INTRODUCTION
Whooping cranes (Grus americana) are an endangered species in North America, with the Aransas-Wood Buffalo population (AWBP) as the only self-sustaining wild population. This population nests in Wood Buffalo National Park, Alberta and Northwest Territories, Canada and winters among coastal marshes at the Aransas National Wildlife Refuge (ANWR) in Texas, USA. During winter 2015–2016, this population was estimated at 329 individuals (95% CI = 293–371; CV = 0·073) (Butler & Harrell, Reference Butler and Harrell2016), a substantial increase from a population low of 15 individuals. Population projections, however, indicate that whooping cranes may not achieve the down-listing criterion of a population size of 1000 individuals (Canadian Wildlife Service & US Fish and Wildlife Service, 2005) until the mid-2060s (Gil-Weir et al. Reference Gil-Weir, Grant, Slack, Wang and Fujiwara2012). Although parasitic infection may be one factor limiting population growth, species management efforts cannot specifically account for infections because very few studies have investigated parasites in the birds (but see Forrester et al. Reference Forrester, Carpenter and Blankinship1978; Bertram et al. Reference Bertram, Hamer, Snowden, Hartup and Hamer2015), likely reflecting the challenges of conducting infectious disease surveillance in an endangered species.
Acute Haemosporida infection has been shown to cause mortality in birds, and has been implicated in the decline and extinction of native Hawaiian bird species (Warner, Reference Warner1968). Due to the short, transient nature of the acute stage of infection and limited mobility of infected hosts during this stage, most studies of Haemosporida infections in wild populations have investigated the chronic phase of infection, which is characterized by low parasitaemias and few or mild clinical signs. Although overt clinical signs are rare, chronic Haemosporida infections are increasingly associated with detrimental effects among infected individuals (Asghar et al. Reference Asghar, Hasselquist, Hansson, Zehtindjiev, Westerdahl and Bensch2015). Prior studies of Haemosporida in cranes of North America are based on examination of blood smears and include descriptions of Haemoproteus antigonis, Haemoproteus balearicae, Plasmodium-polare-like and Leucocytozoon grusi in sandhill cranes (Grus canadensis) (Dusek et al. Reference Dusek, Spalding, Forrester and Greiner2004), and H. antigonis in a small number of non-migratory whooping cranes in Florida (Forrester & Spalding, Reference Forrester and Spalding2003). The majority of chronic Haemosporida infections reported in sandhill cranes have not been reported to cause disease; however, severe anaemia has been associated with acute Haemoproteus balearicae infection in two sandhill crane chicks (Dusek et al. Reference Dusek, Spalding, Forrester and Greiner2004). Previous studies have reported Haemoproteus prevalence of 8–14% in mixed-age sandhill cranes from Florida and from Western North America (Forrester et al. Reference Forrester, Bush, Williams and Weiner1974; Lee et al. Reference Lee, Pence and Gaines1985), and Dusek et al. (Reference Dusek, Spalding, Forrester and Greiner2004) reported a higher prevalence (36%) among sandhill crane chicks in Florida. We have identified H. antigonis infections in AWBP whooping cranes and sympatric sandhill cranes, which comprises a novel molecular clade formed by the parasite species (Bertram et al. unpublished data).
The paucity of health studies on the endangered whooping cranes is a barrier to their effective management and is due in part to the difficulty of obtaining robust sample sizes given permitting and ethical limitations for invasive or disruptive sampling that could negatively impact individuals. The surrogate species approach has been promoted in health studies of endangered taxa to provide otherwise unattainable data for use in assessing potential exposures and disease burden to guide species recovery plans. For example, in order to better understand threats to the endangered Attwater's prairie chicken, sympatric northern bobwhite were assessed for helminthic endoparasites and specific antibodies against infectious agent (Purvis et al. Reference Purvis, Peterson, Dronen, Lichtenfels and Silvy1998), and a similar parasite community was revealed when compared with direct studies of the prairie chickens conducted earlier (Peterson et al. Reference Peterson, Purvis, Lichtenfels, Craig, Dronen and Silvy1998). We suggest that the sandhill crane is a candidate surrogate species for the whooping crane because it is the closest North American relative to the whooping crane, it is abundant, and the Mid-continent population of sandhill cranes has an overlapping range with whooping cranes. We hypothesized that sympatric sandhill cranes will harbour a similar Haemosporida prevalence and diversity to whooping cranes, whereas allopatric sandhill cranes would be associated with a different Haemosporida infection profile. Our objective was to determine the prevalence of infection with Haemosporida (Plasmodium, Haemoproteus and Leucocytozoon) in AWBP whooping cranes and three different wintering populations of sandhill cranes with differing levels of sympatry to the whooping cranes.
