INTRODUCTION
Transmission of infectious diseases in terrestrial beach environments can occur via direct exposure to microbes found in sand or through the flux of microbes from water to sand within the swash or intertidal zone. Exposure to pathogens can include routes such as dermal contact, contact with eyes and ears, inhalation and ingestion. Recent studies suggest that direct exposure to beach sands is a risk factor for infectious disease, particularly in children. An epidemiological study found that gastrointestinal (GI) illness in beach users was associated with exposure to water and intertidal sand (Bonilla et al., Reference Bonilla, Nowosiellski, Cuvelier, Hartz, Grenn, Esiobu, McCorquodale, Fleisher and Rogerson2007; Pinto et al., Reference Pinto, Hachich, Sato, Di Bari, Coelho, Matté, Lamparelli and Razzolini2012a, b; Sabino et al., Reference Sabino, Rodrigues, Costa, Carneiro, Cunha, Duarte, Faria, Ferreira, Gargaté, Júlio, Martins, Nevers, Oleastro, Solo-Gabriele, Veríssimo, Viegas, Whitman and Brandão2014a). A separate epidemiological study (Heaney et al., Reference Heaney, Sams, Dufour, Brenner, Haugland, Chern, Wing, Marshall, Love, Serre, Noble and Wade2009) found that digging in the sand was positively associated with GI illness and was associated with levels of faecal indicator organisms (FIO), enterococci (Heaney et al., Reference Heaney, Sams, Wing, Marshall, Brenner, Dufour and Wade2012). While correlations between beach sand exposure and infectious disease exist, the specific causative mechanisms of infection are yet to be shown including identification of the aetiological agent.
In addition to direct exposure, sand can also serve as a vehicle for transferring pathogenic microbes to and from the adjacent water. Studies on southern Lake Michigan found that densities of Escherichia coli were highest in core samples taken from foreshore sands, often by several logs, but rapidly decreased from that maximum both landwards and lakewards (Whitman & Nevers, Reference Whitman and Nevers2003; Kinzelman et al., Reference Kinzelman, McLellan, Daniels, Cashin, Singh, Gradus and Bagley2004; Whitman et al., Reference Whitman, Nevers and Byappanahalli2006a). Studies at marine beaches in Florida found that the intertidal zone (Figure 1), in particular the upper fringe of this zone, was a hot spot for the accumulation of microbes (Shibata et al., Reference Shibata, Solo-Gabriele, Fleming and Elmir2004; Wright et al., Reference Wright, Abdelzaher, Solo-Gabriele, Elmir and Fleming2011). These microbes can be mobilized from this reservoir by mechanisms such as wave action (Phillips et al., Reference Phillips, Feng, Vogel, Reniers, Haus, Enns, Zhang, Hernandez and Solo-Gabriele2014), pore water transport (Phillips et al., Reference Phillips, Solo-Gabriele, Reniers, Wang, Kiger and Abdel-Mottaleb2011b), and then impact adjacent water quality (Phillips et al., Reference Phillips, Solo-Gabriele, Piggot, Klaus and Zhang2011a). Microbes may also be re-deposited in sand by incoming current and waves (Ge et al., Reference Ge, Nevers, Schwab and Whitman2010). Maximal FIO levels in both marine and freshwater beaches generally occur just behind the highest wave up-rush along the beach. Of particular significance is that the foreshore or intertidal zones are areas where beach goers congregate, and where children tend to play with sand. Thus, the swash zone and foreshore is a dynamic area of the shoreline in terms of microbe accumulation and potential exposures. It is also a zone where the interactions between the water and sand are strongest allowing for an interchange of microbes to and from this zone.

Fig. 1. Zones at the interface between beach sand and water. The terminology differs between tidal marine systems and non-tidal freshwater systems. Mean surface water elevations for marine systems tends to vary with tides. For freshwater systems, in particular within lakes, the mean surface water elevation tends to vary with seiche. For river systems, mean surface water elevations vary with the seasonal elevation of the groundwater table and waves tend to run parallel to the beach as opposed to the perpendicular direction observed in most marine and lake settings. (Image modified from Whitman et al., Reference Whitman, Shively, Pawlik, Nevers and Byappanahalli2014).
To evaluate the current state of knowledge and most pressing research needs in the area of beach sand microbiology, a panel session was convened as part of the TEMPH2014 (Trends in Environmental Microbiology and Public Health, 2014) conference held in Lisbon, Portugal during September 2014. The purpose of the panel was to discuss the potential inclusion of sand quality assessments in monitoring programmes for recreational beaches. The interdisciplinary group of participants held particular expertise in one of two primary categories, recreational beach water quality and environmental mycology. The ideas presented in this article provide an interesting meld of concepts that would benefit beach sand monitoring programmes. Specifically, this work begins by reviewing documents that provide recommendations for changes to existing recreational water quality monitoring guidelines (‘Call-to-Action’ documents), and expands upon these documents by emphasizing the merits of including measures of sand. The manuscript then focuses on describing the spatial distribution and temporal characteristics of microbes in sand, which is necessary for developing general recommendations for sampling programmes. Recommendations for sampling programmes begin by identifying appropriate beach sites and inclusion of beach sanitary assessments. Strategies for sampling and analysis follow, including an emphasis on recommending which microbes to measure and on advances in microbe measurement techniques. We conclude with an identification of research needs and a call for the inclusion of microbial monitoring in sand as an integral part of routine beach health assessments.
OVERVIEW OF ‘CALL-TO-ACTION’ DOCUMENTS
Microbial contamination in recreational waters is monitored through measures of FIOs, including ‘generic’ (non-pathogenic) E. coli and enterococci (EU, 1976, 2006; USEPA, 1986). The enterococci are a group of bacterial species belonging to the genus Enterococcus. FIOs are seldom in themselves pathogenic, but since they are found in faeces of both human and animals they are useful indicators for faecal contamination of water. The ubiquitous distribution of FIOs in faeces stands in contrast to the relatively infrequent occurrence of pathogens, whose detection is complicated by their great diversity. Reliance on FIOs for water quality assessment is thus a matter of practicality, as a set of general targets that are highly concentrated and widely distributed in faeces provides an economical metric, while testing large volumes of water for innumerable pathogens is impractical for monitoring.
The sole reliance on FIO levels in water as a mechanism for classifying recreational waters was challenged by the Annapolis Protocol, a document prepared for the World Health Organization and authored by many of the acknowledged world experts in the field (WHO, 1999), well over a decade ago. The Annapolis Protocol first identified the value of a comprehensive sanitary inspection of recreational waters to identify all sources of potential pathogens, a concept further developed over the past decade (Boehm et al., Reference Boehm, Grant, Kim, Mowbray, McGee, Clark, Foley and Wellman2009a; Gooch-Moore et al., Reference Gooch-Moore, Goodwin, Doresey, Ellender, Mott, Ornelas, Sinigalliano, Vincent, Whiting and Wolfe2011; Abdelzaher et al., Reference Abdelzaher, Solo-Gabriele, Phillips, Elmir and Fleming2013). The Annapolis Protocol introduced the concept of a risk-based approach, and acknowledged that common FIO standards across all waters did not account for the unequal probability of pathogen presence in faecal contamination from different sources. In particular, it identified the probable reduced health risk when FIOs were primarily from non-human sources. Many of the recommendations in the Annapolis Protocol were incorporated into the World Health Organization (WHO) guidelines for recreational waters (WHO, 2003, 2009). Implicit in the WHO approach is the notion that health risks are unacceptable when they exceed a set FIO threshold level.
As understanding of the differential risk inherent in faeces from different sources was accumulating, microbial source tracking (MST) emerged as a discipline. The goal of MST is to determine the host animals responsible for faecal contamination of water. It is accomplished by analysis of host-associated microorganisms (or host genes in the case of mitochondrial DNA) in the faeces of humans and various animals (reviewed in Stoeckel & Harwood, Reference Stoeckel and Harwood2007; Harwood et al., Reference Harwood, Staley, Badgley, Borges and Korajkic2014). In 2005 the US Environmental Protection Agency (EPA) produced a document that outlined the uses of MST to aid in total maximum daily load (TMDL) and risk assessment for recreational waters (USEPA, 2005). Since that time, the ability to discriminate among different sources of contamination in water has steadily improved, and MST has been used to explore FIO sources in beach sand (Russell et al., Reference Russell, Sassoubre, Wang, Masuda, Chen, Soetjipto, Hassaballah and Boehm2013).
The 2003 WHO report has an extensive review of the risk from microbes in sand, but concluded that there was insufficient evidence to support the establishment of a guideline value for indicator or pathogenic microorganisms in beach sand. Neither the subsequent European Union (2006) Bathing Water directive, nor the USEPA (2012) ‘NEEAR’ criteria consider sand contamination, other than acknowledging that it may be a source of FIOs in the adjacent water (EU, 2006; USEPA, 2012). Health Canada recreational water quality guidelines indicate that testing of sand may be warranted in circumstances such as support for sanitary surveys or disease outbreak investigations, though stated that more research was needed before guideline values for sand could be established (Health Canada, 2012). Thus, no regulatory criteria exist currently for microbial levels in sand.
