, PCR and coproantigen
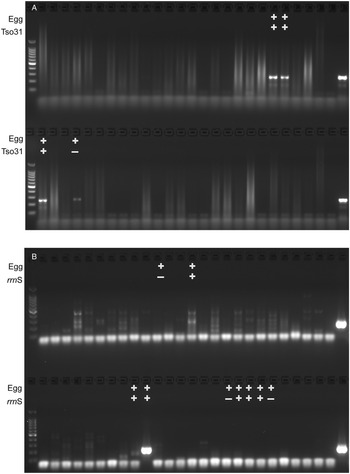
The comparative diagnostic performance of PCR and coproantigen detection for the diagnosis of taeniasis is detailed in Table 1 for those samples for which rrnS PCR, Tso31-nested PCR and coproantigen testing were performed on each. Of the 12 egg-positive samples, 9 (75%) were positive by rrnS PCR, 3 (25%) were positive by Tso31-nested PCR and 9 (75%) were positive by coproantigen testing. Example results obtained in rrnS PCR and the Tso31-nested PCR are shown in Fig. 3. None of the 200 egg-negative fecal samples was positive in either rrnS or Tso31-nested PCR. One egg-positive sample was negative in all other tests and 2 egg-positive samples (8%) that were PCR-negative were found to be positive by coproantigen testing. Two egg-negative samples were positive by coproantigen testing.
Table 1. Comparison of the diagnostic performance of rrnS PCR, Tso31-nested PCR and coproantigen tests for the diagnosis of taeniasis in 12 fecal samples from different persons which were egg-positive for Taenia spp. by the Kato–Katz method. In addition, 200 egg-faecal samples which were negative for Taenia spp. eggs by Kato-Katz were also tested.

KK, Kato–Katz test; CoproAg, coproantigen test; % of KK positives, percentage of all samples positive in Kato–Katz test.

Figure 3. Agarose gel electrophoresis of (A) rrnS PCR and (B) Tso31-nested PCR products obtained from DNA isolated from a subset of 52 human fecal samples that were tested by each method (different fecal samples shown in A and B). Egg+ indicates the fecal sample was positive for taeniid eggs by Kato–Katz. Tso31+ indicates that specific sample tested positive by Tso31-nested PCR, rrnS+ indicates that specific sample tested positive by rrnS PCR. Left hand side lanes, 100 bp markers; right hand side lanes, T. solium DNA positive control.
Discussion
Results of the detection of eggs in the feces by the Kato–Katz method corresponded with those of most egg-positive samples tested by rrnS PCR (Table 1). Of 12 fecal samples in which eggs were detected by Kato–Katz, 9 (75%) were positive in rrnS PCR. Of 9 fecal samples from proven cases of T. solium taeniasis (by egg detection and rrnS DNA sequencing), 7 (78%) were positive by coproantigen. Two of the 12 egg-positive samples were positive by coproantigen but not by PCR. Coproantigen testing identified 2 positive samples among the 200 egg-negative feces which were negative by PCR. It is unknown whether these samples were false positives in the coproantigen test or false negatives in Kato–Katz and PCR. Coproantigens can be detected in fecal samples collected prior to worm patency in both human (Tembo and Craig, Reference Tembo and Craig2015) and animal (Deplazes et al., Reference Deplazes, Gottstein, Stingelin and Eckert1990; Allan et al., Reference Allan, Craig, Garcia Noval, Mencos, Liu, Wang, Wen, Zhou, Stringer, Rogan and Zeyhle1992) cases of taeniasis, providing one possible explanation for these findings.
All taeniasis cases for which the tapeworm species was differentiated were found to be T. solium. A taeniasis survey undertaken in a different region of Madagascar by Rahantamalala et al. (Reference Rahantamalala, Rakotoarison, Rakotomalala, Rakotondrazaka, Kiernan, Castle, Hakami, Choi, Rafalimanantsoa, Harimanana, Wright, Lapierre, Schoenhals, Small, Marcos and Vigan-Womas2022) also identified only cases of T. solium infection. To date, there has been no confirmed case of T. saginata taeniasis described from Madagascar.