METHODS AND MATERIALS
Sample collection
Whooping crane blood samples were collected as part of an ongoing telemetry and health monitoring study of the AWBP whooping cranes between 2010 and 2014 (Pearse et al. Reference Pearse, Brandt, Harrell, Metzger, Baasch and Hefley2015). On both the breeding grounds at Wood Buffalo National Park (Alberta and Northwest Territories, Canada) and the wintering grounds at ANWR (Texas Gulf Coast), birds were captured manually (pre-fledging juveniles) or using remote triggered leg snares that enclosed on the lower tarsus upon triggering. Birds were manually restrained under valid federal, state and provincial permits. Blood was drawn from the jugular vein, and an aliquot of whole blood preserved in Longmire's buffer [0·1 m Tris, 0·1 m EDTA (ethylenediaminetetraacetic acid), 0·01 m NaCl, 0·5% SDS (sodium dodecyl sulphate), pH 8·0] was used in this study. We collected blood samples from hunter-harvested sandhill cranes at necropsy between November 2012 and January 2014 through relationships with the Texas Parks and Wildlife Department, New Mexico Department of Game and Fish, and private hunting clubs and outfitters. Sandhill cranes from the following three populations were sampled: (1) Mid-continent population wintering on the Texas Gulf Coast (harvested in Jackson County, TX). (2) Mid-continent population wintering in the Texas panhandle (harvested in Armstrong and Carson Counties, TX). (3) Rocky Mountain population wintering in New Mexico (harvested in Socorro County, NM). Some birds harvested in New Mexico may also have been part of the Mid-continent population. The Mid-continent population is comprised of Lesser (G. c. canadensis) and Greater (G. c. tabida) subspecies, whereas the Rocky Mountain population is comprised of the Greater subspecies only (Krapu et al. Reference Krapu, Brandt, Jones and Johnson2011). The Rocky Mountain population serves as an out-group for comparison because their breeding, migration and wintering ranges do not overlap with whooping cranes (Fig. 1). All birds were either subjected to necropsy in the field within 6 h post-harvest, or frozen at −20 °C immediately post-harvest and subjected to necropsy in the laboratory at a later date. Each carcass was subjected to a full gross necropsy, at which time we collected whole blood or blood clot, which had pooled in the coelomic cavity. Blood samples were frozen at −20 °C until DNA extraction.
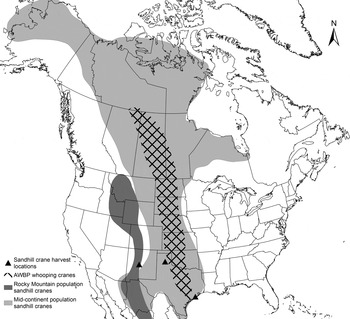
Fig. 1. Ranges of the Mid-continent and Rocky Mountain populations of sandhill cranes in North America and AWBP whooping cranes. Ranges shown include breeding, winter and migration routes (Gil-Weir et al. Reference Gil-Weir, Grant, Slack, Wang and Fujiwara2012; Krapu et al. Reference Krapu, Brandt, Jones and Johnson2011). Locations where sandhill cranes included in this study were harvested are indicated.
Molecular detection of Haemosporida
DNA extraction
DNA was extracted from 100 µL of whole blood or blood clot using the E.Z.N.A Tissue Extraction kit (Omega Biotek, Norcross, GA) following the manufacturer's instructions for tissue extraction with modifications, including an overnight lysis step at 55 °C and elution into 100 µL of elution buffer.
Haemosporida screening
All DNA samples were screened for Haemosporida using three separate PCRs to target different genes and results were interpreted in parallel. A sample was considered positive for overall prevalence estimates, if it met the criteria for positivity on at least one assay. We used negative DNA extraction controls and no template controls in all PCR reactions and samples reported as positive were from clean reactions.