MICROBIAL CHARACTERISTICS OF BEACH SAND
Microbes found in beach sand
Numerous studies have been conducted that document the existence of pathogenic microbes in beach sands, providing evidence for sand as a potential reservoir for aetiological agents of disease (Whitman et al., Reference Whitman, Shively, Pawlik, Nevers and Byappanahalli2014; Sabino et al., Reference Sabino, Rodrigues, Costa, Carneiro, Cunha, Duarte, Faria, Ferreira, Gargaté, Júlio, Martins, Nevers, Oleastro, Solo-Gabriele, Veríssimo, Viegas, Whitman and Brandão2014a). The pathogenic organisms found in sand come from many groups, including bacteria, viruses, protozoa, helminths (worms) and fungi. For example, pathogenic bacteria detected in beach sands include Vibrio vulnificus (Abdelzaher et al., Reference Abdelzaher, Wright, Ortega, Solo-Gabriele, Miller, Elmir, Newman, Shih, Bonilla, Bonilla, Palmer, Scott, Lukasik, Harwood, McQuaig, Sinigalliano, Gidley, Plano, Zhu, Wang and Fleming2010; Shah et al., Reference Shah, Abdelzaher, Phillips, Hernandez, Solo-Gabriele, Kish, Scorzetti, Fell, Diaz, Scott, Lukasik, Harwood, McQuaig, Sinigalliano, Gidley, Wanless, Ager, Lui, Stewart, Plano and Fleming2011), Salmonella (Yamahara et al., Reference Yamahara, Sassoubre, Goodwin and Boehm2012), Campylobacter (Yamahara et al., Reference Yamahara, Sassoubre, Goodwin and Boehm2012), Pseudomonas aeruginosa (Esiobu et al., Reference Esiobu, Mohammed, Echeverry, Green, Bonilla, Hartz, McCorquodale and Rogerson2004) and Staphylococcus aureus including methicillin resistant strains (Plano et al., Reference Plano, Shibata, Garza, Kish, Fleisher, Sinigalliano, Gidley, Withum, Elmir, Hower, Jackson, Barrett, Cleary, Davidson, Davis, Mukherjee, Fleming and Solo-Gabriele2013). Viruses found in sand have included enterovirus by culture (Shah et al., Reference Shah, Abdelzaher, Phillips, Hernandez, Solo-Gabriele, Kish, Scorzetti, Fell, Diaz, Scott, Lukasik, Harwood, McQuaig, Sinigalliano, Gidley, Wanless, Ager, Lui, Stewart, Plano and Fleming2011). Protozoans have included Giardia spp. and Cryptosporidium spp. in nearshore sands (Abdelzaher et al., Reference Abdelzaher, Wright, Ortega, Solo-Gabriele, Miller, Elmir, Newman, Shih, Bonilla, Bonilla, Palmer, Scott, Lukasik, Harwood, McQuaig, Sinigalliano, Gidley, Plano, Zhu, Wang and Fleming2010). Nematode larvae and eggs have also been readily detected in beach sands (Shah et al., Reference Shah, Abdelzaher, Phillips, Hernandez, Solo-Gabriele, Kish, Scorzetti, Fell, Diaz, Scott, Lukasik, Harwood, McQuaig, Sinigalliano, Gidley, Wanless, Ager, Lui, Stewart, Plano and Fleming2011). Similarly, many species of potentially pathogenic yeasts and fungi have been found including Aspergillus sp., Chrysosporium sp., Fusarium sp., Scedosporium sp., Scytalidium sp., Scopulariopsis sp. (Sabino et al., Reference Sabino, Veríssimo, Cunha, Wergikoski, Ferreira, Rodrigues, Parada, Falcão, Rosado, Pinheiro, Paixão and Brandão2011), Candida sp. (Shah et al., Reference Shah, Abdelzaher, Phillips, Hernandez, Solo-Gabriele, Kish, Scorzetti, Fell, Diaz, Scott, Lukasik, Harwood, McQuaig, Sinigalliano, Gidley, Wanless, Ager, Lui, Stewart, Plano and Fleming2011), Penicillium sp., Rhodotorula mucilaginosa (Vogel et al., Reference Vogel, Rogerson, Schatz, Laubach, Tallman and Fell2007), Cladosporium sp., Mucor sp. and Stachybotrys sp. (Gonzales et al., Reference Gonzales, Hanlin, Herrera and Ulloa2000; Migahed, Reference Migahed2003; Gomes et al., Reference Gomes, Cavalcanti, Fernandes, Lima and Passavante2008; Bik et al., Reference Bik, Halanych, Sharma and Thomas2012). Fungi with propensity to infect skin and nails include Trichophyton sp. and Microsporum sp. (Sabino et al., Reference Sabino, Veríssimo, Cunha, Wergikoski, Ferreira, Rodrigues, Parada, Falcão, Rosado, Pinheiro, Paixão and Brandão2011). The presence of black yeasts of the genera Aureobasidium and Exophiala, causing allergies, subcutaneous phaeohyphomycoses and neurotropic infections (de Hoog et al., Reference de Hoog, Guarro, Gene and Figueras2009), have been reported once (Efstratiou & Velegraki, Reference Efstratiou and Velegraki2009), probably reflecting their slow growth and consequential difficulty of detection.
Spatial distribution and temporal characteristics
The spatial and temporal distribution of FIOs and fungi is highly variable in sand over distances as small as a few centimetres (WHO, 2003; Bonilla et al., Reference Bonilla, Nowosiellski, Cuvelier, Hartz, Grenn, Esiobu, McCorquodale, Fleisher and Rogerson2007; Whitman et al., Reference Whitman, Przybyla-Kelly, Shively, Nevers and Byappanahalli2006b). The heterogeneity at this scale may be due to limited transport and mixing of microbes in sand such that once the sand is inoculated under conditions suitable for growth, microbial distribution becomes very patchy. Extreme patchiness can be due to discrete inputs from dog droppings, seabirds and onshore drift, followed by growth. Different zones of the beach may be more or less conducive to microbe growth. Research suggests that the swash zone, and in particular the area just above the maximum up-rush, may also be conducive for regrowth of FIOs through distinct wetting and drying actions and unique characteristics afforded by wrack, which is defined to include seaweed, algae, Sargassum, kelp, Cladophora, macrophytes and other vegetation-like accumulations (Alm et al., Reference Alm, Burke and Spain2003; Whitman et al., Reference Whitman, Harwood, Edge, Nevers, Byappanahalli, Vijayavel, Brandão, Sadowsky, Alm, Crowe, Ferguson, Zhongfu, Halliday, Kinzelman, Kleinheinz, Przybyla-Kelly, Staley, Staley and Solo-Gabriele2003; Olapade et al., Reference Olapade, Depas, Jensen and McLellan2006; Ishii et al., Reference Ishii, Hansen, Hicks and Sadowsky2007; Yamahara et al., Reference Yamahara, Walters and Boehm2009). Although sand moisture content of approximately 8% is sufficient to permit the survival of bacteria, yeasts and nematodes (Whitman et al., Reference Whitman, Shively, Pawlik, Nevers and Byappanahalli2014), there is a lack of consistency in the literature over the distribution of organisms in sand and their relation to moisture content. Generally, a greater density of FIOs in wet foreshore sand has been observed compared with either submerged, backshore, sand at depth, or dry sand (Whitman et al., Reference Whitman, Shively, Pawlik, Nevers and Byappanahalli2014). Conversely, studies at a Florida marine beach found higher concentrations of E. coli and enterococci in supratidal sand (above the high water mark) than in intertidal sand (Abdelzaher et al., Reference Abdelzaher, Wright, Ortega, Solo-Gabriele, Miller, Elmir, Newman, Shih, Bonilla, Bonilla, Palmer, Scott, Lukasik, Harwood, McQuaig, Sinigalliano, Gidley, Plano, Zhu, Wang and Fleming2010).
The accumulation of wrack in the swash zone also serves to maintain FIO populations by serving as a source of nutrients (Byappanahalli et al., Reference Byappanahalli, Shively, Nevers, Sadowsky and Whitman2003; Imamura et al., Reference Imamura, Boehm, Thompson and Jay2011), providing protection from UV light (Feng et al., Reference Feng, Reniers, Haus and Solo-Gabriele2013), and regulating the temperature and moisture conditions in sands located immediately below them. Studies have shown that E. coli and enterococci can survive for over 6 months in sun-dried algal mats (Cladophora) stored at 4°C, and the residual bacteria in the dried alga readily grew upon rehydration (Whitman et al., Reference Whitman, Harwood, Edge, Nevers, Byappanahalli, Vijayavel, Brandão, Sadowsky, Alm, Crowe, Ferguson, Zhongfu, Halliday, Kinzelman, Kleinheinz, Przybyla-Kelly, Staley, Staley and Solo-Gabriele2003). Experimental work in the UK found that FIOs are liable to persist, and possibly proliferate, in supra-littoral wrack piles on a beach (Ward, Reference Ward2009; Dunhill et al., Reference Dunhill, Nicholson, Long, Porter and Rahman2013). In addition to the unique hydrodynamics of this area, which is conducive to the accumulation of wrack, the swash zone also attracts shorebirds that feed and roost in this area and may contribute to the microbial load through their faeces (Lévesque et al., Reference Lévesque, Brousseau, Simard, Dewailly, Meisels, Ramsay and Joly1993; Fogarty et al., Reference Fogarty, Haack, Wolcott and Whitman2003; Wright et al., Reference Wright, Solo-Gabriele, Elmir and Fleming2009; Edge & Hill, Reference Edge and Hill2007; Lu et al., Reference Lu, Hodon, Hill, Schoen, Ashbolt, Edge and Santo Domingo2011). Bird faeces may contribute directly to beach water contamination, although microbial source tracking techniques have found that beach sand (with bird-derived E. coli) can be a more significant secondary source of contamination to adjacent beach water than directly from the bird droppings themselves (Edge & Hill, Reference Edge and Hill2007).
The growth of microbes in sand is not limited to bacteria. It is well recognized that fungi survive, and even grow in sand (Anderson, Reference Anderson1979). This has been demonstrated using both culture and microscopic analyses. For example, Khiyama & Makemson (Reference Khiyama and Makemson1973) reported that culturable fungi in 42 Mediterranean beaches can reach as high as ~7 × 106 CFU g−1 (Larrondo & Calvo, Reference Larrondo and Calvo1989). Fungi levels at beaches have been observed to vary temporally with extreme events. In the volcanic islands of Madeira and Porto Santo, an archipelago of Portugal, pathogens in the beach sands have been associated with intense rainfall events, flash floods and debris flow (Pereira et al., Reference Pereira, Figueira, Aguiar, Vasconcelos, Vasconcelos, Silva, Brandao and Prada2013; Marzol et al., Reference Marzol, Yanes, Romero, Brito de Azevedo, Prada, Martins, Cuadrat Prats, Saz Sánchez, Vicente Serrano, Lanjeri, De Luis Arrillaga and González-Hidalgo2006a, Reference Marzol, Yanes, Romero, Brito de Azevedo, Prada and Martinsb). In a study of 15 Portuguese Atlantic Coast beaches, the highest number of viable fungal colony forming units in sand was in supratidal sand, at around 500 CFU g−1 (Brandão et al., Reference Brandão, Wergikosky, Rosado, Noronha, Verissimo, Falcão, Giraldes, Simões and Rebelo2002).