Analysis of the quantity of DNA obtained from the feces of 12 egg-positive samples revealed that the 4 samples with the smallest quantity of DNA were all negative in rrnS PCR utilizing 2 μL fecal DNA. Increasing the quantity of DNA to 20 μL led to 2 of these 4 samples being detected as PCR-positive. The relationship between the quantity of DNA from egg-positive samples used in rrnS PCR and the outcome of the test is shown in Fig. 4. One sample which was negative by both PCR and coproantigen tests was not among those with very low quantities of DNA.

Figure 4. Relationship between the quantity of DNA isolated from 12 individual, egg-positive fecal samples and the outcome of rrnS PCR for taeniasis. Closed symbols: PCR-positive samples; open symbols: PCR-negative samples; open circles: samples positive by coproantigen test; open triangle: samples negative by PCR and coproantigen tests.
We were unable to replicate the sensitivity of the Tso31-nested PCR described by Mayta et al. (Reference Mayta, Gilman, Prendergast, Castillo, Tinoco, Garcia, Gonzalez and Sterling2008) who recorded the test to have a sensitivity with purified T. solium genomic DNA as low as 200 fg, whereas in our case the test was positive at 500 fg but negative at 200 fg (Fig. 1). Variations in DNA quantitation methods are one possible reason to explain this difference. Direct comparison of the rrnS and Tso31-nested PCRs with dilutions of fecal DNA from proven cases of T. solium taeniasis (proven by egg detection followed by rrnS PCR and DNA sequencing) found that the rrnS PCR was superior in sensitivity to Tso31-nested PCR in identifying cases of taeniasis (Fig. 2). As the quantity of fecal DNA decreased, the quantity of rrnS PCR-amplified DNA product declined but remained detectable. However, with the Tso31-nested PCR, DNA products were either abundant or completely absent, even when the rrnS PCR remained positive. While the Tso31 PCR involved a pair of nested PCRs, the target (Tso31) is a single-copy gene in the genome (Mayta et al., Reference Mayta, Hancock, Levine, Gilman, Farfan, Verastegui, Lane, Garcia, Gonzalez and Tsang2007). The rrnS PCR targets a segment of the mitochondrial genome. It is unclear just how many copies of the mitochondrial genome are there per cell in T. solium. Depending on cell type, human cells contain between 100 and 10 000 copies of the mitochondrial genome (Wai et al., Reference Wai, Ao, Zhang, Cyr, Dufort and Shoubridge2010). Ultrastructural investigations clearly identify multiple mitochondria in T. solium cells (Willms et al., Reference Willms, Robert and Caro2003) and those of other taeniids (Willms et al., Reference Willms, Robert and Caro2003; Jabbar et al., Reference Jabbar, Crawford, Mlocicki, Swiderski, Conn, Jones, Beveridge and Lightowlers2010). The higher copy number of mitochondrial DNA targets for the rrnS PCR may explain the greater sensitivity of this technique with fecal DNA extracts from T. solium tapeworm carriers compared with the Tso31-nested PCR, despite the latter being a nested PCR which could have been expected to have greater analytical sensitivity. With a single sample of fecal DNA from a case of T. asiatica taeniasis, the Tso31 PCR amplified a product of the same size as that obtained with T. solium, the sequence of which was similar to the sequence of the Tso31 gene in T. solium (Fig. 1), suggesting that the Tso31-nested PCR may not be species-specific where T. solium and T. asiatica are sympatric.