Antimicrobial resistance
Environmental reservoirs of both antibiotic resistant bacteria (Francino, Reference Francino2012; Wellington et al., Reference Wellington, Boxall, Cross, Feil, William, Hawkey, Johnson-Rollings, Jones, Lee, Otten, Thomas and Williams2013) and antifungal resistant fungi have been emerging. The causes may be associated with the release of antibiotic and antifungal residues, from agriculture, animal feeding, aquaculture and also hospital wastewater (Jiang et al., Reference Jiang, Hu, Yin, Zhang and Yu2011; Suzuki & Hoa, Reference Suzuki and Hoa2012; Diwan et al., Reference Diwan, Stålsby and Tamhankar2013). A variety of antibiotic resistant bacteria have been isolated from sand and beach water, which can be in contact with humans (Velonakis et al., Reference Velonakis, Dimitriadi, Papadogiannakis and Vatopoulos2014). Examples include MRSA (methicillin-resistant Staphylococcus aureus) which has been detected in correlation with the quality of water and sand, showing a relationship with beach-user overcrowding, the concentration of other microorganisms, the presence of yeasts from human origin, as well as water temperature (Papadakis et al., Reference Papadakis, Mavridou, Richardson, Lampiri and Marcelou1997; Plano et al., Reference Plano, Garza, Shibata, Elmir, Kish, Sinigalliano, Gidley, Miller, Withum, Fleming and Solo-Gabriele2011; Roberts et al., Reference Roberts, Olusegun and No2013).
Mudryk et al. (Reference Mudryk, Kosiorek and Perlinski2013) showed that Vibrio species inhabiting sand were more resistant to antibiotics than those isolated from seawater; in addition, more than 90% of planktonic and benthic Vibrio-like bacteria could present multiple antibiotic resistance. Also multidrug resistant Enterococcus faecium from beach sand were identified with similar features to those from clinical human isolates (Heikens et al., Reference Heikens, van Schaik, Leavis, Bonten and Willems2008) indicating that enterococci can be included in the monitoring of sand, with the respective characterization of antibiotic resistance and virulence factors (Pinto et al., Reference Pinto, Hachich, Sato, Di Bari, Coelho, Matté, Lamparelli and Razzolini2012a, Reference Pinto, Ruivo, Vandamme and Lopesb).
Fungi can have an intrinsic antifungal resistance to certain antifungal substances (primary resistance) but initially susceptible microorganisms can also develop resistance (secondary resistance). In the first case, we have examples such as Candida krusei, resistant to fluconazole (Orozco et al., Reference Orozco, Higginbotham, Hitchcock, Parkinson, Falconer, Ibrahim, Ghannoum and Filler1998) or specific Fusarium species (Carneiro et al., Reference Carneiro, Coleman, Restrepo and Mylonakis2011), resistant to the majority of antifungals used in clinical practice (Alastruey-Izquierdo et al., Reference Alastruey-Izquierdo, Cuenca-Estrella, Monzón, Mellado and Rodríguez-Tudela2008). These species have coded in their genome molecular mechanisms that enable them to survive in presence of those antifungals. Candida spp. and Fusarium spp. are frequently found in sand samples and are considered as parameters to evaluate the microbiological quality of a given sample (Sabino et al., Reference Sabino, Veríssimo, Cunha, Wergikoski, Ferreira, Rodrigues, Parada, Falcão, Rosado, Pinheiro, Paixão and Brandão2011). Nevertheless, the number of fungi showing antifungal resistance has been rising over the years. Aspergillus is one of the major fungal threats showing high rates of resistance to azoles, especially in Europe (Sabino et al., Reference Sabino, Veríssimo, Cunha, Wergikoski, Ferreira, Rodrigues, Parada, Falcão, Rosado, Pinheiro, Paixão and Brandão2011) with the environment serving as one of the possible sources of resistant strains (Snelders et al., Reference Snelders, van der Lee, Kuijpers, Rijs, Varga, Samson, Mellado, Donders, Melchers and Verweij2008; Verweij et al., Reference Verweij, Snelders, Kema, Mellado and Melchers2009; Mortensen et al., Reference Mortensen, Mellado, Lass-Florl, Rodriguez-Tudela, Johansen and Arendrup2010; Chowdhary et al., Reference Chowdhary, Kathuria, Xu and Meis2013). In a recent study (Sabino et al., Reference Sabino, Veríssimo, Parada, Brandão, Viegas, Carolino, Clemons and Stevens2014b), the cryptic species of A. fumigatus, A. lentulus and Neosartorya pseudofischeri were detected in samples from beaches. These species have been reported to be resistant in vitro to the azole antifungals (Balajee et al., Reference Balajee, Gribskov, Hanley, Nickle and Marr2005; Yaguchi et al., Reference Yaguchi, Horie, Tanaka, Matsuzawa, Ito and Nishimura2007; Alcazar-Fuoli et al., Reference Alcazar-Fuoli, Mellado, Alastruey-Izquierdo, Cuenca-Estrella and Rodriguez-Tudela2008).
The rapid increase of antibiotic and antifungal resistance compromises the treatment of several bacterial (ECDC, 2013; WHO, 2014) and fungal infections (CDC, 2015). Antibiotic and antifungal resistance is considered a serious threat to human health because of the limited treatments available resulting in greater costs (Finley et al., Reference Finley, Collignon, Larsson, McEwen, Li, Gaze, Reid-Smith, Timinouni, Graham and Topp2013) and increased morbidity and mortality (Ashbolt et al., Reference Ashbolt, Amézquita, Backhaus, Borriello, Brandt, Collignon, Coors, Finley, Gaze, Heberer, Lawrence, Larsson, McEwen, Ryan, Schönfeld, Silley, Snape, Van den Eede and Topp2013; World Health Organization, 2014). MRSA is particularly notorious among the antibiotic resistant bacteria as it causes life-threatening skin ailments that are difficult to treat. Escherichia coli, although a natural inhabitant of the human intestine, has several pathogenic forms causing extreme gastrointestinal infection, some of which exhibit cephalosporin resistance (de Kraker et al., Reference de Kraker, Davey and Grundmann2011). Antibiotic resistant bacteria are found in bathing waters and studies have shown that risks are related to the type of water activity (Leonard et al., Reference Leonard, Zhang, Balfour, Garside and Gaze2015). Regarding fungi, invasive Candida infections are the fourth leading cause of hospital-acquired bloodstream infections, and they are associated with a high mortality (>40%) (Sipsas et al., Reference Sipsas, Lewis, Tarrand, Hachem, Rolston, Raad and Kontoyiannis2009). Candida and Aspergillus species cause a majority of serious infections in non-HIV patients. Because of the high risk of fungal infections in immunocompromised individuals, antifungal prophylaxis is often used to treat these patients. However, the expanding use of antifungal drugs has been associated with increasing incidence of antifungal drug resistance resulting from inherently less sensitive species and/or acquisition of drug class-specific resistance mechanisms (Pfaller et al., Reference Pfaller, Moet, Messer, Jones and Castanheira2011). Most alarming in recent years, resistant strains of certain Candida or Aspergillus species have emerged that are resistant to azoles, especially due to antifungal prophylaxis. Nevertheless, in Aspergillus, and considering the molecular mechanisms underlying their high rates of resistance to azoles, especially in Europe (Chowdhary et al., Reference Chowdhary, Kathuria, Xu and Meis2013) (TR34/L98H and TR46/Y121F/T289A mutations of the CYP51A gene), it was hypothesized that one of the possible sources of resistant strains is the environment (Snelders et al., Reference Snelders, van der Lee, Kuijpers, Rijs, Varga, Samson, Mellado, Donders, Melchers and Verweij2008; Verweij et al., Reference Verweij, Snelders, Kema, Mellado and Melchers2009; Mortensen et al., Reference Mortensen, Mellado, Lass-Florl, Rodriguez-Tudela, Johansen and Arendrup2010; Chowdhary et al., Reference Chowdhary, Kathuria, Xu and Meis2013).
RECOMMENDATIONS FOR SAND MONITORING PROGRAMMES
Criteria for selecting designated recreational beaches
The European Union under the 1976 directive has a specific two-part definition for bathing waters. It defines bathing waters as areas where bathing is explicitly authorized by the competent authorities of each member State, or where bathing is not prohibited and is traditionally practiced by a large number of bathers; The second criteria means that the public largely self select what will become a bathing area and it is then up to the authorities to ensure suitable microbial quality.
For purposes of this review, we consider a recreational beach to serve as a bathing water. We also consider a recreational beach to be a designated shore and water complex largely used for recreation. Waters may be marine or fresh water (fluvial or lentic), and sands may be calcareous, basaltic or siliceous in origin. Indeed, some very well-known beaches are made up of cobble or even bedrock, but here we restrict our discussion to sands. Biologically, beaches like all shorelines are ecotonal, where the terrestrial and nearshore ecosystems interact and overlap (Pennak, Reference Pennak1951). Here organisms interact at many different trophic levels including the microbes, which are found at the lowest levels. Little is known about obligate or specific bacteria, fungi or viruses in sand but we describe in this paper a diverse array of common indicators or pathogenic species known to occur in marine and freshwater sands.
Beach characteristics
At the local level, the beach itself is divided into zones (Figure 1) largely influenced by hydrology: the swash – where wave run-up and return occurs; intertidal – the horizontal extent of tides; the berm – a raised sand ridge of sand deposited by maximum wave run up; foreshore – area under influence of waves and tides; and backshore – landward side of the beach generally not affected by water except during storms. The berm tends to have the highest concentration of microbes due to filtering waves as they infiltrate the sand. The berm may also be a significant source of bacteria to bathing waters when waves or tides re-suspend stored material and return it to the nearshore (Whitman et al., Reference Whitman, Shively, Pawlik, Nevers and Byappanahalli2014). The back- and foreshore may also have significant input from animals especially seabirds. More research is needed on the transport of bacteria or fungi to or from the beach via groundwater but it is presumed that some microbes, especially the smaller ones (<5 microns) (Solo-Gabriele et al., Reference Solo-Gabriele, Neumeister, Dubon and Palmer1998), can pass through shallow groundwater relatively easily. Boehm et al. (Reference Boehm, Shellenbarger and Paytan2004) found that microbes could be potentially transported to the surf zone through tidally driven exchange of groundwater. de Sieyes et al. (Reference de Sieyes, Yamahara, Layton, Joyce and Boehm2008) hypothesized that the transport of nutrients via groundwater promotes the persistence and population replication of bacteria within the surf zone.
Sanitary assessments and sand remediation methods
A sanitary survey is the first step in evaluating pollution sources of a beach. This requires that the beach be viewed within the context of its beachshed. A ‘beachshed’ is ‘a defined stretch of shoreline and the biogeochemical factors that influence it’ (Whitman et al., Reference Whitman, Shively, Pawlik, Nevers and Byappanahalli2014). The extent of the beachshed and its potential influence on a beach should be considered before conducting a sanitary survey, developing a monitoring programme, estimating risk or conducting a microbial source tracking exercise. This would include groundwater, runoff, incoming streams, anthropogenic and natural faecal input, surface water dynamics, offshore influences and general water quality. A common mistake is to seek a single cause of poor sand quality or to finalize a survey after discovering obvious or superficial factors. Multiple sources of contamination are illustrated by the 63rd Street Beach, Chicago (Whitman et al., Reference Whitman, Horvath, Goodrich, Nevers, Wolcott and Haack2001). Investigation has shown that FIOs may be introduced into the foreshore from (1) direct defecation from birds, (2) accumulation of sand wave infiltration, (3) shoreward drift from re-suspended bacteria, (4) growth in stranded green algae Cladophora and (5) in situ growth of bacteria in moist sand (Halliday & Gast, Reference Halliday and Gast2011; Whitman et al., Reference Whitman, Shively, Pawlik, Nevers and Byappanahalli2014). Human faecal sources should be given high priority because they not only pose the greatest health risk, but may lend themselves to engineering and management solutions.
One of the obvious benefits of conducting a sanitary survey is discovering the source or factors contributing to sand contamination. Practical beach management and visitor education are good first steps. An adequate number of animal-proof rubbish receptacles will reduce disease-carrying wildlife, the spread of spoiled, discarded food and waste, and the associated vectors on the beach. Wrack management may help in minimizing the persistence of indicator microbes and some pathogens. Removal of nearby air pollution sources (in the case of airborne fungal spores) could help in the management of fungi in beach sands. As mentioned above, humans are sources of pathogenic microbes and limiting the number of people at a beach to prevent overcrowding will also avoid excessive microbial contributions within a congested environment (Brandão et al., Reference Brandão, Wergikosky, Rosado, Noronha, Verissimo, Falcão, Giraldes, Simões and Rebelo2002). The number of beach visitors could possibly be controlled by limiting access and, in some cases, through the availability of parking. Encouraging visitors to shower before and after returning from the beach and hand washing before eating may reduce illnesses. One study showed that even just rinsing hands in the beach water greatly reduces bacterial adhering on hands (Whitman et al., Reference Whitman, Shively, Ting, Tseng and Nevers2009). If birds contribute to poor sand quality an assessment would be necessary to determine whether deterrence is in line with local ecosystem preservation efforts. If so, there are techniques such as landscaping, sand grooming, and even the use of dogs and other tactics that can be used to deter birds. Nearby streams and margins may also contain high bacteria levels and swimmers should be encouraged avoid these areas. Often break-walls direct contaminants shoreward and managers may wish to have visitors avoid these areas (Byappanahalli et al., Reference Byappanahalli, Nevers, Whitman, Ge, Spoljaric, Shively and Przybyla-Kelly2015). Seepage or runoff on to the beach might also increase contamination. Thus, comprehensive sanitary surveys coupled with visitor education, adaptive management, and well-designed monitoring, will go far in providing safer enjoyment of recreational beaches.
In cases where a sanitary assessment and prevention is not enough, remediation may be needed. Remediation technologies include sand grooming (Kinzelman et al., Reference Kinzelman, Whitman, Byappanahalli, Jackson and Bagley2003; Kinzelman & McLellan, Reference Kinzelman and McLellan2009), sand re-nourishment (Hernandez et al., Reference Hernandez, Hernandez, Jimenez, Piggot, Klaus, Feng, Reniers and Solo-Gabriele2014) and treatment through chemical disinfectants and physical sterilization. Iodine spraying is one of the options currently employed in Portugal (Costa et al., Reference Costa, Bértolo, Bagulho and Duarte2009) but theoretically other non-hazardous options exist such as sonication and high-energy light bathing (such as UV and infra-red radiation, ozone). As harmless for beach users as all these possibilities may be, the downstream pollution cleanup procedures inevitably will act both upon harmful contaminants and normal innocuous flora. Care should be taken that these methods are used only in extreme pollution events rather than as routine procedures.
General considerations for developing a monitoring programme
Designing a sampling programme begins with several preliminary questions: where should we sample, how do we sample, and how often should we sample? Deciding where to monitor can be difficult; experience shows that contaminants arising from the water accumulate along the foreshore but substantial contamination may be occurring from surrounding areas, wildlife, pets and humans themselves higher on the beach. A programme that encompasses areas where visitors might encounter pathogens, ranging from backshore sand to the swash zone, may be appropriate at many beaches. Longitudinal transects along the beach at pre-selected intervals are preferred from a statistical standpoint, but known ‘hotspots’ should not be excluded from these studies.
Achieving representative sampling at beaches is difficult due to diverse inputs of microorganisms that create a heterogeneous community ‘landscape’. Sand is arguably a more problematic matrix than water, as it is relatively less prone to mixing than water. Sample replication is essential since microbial distribution in sand is patchy. An alternative or companion strategy to replication is to collect many individual samples and mix them to create a composite sample, keeping in mind that pseudo-replicate sampling of composite samples should also be carried out to avoid placing undue weight on data based on a very small fraction of the sample. For example, Phillips et al. (Reference Pinto, Hachich, Sato, Di Bari, Coelho, Matté, Lamparelli and Razzolini2011a) collected 60 shallow core samples (each 2.5 cm diameter and 2.5 cm deep) along target transects. They combined these core samples, mixed them thoroughly, and utilized an aliquot for analysis.
Temperature and irradiation of exposed sand can vastly reduce levels of surface microbes. While deep within the sand, communities change due to more negative redox potential and lack of oxygen. Core samples are essential in order to provide an integrated survey of potential microbial pathogens. Sampling depths to 20–30 cm are the most practical and protective for the casual beach visitor. Once collected samples require extraction or elution prior to analysis (Boehm et al., Reference Boehm, Ashbolt, Colford, Dunbar, Fleming, Gold, Hansel, Hunter, Ichida, McGee, Soller and Weisberg2009b).
Decisions on the frequency of monitoring should be based on the amount of beach use, susceptibility to contamination, and also on cost. In temperate climates, beach use is very limited during cold weather, and sampling programmes may be minimized. It is known that sand microbial quality changes much less rapidly than the frequent temporal variability observed in water (Boehm et al., Reference Boehm, Griffith, McGee, Edge, Solo-Gabriele, Whitman, Cao, Getrich, Jay, Ferguson, Goodwin, Lee, Madison and Weisberg2002; Enns et al., Reference Enns, Vogel, Abdelzaher, Solo-Gabriele, Plano, Gidley, Phillips, Klaus, Piggot, Feng, Reniers, Haus, Elmir, Zhang, Jimenez, Abdel-Mottaleb, Schoor, Brown, Khan, Dameron, Salazar and Fleming2012). On the other hand, studies have shown correlations between water and sand FIO content, especially at the foreshore (Whitman & Nevers, Reference Whitman and Nevers2003; Phillips et al., Reference Phillips, Solo-Gabriele, Piggot, Klaus and Zhang2011a, Reference Pinto, Ruivo, Vandamme and Lopesb). Generally, any monitoring programme should account for tradeoffs between visitor risk, budget, effectiveness, and accounting for expected variations in targeted microbes. Beach sand should be routinely sampled at least annually and whenever there has been an event such as a sewage release, major storm events or known seasonal events. Pre- and post-event monitoring where large crowds are anticipated might be considered. High-risk beaches, such as those potentially impacted by human sewage, require more frequent monitoring.
Sampling strategies based upon desired outcomes and integrating traditional approaches
Development of a monitoring programme for beach sand requires first that the desired outcomes of such a programme are clearly defined. An example of a two-part outcome is protecting public health while allowing maximal use of the important resource represented by the beach. Another outcome might be to identify the source of the contamination so that mitigation of the contamination can occur. These outcomes require different sample collection and analysis approaches. For example, if public health is the main criteria, measurements should focus on the primary aetiological agent of disease or an indicator of the aetiological agent. Similarly if the focus is to identify the source of bacterial contamination, then MST should be the focus of sampling efforts. In most cases the aetiological agents and sources are inter-related and so monitoring programmes may be focused on identifying both.