Many PCR-based methods have been described for the diagnosis of T. solium taeniasis using fecal DNA; however, few have been validated using parasitologically proven cases of infection (Lightowlers et al., Reference Lightowlers, Garcia, Gauci, Donadeu and Abela-Ridder2016). Of those that were well validated (e.g. Yamasaki et al., Reference Yamasaki, Allan, Sato, Nakao, Sako, Nakaya, Qiu, Mamuti, Craig and Ito2004; Mayta et al., Reference Mayta, Gilman, Prendergast, Castillo, Tinoco, Garcia, Gonzalez and Sterling2008; Praet et al., Reference Praet, Verweij, Mwape, Phiri, Muma, Zulu, van Lieshout, Rodriguez-Hidalgo, Benitez-Ortiz, Dorny and Gabriel2013; Rodriguez-Hidalgo et al., Reference Rodriguez-Hidalgo, Geysen, Benitez-Ortiz, Dorny, Saa and Brandt2015), they include procedures such as restriction fragment length polymorphism on the PCR amplicons (e.g. Rodriguez-Hidalgo et al., Reference Rodriguez-Hidalgo, Geysen, Benitez-Ortiz, Dorny, Saa and Brandt2015) or quantitative PCR (qPCR) (e.g. Praet et al., Reference Praet, Verweij, Mwape, Phiri, Muma, Zulu, van Lieshout, Rodriguez-Hidalgo, Benitez-Ortiz, Dorny and Gabriel2013; Gordon et al., Reference Gordon, McManus, Acosta, Olveda, Williams, Ross, Gray and Gobert2015; Ng-Nguyen et al., Reference Ng-Nguyen, Stevenson, Dorny, Gabriel, Vo, Nguyen, Phan, Hii and Traub2017). A comparison has not been made previously between the performance of different PCR-based methodologies using the same fecal DNA samples from confirmed cases of T. solium taeniasis. Rahantamalala et al. (Reference Rahantamalala, Rakotoarison, Rakotomalala, Rakotondrazaka, Kiernan, Castle, Hakami, Choi, Rafalimanantsoa, Harimanana, Wright, Lapierre, Schoenhals, Small, Marcos and Vigan-Womas2022) used an approach to species-specific diagnosis of taeniasis involving PCR with non-specific cox1 primers followed by sequencing the DNA product in positive reactions, similar to the one we used. An advantage of the methodology used here, which targeted mitochondrial rrnS, is the small size of the target DNA sequence (267 bp) and that the 3 taeniid species infecting humans can be differentiated simply and unequivocally due to there being numerous sequence differences between the species across a region of only 30 bp. These differences can be determined from single-strand sequencing and without necessarily using sequence comparison software.
Full transmission of T. solium only occurs where pigs have access to materials contaminated with human feces. For this reason, transmission is limited to poor communities in developing countries. It is in these regions and countries where accurate diagnostic methods are needed for T. solum taeniasis. Techniques such as qPCR are poorly suited to these places. Coproantigen testing could offer a simple, potentially inexpensive diagnostic method; however, the currently used methods are neither species-specific nor are the reagents available commercially. A publication has described a species-specific coproantigen test (Parkhouse et al., Reference Parkhouse, Rojas, Aguilar, Medina, Ferrer and Cortez Alcovedes2020), which was evaluated with feces from 2 cases of T. solium taeniasis and 5 cases of T. saginata taeniasis, among other samples; however, there have been no further data published about this test in the ensuing 3 years since its description.
The data presented here suggest that a suitable method for species-specific diagnosis of T. solium taeniasis in fecal samples, which may be relatively simple and suitable for adoption in endemic countries, is an evaluation of fecal samples using a non-specific method such as egg detection or, where available, coproantigen testing, followed by a method where the positive samples are confirmed and speciated by rrnS PCR and DNA sequencing.
Data availability
Data supporting results are provided within the article.
Acknowledgements
The authors thank Davidra Rajaonatahiana, Pascaline Ravoniarimbinina and all team members of the schistosomiasis laboratory's programme at the Ministry of Public Health and in the field for their contributions to Kato–Katz testing.
Author's contribution
M. W. L. and M. D. conceived the study, obtained the funding and undertook data analyses. D. E. A. M., Mi. R., V. C. R. and Mo. R. coordinated the field activities. J. A. N. and N. S. R. organized and supervised field activities and coprological testing. H. A. R. advised on the study design. M. W. L. undertook the lab work for PCRs and DNA sequencing. C. G. G. and A. J. advised on PCR methods. K. E. M. undertook the coproantigen testing. M. W. L. drafted the manuscript and all authors contributed to the final manuscript.
Financial support
Funding for the project was provided by the Livestock Vaccine Innovation Fund, which is supported by the Bill and Melinda Gates Foundation (BMGF), Global Affairs Canada (GAC) and Canada's International Development Research Centre (IDRC), grant number 109273-001.
Competing interest
None.
Ethical standards
Ethics approval for the study was granted by the Madagascar Ministry for Health Ethics Committee for Biomedical Research No. 088-MSANP/SG/AMM/CERBM.