Other factors that contribute towards different sand monitoring approaches include historical factors. When considering the two main categories of application, recreational water quality monitoring vs. environmental mycology, the approaches have been very different. In the USA, the modern era of recreational water quality monitoring was initiated by the amendment to the Federal Water Pollution Control Act of 1948, the Clean Water Restoration Act of 1966. Recreational water quality monitoring using FIO has been guided by a National Academy of Science initiative (NAS, 1972) with its inclusion within regulatory language by 1976 (EU, 1976; USEPA, 1976). The original purpose of beach monitoring during the 1970s was to detect whether the beach was impacted by sewage. This focus was initiated during an era when sewage was disposed to coastal areas with minimal treatment, resulting in direct impacts on local beach water quality from sewage outfalls. In this case, the concept of an indicator microbe, one found in human sewage, made perfect logical sense as indicator microbes are suited to track the aetiological agents of disease from human sewage, especially if the contamination is nearby. However, with improvements to sanitary infrastructures in most developed countries, direct impacts from human sewage have become less significant and other sources of contamination now make a larger contribution to overall beach water quality. These other sources include faeces from animals, wash-off of microbes from human bathers (Elmir et al., Reference Elmir, Wright, Solo-Gabriele, Abdelzaher, Fleming, Miller, Rybolowik, Shih, Pillai, Cooper and Quaye2007, Reference Elmir, Shibata, Solo-Gabriele, Sinigalliano, Gidley, Miller, Plano, Kish, Withum and Fleming2009) and potential regrowth of indicator microbes in beach sands. As a result, the recreational water quality research community argues that the FIO concept is not meeting its originally intended purpose, particularly when the source of contamination is not human sewage. The scientific community has widely acknowledged the need to expand assessment to include additional measures of potential disease-causing agents due to the recognition that there are many other sources of microbes besides human sewage (Fujioka et al., submitted). These additional measures can include alternative indicators such as C. perfringens and Bacteroidales which, unlike the traditional FIO, are unable to multiply in the environment under aerobic conditions.
In contrast, applications in environmental mycology have taken a much different approach. Environmental mycology is generally based upon an underlying notion that fungi are ubiquitous and are derived from many different sources. As such, studies focused on characterizing health threats from fungi in the environment typically include analyses of many fungi, usually as many as can practically be measured. This results in very cumbersome and time-consuming analyses, which in many cases are difficult to assess in terms of human health outcomes; in particular due to the lack of dose response data between fungal exposures and human health outcomes. Ultimately, each individual has its exposure limit to fungi, which is dependent in part upon immune status. As a result, the environmental mycology community recognizes the need to simplify its approach for assessing the potential for transmission of infectious disease within beach settings. The ‘tiered’ approach described below represents a compromise between the recreational water quality community and those whose primary focus area is in environmental mycology.
A tiered approach for beach sand monitoring programmes
The ‘first pass’ of most monitoring programmes is culturable microbes. From the recreational monitoring community these microbes are normally FIOs. But from the environmental mycology perspective these may include total culturable fungi. The methods for analysing culturable microbes are relatively inexpensive, can be performed in laboratories with minimal specialized equipment and expertise, and in many cases have extensive historical use, providing context to new measurements. However, enumerating traditional FIOs, in particular, provides no information about contamination source in sand or water (recently reviewed in Harwood et al., Reference Harwood, Staley, Badgley, Borges and Korajkic2014), and the dearth of sand-related epidemiology studies leaves a substantial knowledge gap about human health risks (Whitman et al., Reference Whitman, Shively, Pawlik, Nevers and Byappanahalli2014).
In contrast to traditional regulatory approaches, the classic approach in environmental mycology has been to measure all fungi and FIO present in sand together with total counts of viable colony-forming units, as described in Brandão et al. (Reference Brandão, Wergikosky, Rosado, Noronha, Verissimo, Falcão, Giraldes, Simões and Rebelo2002). More recently, however, environmental mycologists have started to focus on measures of specific microorganisms, more relevant in terms of public health (i.e. black moulds and keratinophilic fungi) (Sabino et al., Reference Sabino, Rodrigues, Costa, Carneiro, Cunha, Duarte, Faria, Ferreira, Gargaté, Júlio, Martins, Nevers, Oleastro, Solo-Gabriele, Veríssimo, Viegas, Whitman and Brandão2014a). This change is due to the lengthy and expensive practice of identifying all possible organisms. Thus, Environmental Mycology is gradually migrating towards the concept of measuring representative microbes or a fungal indicator microbe. As such the first tiered approach for monitoring the microbiological quality of water and/or beach sand should focus on measures of indicator microbes. This is consistent with current regulatory approaches used to assess recreational water for faecal contamination and is also consistent with the more recent evolving approaches in environmental mycology. In the case of fungi, the first tier analysis approach could include measures of total culturable fungi.
A second-tier approach using source-specific testing (microbial source tracking; MST) may be undertaken if information on the source of faecal contamination or fungi is required, either for mitigation or for risk assessment. Recent studies have estimated very different human health risks from faecal contamination originating from different host animals (Soller et al., Reference Soller, Bartrand, Ashbolt, Ravenscroft and Wade2010, Reference Soller, Schoen, Varghese, Ichida, Boehm, Eftim, Ashbolt and Ravenscroft2014) and so acceptable levels of indictor microbes should consider the potential source of the FIO. For fungi, species identification may help in identifying sources. For example, some fungi are associated with superficial infection, like the contagious Tinea corporis (ringworm). The Trichophyton and Microsporum genera include species of human, animal or soil origin (Badillet, Reference Badillet1973). Thus, identification of species may point to the contamination source for fungi in particular.
A third-tier approach would generally be most useful where many people are likely to be exposed (e.g. a crowded beach with human densities greater than 1 per square metre of beach sand area), and where MST has indicated the likely presence of high-risk faecal or fungal sources. The information provided by MST could be used to target particular pathogens that are known to be shed by the indicated host types. It can also require a great number of separate tests if several host types contribute to contamination. Alternatively, one can use a microarray approach, where hundreds of nucleic acid sequences representing FIOs, pathogens and MST markers can be simultaneously queried (Weidhaas et al., Reference Weidhaas, Li and Harwood2014). An issue with microarray is that sample sizes are very small, so very efficient concentration methods that do not result in interference with nucleic acid hybridization are necessary, and these are still under development.
The second tier and third tier of analyses may be left for reference laboratories, laboratories with the technical expertise to analyse MST markers and pathogens from environmental samples, including capabilities for molecular analyses. These reference laboratories should assess regional specificities of indicator microorganisms and support or validate laboratories capable of basic analysis. For this level of reference, accreditation by ISO 17025 (ISO/IEC 17025, 2005) will ensure the technical proficiency of a laboratory and technical personnel. Inter-laboratory quality assessment schemes will capacitate the laboratories at this level to the point where they can validate first tier analytical approaches and provide expertise for non-standardized analytical methods. In this case, reproducibility and repeatability will ensure results independent of laboratories and technicians.
What should be measured?
The rapid pace of technological advances in the environmental detection and quantification of microbial targets has created what might be considered an embarrassment of riches. It engenders questions such as, should we test for a representative pathogen or two, or a broad suite of pathogens? Does testing need to be quantitative, or do binary results (plus/minus) (presence/absence) suffice? Should the focus be on one microbial type, such as viruses, or should the group of targets be broadened? Ideally, monitoring methods for beach sand monitoring should be inexpensive, provide instant, or at least same-day results, and be directly connected with human health outcomes (Figure 2). The current reality of monitoring methods is that there is no ready protocol that leads precisely to such an elegant outcome. Instead, compromise on one or more aspects of the ideal indicator is necessary and for this reason a tiered approach is recommended by the authors of this review as described above.

Fig. 2. Conceptual triangle for ideal characteristics of an indicator organism used for the first tier of screening sand quality at beaches.
Over the past 60 years FIOs have proved a useful surrogate for measuring pathogens. The FIOs most commonly used for regulatory purposes are enterococci for marine waters (WHO, 2003) and E. coli for fresh water (USEPA, 2012). Other alternative indicator microbes that have been recommended include Clostridium perfringens (Fujioka & Shizumura, Reference Fujioka and Shizumura1985; Roll & Fujioka, Reference Roll and Fujioka1997; Boehm et al., Reference Boehm, Grant, Kim, Mowbray, McGee, Clark, Foley and Wellman2009a), Bacteroidales (Boehm et al., Reference Boehm, Grant, Kim, Mowbray, McGee, Clark, Foley and Wellman2009a) and coliphage (Havelaar et al., Reference Havelaar, van Olphen and Drost1993; Luther & Fujioka, Reference Luther and Fujioka2004; Boehm et al., Reference Boehm, Grant, Kim, Mowbray, McGee, Clark, Foley and Wellman2009a). Although FIOs are utilized extensively worldwide, their limitations have been recognized (EU, 2006). Limitations include the fact that commonly used FIOs are invariably bacterial species, whereas the majority of the reported illnesses are believed to be caused by viruses (particularly norovirus), and analytical techniques suitable for routine use are poor at distinguishing between human and animal sources of bacterial FIOs. Data from the USA identified the following eight faecal pathogens as dominating waterborne illness: norovirus, rotavirus, adenovirus, Cryptosporidium spp., Giardia lamblia, Campylobacter jejuni, Salmonella enterica and E. coli O157:H7 (Mead et al., Reference Mead, Slutsker, Dietz, McCaig, Bresee, Shapiro, Griffin and Tauxe1999; Vital et al., Reference Vital, Hammes and Egli2008). Investigations at beaches in Miami, FL, USA support the hypothesis that existing indicator microbes indirectly monitor several pathogens through common factors, at least in sand. However, for pathogens such as Cryptosporidium spp., Giardia spp. and enterovirus, generalizations about the predictive ability of indicator microbes must be treated with caution owing to the sparseness of data.
Soller et al. (Reference Soller, Bartrand, Ashbolt, Ravenscroft and Wade2010) concluded that in fresh water, enteric viruses and Giardia appear to account for the vast majority of the observed swimming-associated GI illnesses, and when treated sewage effluent predominates, norovirus alone may represent the primary concern. The pre-eminence of noroviruses is supported by work by Public Health England, where norovirus dominated the identified cause of illness from consuming sewage-contaminated shellfish (Figure 3). Evaluating trends from 1991 through 2011, the aetiological agent most frequently identified as the cause of an outbreak was norovirus. This is particularly apparent for more recent years where detection technologies have been capable to identify the aetiological agent responsible for the outbreaks (David Lees, personal communication, CEFAS UK).

Fig. 3. Aetiology of bivalve shellfish associated infections as reported by Public Health England. Data are unpublished. The upper image provides the distribution of aetiological agents of disease for England and Wales Health Protection Agency, UK as provided by Craig Baker-Austin from the Centre for Environment, Fisheries and Aquaculture Science (CEFAS) Laboratory in Weymouth. DSP is Diarrhetic Shellfish Poisoning. The lower image provides the distribution of aetiological agents from 1991 through 2011 showing that among the aetiological agents identified, the one responsible for the majority of the outbreaks is norovirus. The data for the lower plot was provided by David Lees, CEFAS, UK.
The European Union has sponsored an investigation (Virobathe) into analytical methods for viruses for possible incorporation into the 2020 revision of the European Union's (2006) Bathing Water Directive. The report of this work concluded that whilst adenoviruses were a possible control parameter, noroviruses were encountered too infrequently to be considered (EU, 2009; Wyer et al., Reference Wyer, Wyn-Jones, Kay, Au-Yeung, Gironés, López-Pila, de Roda Husman, Rutjes and Schneider2012). However the authors of Virobathe have since recommended to the European Commission that a viral pathogen standard should not be adopted on both analytical and public health grounds. They recommend instead that future risk continues to be managed through demonstrating connectivity to faecal sources, rather than proving that a pathogen is being excreted by the contributing population (Kay, Reference Kay, Wyer, Crowther and Fewtrell2015). Overall, because of their link to gastrointestinal disease and/or detection in recreational waters, viruses that should be considered when evaluating potential aetiological agents in sand include norovirus, adenovirus, rotavirus, enterovirus and hepatitis.
The main fungi pathogenic to man and other mammals are found within the anamorphic (asexual reproductive phase) group. These fungi are saprophytic and occasionally pathogenic, and can be isolated from water, soil, animals and humans (Gomes et al., Reference Gomes, Cavalcanti, Fernandes, Lima and Passavante2008). The presence of certain ubiquitous fungal genera, such as Alternaria, Acremonium, Aspergillus, Candida, Chaetomium, Cladosporium, Fusarium, Mucor, Penicillium, Phoma, Rhodotorula and Trichoderma can be clinically important (Velonakis et al., Reference Velonakis, Dimitriadi, Papadogiannakis and Vatopoulos2014; Wang et al., Reference Wang, Singh, Gao, Zhang, Johnson and Wang2014) due to their involvement in human diseases (Dolenc-Voljč, Reference Dolenc-Voljč2005; The Fungal Research Trust, 2011; Kaštelan et al., Reference Kaštelan, Utješinović-Gudelj, Prpić-Massari and Brajac2014). Among these Aspergillus, Candida, Fusarium and dermatophytes like Microsporum and Trichophyton were identified as representing the majority of fungal isolates from clinical samples important for human health, and that these were appropriate for inclusion in beach sand quality legislation (Rees et al., Reference Rees, Pinner, Hajjeh, Brandt and Reingold1998). These genera should be included in beach sand quality legislation, as should allergenic airborne spore releasing moulds to protect those with respiratory disorders such as cystic fibrosis, asthma and reactive bronchitis. Beach specific studies in Egypt suggest that Candida and Scopulariopsis may be widespread (Migahed, Reference Migahed2003). Melanized fungi, such as black yeasts from the family Herpotrichiellaceae are also recognized as new emerging pathogens (de Hoog et al., Reference de Hoog, Guarro, Gene and Figueras2009). Fungi from genera Cladophialophora, Exophiala and Fonsecaea are causative agents of chromoblastomycosis in subtropic and tropic regions and since they have been detected on wood, soil, plant material and in environments polluted with oil or creosote, their presence should also be evaluated in sand (Vicente et al., Reference Vicente, Attili-Angelis, Pie, Queiroz-Telles, Cruz, Najafzadeh, de Hoog, Zhao and Pizzirani-Kleiner2008), especially on inland beaches. Since they are often detected in beach sand, species like F. pedrosoi, F. monophora, C. bantiana and E. dermatitidis should be included in future legislations of beach sand quality. Organisms of biosafety level 3 like Cladophialophora bantiana should also be considered. Figure 4 brings together data on fungi found in water and sand environments and those in clinical experiments to identify those most of concern in studies of recreational waters.

Fig. 4. Presence of fungal genera in environmental and clinical studies. Blue circle includes fungal genera reported from seawater and ocean studies, orange circle presents fungi isolated from sand. In red circle there are genera reported as causative agents for human disease. The intersection of the circles includes fungi, isolated from two (seawater – beach sand, beach sand – clinical samples, clinical samples – seawater) or all three sampled sites (seawater – beach sand – clinical samples).
Technological advances in microbe measurement techniques
Improving technology has provided several new, but relatively expensive, methodologies for determining the safety of beach sand, e.g. quantitative PCR for specific pathogens or host-specific gene markers, or multi-target methods such as microarray or next-generation nucleic acid sequencing. However, it is important to note that molecular analysis will only reveal the presence of microbial genetic material, which does not always represent viable microbes. Culturable organisms must be capable of replication in order to be detected, a condition that is closer to an infective state that simply possessing genetic material. An example of this discrepancy is the 2012 US Environmental Protection Agency criteria for recreational water quality, which estimates 36 cases of gastroenteritis per 1000 exposed individuals in waters containing a geometric mean level of 35 culturable enterococci per 100 mL, but 470 ‘cell equivalents’ per 100 mL by qPCR (USEPA, 2012). Conversely, some researchers have reported that FIO can enter a viable but non-culturable (VBNC) state in water from which they may be infective (Heim et al., Reference Heim, Del Mar Lleo, Bonato, Guzman and Canepari2002; Lleò et al., Reference Lleò, Bonato, Benedetti and Canepari2005; Boehm & Sassoubre, Reference Boehm, Sassoubre, Gilmore, Clewell, Ike and Shankar2014), and these forms can be detected by molecular methods. Thus although molecular methods can detect specific microbes that are associated with human health outcomes, what they detect is different from culture-dependent methods, which adds to the complexity of adopting new methods based upon knowledge gained from older technologies. Few epidemiology studies have been conducted to determine whether new methods to detect specific pathogens are better assessments of human health outcomes than the century-old FIO paradigm. So a knowledge void exists that fosters indecision about which methods(s) of assessment should be used. Although more information through epidemiology studies may be deemed better, economic constraints become quite important to regulatory agencies and the citizens who must ultimately pay for the testing.
Given the advent of new genomics approaches, measurements of the entire microbial community represent a potential new approach (see Application of Metagenomics to Assess Microbial Communities in Water and Other Environmental Matrices by Staley and Sadowsky in this issue). Many of the microbes living in sand have not been cultured and may not be culturable; thus a complete understanding of the microbial ecology of sand communities has not been possible. Furthermore, the lack of detection of unculturable and potentially infectious microorganisms has confounded monitoring efforts to protect public health. Consequently, metagenomic and 16S-amplicon-based studies to characterize microbial communities in sand, water and sediment habitats will offer great insight into the ecology of these systems (Lozupone & Knight, Reference Lozupone and Knight2007; Staley et al., Reference Staley, Gould, Wang, Phillips, Cotner and Sadowsky2015). In addition, characterization of these communities will provide a context for the role and relative abundance of potential pathogens, which has previously only been assessed using a relatively small number of ephemeral molecular targets (Aw & Rose, Reference Aw and Rose2012). While in its infancy, this type of approach has been taken by Cui et al. (Reference Cui, Yang, Pagaling and Yan2013) who used 454 sequencing to find backshore sands in Hawaii had a more diverse community and contained different populations than other beach zones. Piggot et al. (Reference Piggot, Klaus, Johnson, Phillips and Solo-Gabriele2012) found that the Proteobacteria and Bacteroidetes dominate biofilm communities in South Florida beach sand, with microbial communities that vary by location within the tidal zones and in relation to water activity. More recently, Halliday et al. (Reference Halliday, McLellan, Amaral-Zettler, Sogin and Gast2014) reported that sand at the high tide line, intertidal sand and adjacent water samples contained different overall bacterial communities, that there was some similarity in community composition between coastal water samples from two distant sites, and there was dissimilarity between bacterial communities from high tide and intertidal sands.
Recently, 16S rDNA amplicon analysis, using the Illumina HiSeq and MiSeq platforms was used to examine microbial communities in sands obtained from an estuarine beach and a marine site in Tampa, FL; a freshwater lake in Saint Paul, MN; and Lake Michigan, near Chicago, IL (Whitman et al., Reference Whitman, Shively, Pawlik, Nevers and Byappanahalli2014). Among all three sites, Proteobacteria, Bacteroidetes, Firmicutes and Actinobacteria were the most abundant phyla, with families at all sites including Rhodobacteraceae, Flavobacteriaceae, Flammeovirgaceae and Campylobacteraceae. Sand from the marine sites had greater richness and higher non-parametric diversity indices than the other sites examined (Figure 5). More recently a programme ‘sands of the world’ has been initiated which utilizes, 16S amplicon sequencing and Illiumina HiSeq to examine spatial and temporal diversity of bacterial as well as fungal communities in beach sands collected from fresh (Great Lakes) and salt water beaches around the world. More specifically, this project will characterize microbial diversity in sands from four beaches along the Great Lakes as well as marine beaches on both US coasts, the Gulf of Mexico, Hawaii, Japan and Korea. This information will be useful to determine what environmental factors control beach microbial communities and whether sands harbour unique or similar bacteria, archeal and fungal microbial communities that vary in some predictable manner. In addition, these data may give us insight into which microbial taxa are related to specific sand habitats.

Fig. 5. Most abundant phyla found in beach sand at (A) freshwater, temperate beaches (MN and IL) and (B) marine, tropical beaches (FL). A total of 36 phyla were identified by sequencing of the V6 hypervariable region of the 16S rDNA among all samples.
SCIENTIFIC RESEARCH NEEDS
The research community recognizes that the traditional FIO paradigm is limited. This paradigm has served the public health community well for more than a century, especially in areas directly impacted by sewage. In developed countries, direct sewage impacts on beaches are the exception rather than the rule. In beaches impacted by non-point source contamination, the relationships between FIOs and human health outcomes are not well understood; therefore it is unclear whether continued monitoring for FIOs in these cases is relevant. Regardless of the source of contamination, the incidence of illness is higher for beach bathers relative to non-bathers (Colford et al., Reference Colford, Wade, Schiff, Wright, Griffith, Sandhu, Burns, Sobsey, Lovelace and Weisberg2007; Fleisher et al., Reference Fleisher, Fleming, Solo-Gabriele, Kish, Sinigalliano, Plano, Elmir, Wang, Withum, Shibata, Gidley, Abdelzaher, He, Ortega, Zhu, Wright, Hollenbeck and Backer2010; Sinigalliano et al., Reference Sinigalliano, Fleisher, Gidley, Solo-Gabriele, Shibata, Plano, Elmir, Wanless, Bartkowiak, Boiteau, Withum, Abdelzaher, He, Ortega, Zhu, Wright, Kish, Hollenbeck, Scott, Backer and Fleming2010) and also for beach users who play in the sand relative to those who do not play in the sand (Heaney et al., Reference Heaney, Sams, Wing, Marshall, Brenner, Dufour and Wade2012). It is also recognized that sand can serve as a reservoir of pathogenic microbes, including faecal pathogens of human or animal source, as well as fungi, which are generally considered to be associated with environmental sources. Thus, pathogens may be present and transmitted in the beach environment, even in the absence of sewage contamination. We recommend research to establish a cause and effect relationship for infectious disease contracted within beach environments. First and foremost is the identification of the aetiological agent of disease followed by establishing stronger links between environmental monitoring parameters and human health risk. Specific recommendations include:
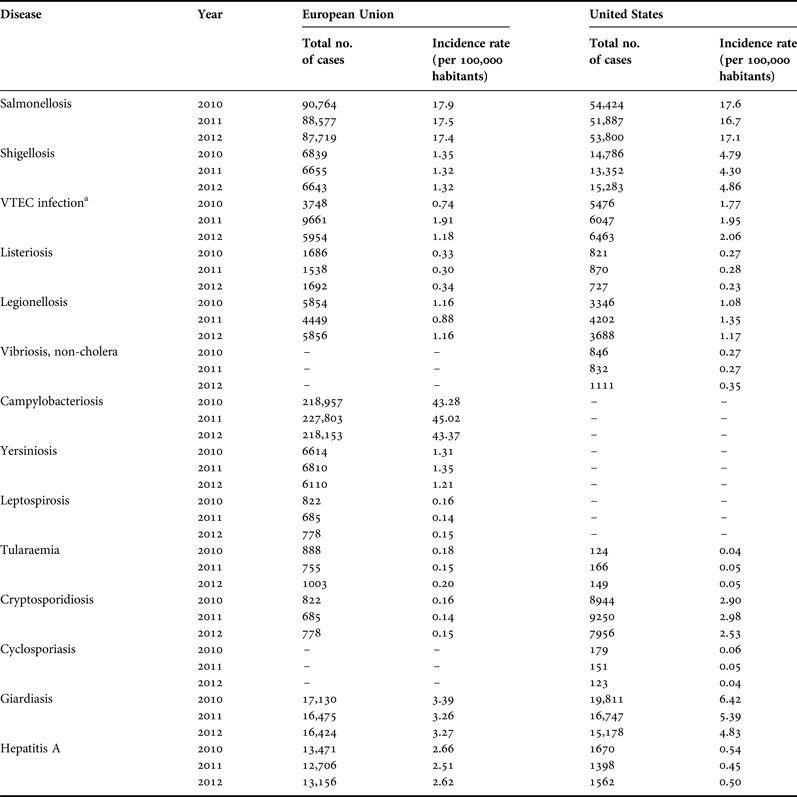
(1) Identification of the aetiological agent(s) of disease. A better understanding is needed of the aetiological agents responsible for the majority of disease attributed to recreational water and sand contact. Identifying the aetiological agent will provide a stronger mechanistic understanding for disease transmission in beach settings. With this understanding, effective control and monitoring programmes can be implemented. To narrow the list of possible aetiological agents, public health data should be examined. A preliminary assessment of reportable diseases in the EU and USA (Table 1) suggest that significant pathogens that have potential sand reservoirs include GI pathogens Salmonella, Shigella, verotoxin-producing E. coli, Campylobacter, Cryptosporidium, Cyclospora, Vibrio, Giardia, hepatitis A and Listeria. Those associated with sediment and water reservoirs include Yersinia, Leptospira and Tularaemia. Although a list of reportable diseases is available, low-level self-limiting diseases (e.g. GI illness, mild skin infections and mild respiratory infections) are usually not reported and the aetiological agents are typically not measured. Wheeler et al. (Reference Wheeler, Sethi, Cowden, Wall, Rodrigues, Tompkins, Hudson and Roderick1999) found that the incidence rate for mild gastroenteritis was under-reported by a factor of 31. Thus numbers listed in Table 1 may significantly underestimate the incidence of reportable diseases. Concerted efforts are needed to encourage clinical practitioners to more often request an evaluation of etiological agents of disease to better track them. Because of unique clinical manifestations, the tracking of fungal infections may be more easily accomplished as opposed to GI infections.
(2) Quantitative microbial risk assessment (QMRA). QMRA methods should be utilized to specifically estimate public health risks from various pathogens (bacteria, fungi, viruses, protozoa, helminths) in beach sand, which can transmit diseases by various exposure routes (contact, ingestion, inhalation). QMRA methods are generally less expensive and less time consuming than epidemiological studies; however, in some cases relationships needed in calculating risks and disease rates are not available (e.g. dose-response relationships for some microbes). An assessment must be made as to which pathogens in beach sand can and cannot be evaluated by QMRA. Preliminary assessments (Shibata & Solo-Gabriele, Reference Shibata and Solo-Gabriele2012) have identified the need for dose-response estimates for fungi and helminths. Moreover, to obtain a better estimate of skin-related ailments, the impacts of wounds should be evaluated on the dose-response of various aetiological agents known to cause skin disease. In some cases, such as for helminths, larvae can penetrate the skin without requiring a wound for entry.
(3) Epidemiological studies. Epidemiological studies measure disease in the exposed population and are the method of choice for establishing the link between human health (GI illness, acute febrile respiratory illness, skin ailments, ear and eye infections) and environmental factors. Since this method is expensive, work intensive and time consuming, it should be used at selected beach sites, bearing in mind the slow onset of infections caused by fungi and parasites. Epidemiologists should determine if study designs can specifically measure pathogen disease rate and measure exposure to the sand by three separate routes (contact, ingestion, inhalation). Controlled cohort or randomized trial studies – similar in aim to those undertaken in water – are needed to better quantify disease risk from exposure to sand.
(4) Evaluate alternatives to FIO for beach monitoring programmes. Although measurements of FIOs should not be discontinued because many beaches are susceptible to sewage contamination, their usefulness needs to be reassessed. The most common FIOs (E. coli, enterococci) fail to fulfil the following three scientifically based ideal criteria or cellular properties of the indicator bacteria, which are required to ensure that the numbers of FIO will correspond to the numbers of sewage-borne pathogens in the water samples tested (Bonde, Reference Bonde1966; Yates, Reference Yates2007): (a) The indicator should be consistently and exclusively associated with a source of human pathogens (e.g. human faeces/sewage); (b) FIO should not be able to multiply under environmental conditions because they would no longer track some sewage-borne pathogens (viruses, protozoa), which presumably do not multiply in the environment; (c) their resistance or survival characteristics to environmental conditions and to wastewater treatment processes should be similar to that of pathogens. Without these cellular characteristics and multiplication in human intestinal sources, the FIO would not track pathogens and subsequently human health. Research is needed to evaluate the reliability and feasibility of monitoring sand for alternative faecal indicators that meet the above three ideal criteria and thus appropriately track pathogens. Since noroviruses are one of the more likely aetiological agents for water-borne transmission of diseases during recreational uses of water, there is a pressing need to develop appropriate viral indicator(s). Viral indicators could include bacteriophages (bacterial viruses), which have similar size, chemical composition and survival characteristics as human enteric viruses. Phages, which are considered promising indicators of human enteric viruses, are F-specific RNA bacteriophages (Havelaar et al., Reference Havelaar, van Olphen and Drost1993; Luther & Fujioka, Reference Luther and Fujioka2004), phages of enterococci bacteria (Santiago-Rodriguez et al., Reference Santiago-Rodriguez, Marcos, Monteiro, Urdaneta, Santos and Toranzos2013) or phages of Bacteroides (Ebdon et al., Reference Ebdon, Muniesa and Taylor2007; McMinn et al., Reference McMinn, Korajkic and Ashbolt2014). In addition, it will be important to evaluate beach sand for the alternative FIO (C. perfringens) because it is a conservative indicator of sewage contamination (Fujioka & Shizumura, Reference Fujioka and Shizumura1985; Roll & Fujioka, Reference Roll and Fujioka1997). Because of their persistence as spores, the use of C. perfringens may be most useful in areas where currents dilute and remove existing contamination.
(5) Develop techniques for detection and quantification of microbe levels. There is a need for improved cultivation methods for detection of fungi, viruses, helminths, protozoa and bacteria in environmental samples. There is a need to determine if the method to detect a specific pathogen or class of pathogens is feasible and reliable for monitoring purposes. If a pathogen detection method is not feasible, then a feasible and reliable indicator monitoring method should be implemented, which should provide data on the quantity and infectivity for that pathogen or that class of pathogens. In this regard, culturable methods provide information on the theoretical infectivity of the pathogen and this kind of data can be used to determine public health risks. Currently, many molecular methods have been developed to rapidly and reliably detect specific pathogens. The limitation of this method is that it does not differentiate between dead and living pathogens. As a result, public health assessments must be based on some assumptions. The value of molecular methods is that they can be used to confirm the presence or absence of specific pathogens in beach sand, regardless of their viability. Improved molecular techniques for the detection of medically important fungi in sand are needed. Also, the ecological role of fungi in coastal reservoirs such as beach sand is little understood (Migahed, Reference Migahed2003) and needs to be investigated. In securing better protection, unnecessary complexity in monitoring is to be avoided. Concern has been expressed on the cost burden of monitoring – particularly in developing regions – of even the existing criteria (WHO, 2003). Against this must be balanced a better cost-benefit balance of targeted improvements and the avoidance of expenditure on ineffective measures undertaken simply to meet flawed criteria (Kay et al., Reference Kay1999).
(6) Pathogen levels and survival in sands. A more complete picture of the types and levels of pathogens in sand is needed, including a focus on evaluating their geographic, spatial and temporal distribution. Multiple studies have documented E. coli and enterococci reservoirs in sand, but few studies have concurrently measured pathogens to determine if E. coli and enterococci are indicative of faecal pollution that carries pathogens, or uncoupled from their original source through prolonged survival or growth. The general consensus among researchers is that pathogens do not multiply in the environment. So prolonged survival or growth of E. coli and enterococci would result in their presence in the absence of pathogens. New research is needed that can provide tools to determine whether FIO are indicative of pathogens within sand environments. The sources of faecal pollution will largely determine the types of pathogens that may be present in sand. Faecal pollution can be deposited directly in sand through outfall runoff or wildlife, or may be delivered through contaminated water. Understanding how faecal pollution and its co-occurring pathogens are modulated in both the sand and water environment can guide the types of indicators or pathogens chosen. Further, gaining a more complete picture of the pathogens that persist in the beach sand and the causative agents for disease, will guide choices of indicators for monitoring and improve assessments of risk.
(7) Develop tools to identify sources. Although considerable advances have been made through MST, more work is needed to identify and approve/agree methods that distinguish between human and non-human sources. There is a need to understand differences in risk among these sources. An improved understanding of the relative risk of faecal contamination from human and other sources is necessary to establish acceptable levels of FIOs in the environment.
(8) Regulations are to reflect microbial sources. Through application of the Annapolis Protocol the WHO has confirmed the need to consider all potential sources of pathogens, not only those from faecal point sources, an approach endorsed by the Rotorua declaration of 2011 (IWA, 2011). The beach environment is an important contributor to water, both through the retention, and possible regrowth, of FIOs within the sand matrix and beach wrack, but also for the presence in sand of non-faecal pathogens including fungi, protozoa and parasites. Forthcoming developments of regulatory standards need to reflect this evolving understanding of microbial sources, the pathogens they contain, and the associated health risks.
(9) Develop reliable sand collection methods designed to recover average pathogen loads for a given beach site or at a specific site where pathogens are suspected. Since pathogen contamination at sandy beaches is expected to be patchy, sand samples from multiple sites should be collected and pooled to determine average concentrations of pathogens in sandy areas. However, targeted sampling (within decaying algae, bird roosts, swash zone, public showers, land-based discharges onto beach sand) should also be conducted where pathogen contamination is suspected. These contaminated patches of sand are good sources for microbial source tracking analysis.
(10) Determine beach sand quality at freshwater vs. marine beaches. Fresh and marine beach sands have been reported to be contaminated by different sources of FIO and pathogens (Whitman et al., Reference Whitman, Shively, Pawlik, Nevers and Byappanahalli2014). For example, different species of algae have been reported to contaminate shorelines of freshwater beaches (Byappanahalli et al., Reference Byappanahalli, Shively, Nevers, Sadowsky and Whitman2003) vs. marine beaches (Imamura et al., Reference Imamura, Boehm, Thompson and Jay2011). In this regard, decay of Cladophora in freshwater beaches has been reported to allow for the growth of FIO and other pathogenic bacteria (Ishii et al., Reference Ishii, Yan, Shively, Byappanahalli, Whitman and Sadowsky2006). Therefore, sand at freshwater beaches and sand at marine beaches can be expected to differ with respect to sources of contamination, types of pathogens and their survival characteristics.
(11) Assess beach sand quality based on contamination by land- and air-based discharges, which are known to be major factors that determine the sources as well as persistence of microorganisms in beach sand. High rainfall patterns cause land-based discharges such as streams and storm drains and may include discharges from agricultural and animal raising facilities. Even beaches with low rainfall can receive substantial urban discharges (storm drains) that include effluents from sewage and industrial facilities as well as discharges of human faeces directly into storm drains. The impacts of these land-based discharges affect the quality of beach sand differently at different beaches and must be assessed as site-specific factors. Air transportation plays the same role for sporulating microorganisms.
(12) Assess standardized methods to recover and disinfect FIO and pathogens from silica-based vs. calcium carbonate-based sands. Both silica sand and calcium carbonate sand are chemically stable sand particles. However, calcium carbonate sand is more reactive and dissolves in dilute acid more readily compared with silica sand. As a result, each may not react similarly to all reagents and may have different influences on survival of microorganisms. There is a need to determine the impact of silica-based and calcium carbonate-based sands on survival characteristics of microbes, on the use of reagents to recover microbes from sand and in the use of chemical reagents to disinfect these two types of beach sand. As a corollary to disinfection, efforts are needed to evaluate the impacts of sand disinfection on the microbial ecosystems and at upper trophic levels.
Table 1. Number of cases of reportable diseases and incidence rates in the EU and US. Data from ECDC (2011a, b, 2012, 2014a, b) and CDC (2013, 2014). Incidence rates based upon population estimates of 506–503 million and 309–314 million for the EU and US, respectively.

a VTEC, verotoxin-producing Escherichia coli, the Shigella-like toxin (includes toxin producing O157). Also known as shiga toxin producing E. coli (STEC).
SUMMARY AND CONCLUSIONS
There is compelling scientific evidence that beaches, through their sands, are a significant contributor to the pathogen load to which beach users are exposed. Many beach epidemiological studies have focused on the impacts of bathing. At beaches that are not impacted by sewage effluent, the source of pathogens originates from the local beach site itself and includes human visitors at the beach, animals, local runoff and the release of microbes from sand. The microbes released from sand can include native microbes (autochthonous) or those that have been deposited from outside sources (allochthonous). Studies have identified the presence of pathogenic microbes in beach sand and have identified factors other than point source pollution that contribute to their presence (e.g. moisture, wrack, wildlife, domestic animals, beach morphology, currents). More recent epidemiological studies have shown that children who play in sand are subject to higher rates of illness relative to those who do not play in the sand. Thus beach sand can serve as a vehicle for disease transmission, either through direct sand contact containing microbes or indirectly through contact with water containing microbes washed off from sand. Given the ability of sand to harbour microbes, we recommend the inclusion of sand measurements in all beach monitoring programmes.
We provide a series of recommendations for beach monitoring programmes that begin by identifying designated recreational beach areas, beach sanitary surveys inclusive of remediation methods, general considerations for monitoring programmes, and a sampling strategy based upon desired outcomes. Given the large number of potential aetiological agents of disease, a tiered approach is recommended for beach sand monitoring. The approach should begin with measures of FIOs and/or total culturable fungi followed by microbes with potential for source tracking. For microbes transmitted via faecal-oral routes, sources should be identified through microbial source tracking. For fungi, specific species can be used to help identify sources. For the third tier, the specific aetiological agent responsible for disease should be measured. No beach epidemiological study to date (whether focused on water or sand) has directly measured pathogens in human subjects to confirm the aetiological agent of disease. According to QMRA methods, the most likely aetiological agents for faecal-oral beach illnesses include norovirus and Giardia. So far, measurements of fungi have not been included in beach epidemiological studies. However, given their propensity in beach sands, agents that we recommend for inclusion in the third tier of measurements are pathogenic Aspergillus sp., Candida sp., Microsporum sp., and Trichophyton sp. Measurement techniques include culture-based methods and quantitative PCR. An alternative approach can include the measure of the beach metagenome as a means of assessing the microbial ecological factors that may facilitate the presence of pathogens.
Considerable evidence exists that sand can serve as a reservoir of enteric microorganisms and fungi, which can be vehicles of disease transmission at beach sites. Current policies worldwide, at both national and international levels, give scant regard to the impact of sands on the health of users of beaches. We recommend that sand quality measures should be considered with some urgency for inclusion in regulatory programmes aimed at protecting recreational beach user health. Contaminated sands present health and economic costs that can and should be known by decision makers, communities and by individuals. Available evidence should be evaluated by both scientists and regulators with a view to filling the data gaps outlined here, which should be followed by sound policy development for safeguarding public health.
ACKNOWLEDGEMENTS
The authors acknowledge the hard work and investment of Eruditus, Relações Publicas e Serviços for the logistics involved in organizing the round table at TEMPH2014 which originated this paper. We acknowledge Katsia Kprzybyla-Kelly for providing valuable suggestions. We thank the ECDC for providing data reported in Table 1 on notifiable infections within Europe. Data provided by ECDC extracted from The European Surveillance System – TESSy. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the ECDC. The accuracy of the authors’ statistical analysis and the findings they report are not the responsibility of ECDC. ECDC is not responsible for conclusions or opinions drawn from the data provided. ECDC is not responsible for the correctness of the data and for data management, data merging and data collation after provision of the data. ECDC shall not be held liable for improper or incorrect use of the data.
FINANCIAL SUPPORT
We thank the Fulbright Foundation for support of Dr Harwood. Participation by H. Solo-Gabriele is associated with the University of Miami Center for Oceans and Human Health (NSF 0CE0432368/0911373/1127813 and NIEHS P50 ES12736). Raquel Sabino was financially supported by a fellowship from Fundação para a Ciência e Tecnologia (FCT) Portugal (contract SFRH/BPD/72775/2010).