First, Plasmodium and Haemoproteus infections were detected using a nested PCR reaction targeting an approximately 500 bp region of the 3′ end of the mitochondrial cytochrome b (cytb) gene. The first PCR reaction used the primers 3932F (Fecchio et al. Reference Fecchio, Lima, Svensson-Coelho, Marini and Ricklefs2013) and DW4 (Perkins & Schall, Reference Perkins and Schall2002) at a concentration of 0·2 µ m in a 15 µL reaction. Remaining reaction components consisted of 1× FailSafe PCR Premix E (Epicentre, Madison, WI), 0·15 µL FailSafe enzyme, 0·1 µg µL−1 BSA and 1 µL of sample template. The second PCR reaction used the primers 413F and 926R (Ricklefs et al. Reference Ricklefs, Swanson, Fallon, Martinez-Abrain, Scheuerlein, Gray and Latta2005) at a concentration of 0·2 µ m in a 15 µL reaction. Remaining reaction components were identical to the first PCR, except 1 µL of the product from the first PCR was used as the template. In both rounds of PCR, cycling parameters were as described by Fecchio et al. (Reference Fecchio, Lima, Svensson-Coelho, Marini and Ricklefs2013). A sample collected from a northern cardinal (Cardinalis cardinalis) and known to be infected with Plasmodium was used as a positive control (Medeiros et al. Reference Medeiros, Hamer and Ricklefs2013).
Second, we used a nested PCR reaction targeting an approximately 900 bp region of the mitochondrial cytochrome oxidase subunit I (coI) gene. The first PCR reaction used the primers coI/outerF and coI/outerR (Martinsen et al. Reference Martinsen, Perkins and Schall2008) at a concentration of 0·3 µ m in a 15 µL reaction with remaining reaction components as outlined above. The second PCR used the primers coI/nestedF and coI/nestedR (Martinsen et al. Reference Martinsen, Perkins and Schall2008) at a concentration of 0·3 µ m in a 15 µL reaction. Remaining reaction components were identical to the first PCR, except 1 µL of the product from the first PCR, diluted 1:20, which was used as the template. In both rounds of PCR, cycling parameters were as described by Martinsen et al. (Reference Martinsen, Perkins and Schall2008). The same positive control used for cytb PCR reactions was also used for coI PCR reactions.
Third, DNA samples were screened specifically for Leucocytozoon infections using a nested PCR reaction targeting an approximately 700 bp region of the mitochondrial cytb gene. The first PCR reaction used the primers DW2 and DW4 (Perkins & Schall, Reference Perkins and Schall2002) at a concentration of 0·4 µ m in a 15 µL reaction. Remaining reaction components consisted of 1× FailSafe PCR Premix B (Epicentre, Madison, WI), 0·15 µL FailSafe enzyme and 1 µL of sample template. The second PCR used the primers LeucoF and LeucoR (Sehgal et al. Reference Sehgal, Hull, Anderson, Valkiunas, Markovets, Kawamura and Tell2006) at a concentration of 0·4 µ m in a 20 µL reaction. Remaining reaction components consisted of 1× FailSafe PCR Premix B (Epicentre, Madison, WI), 0·2 µL FailSafe enzyme, and 2 µL of the product from the first PCR, diluted 1:20. In both rounds of PCR, cycling parameters were as described by Sehgal et al. (Reference Sehgal, Hull, Anderson, Valkiunas, Markovets, Kawamura and Tell2006). DNA extracted from bird blood known to be positive for Leucocytozoon was obtained from Ravinder Sehgal at San Francisco State University, San Francisco, CA, and used as a positive control.
Sequencing and phylogenetic analyses
Amplicons were purified using ExoSAP-IT (Affymetrix, Santa Clara, CA) according to manufacturer's instructions. Purified samples were submitted for bi-directional sequencing to Eton Bioscience Inc. (San Diego, CA). Chromatographs were examined manually for quality and to assess congruence of forward and reverse strands using Clustal W within Mega 6·0 (Tamura et al. Reference Tamura, Stecher, Peterson, Filipski and Kumar2013). Sequences with double nucleotide peaks were separated using phasing. In this process, sequences with double nucleotide peaks were manually separated into all possible combinations of nucleotides that could have resulted in both nucleotides at a single polymorphic site. The resulting sequences were retained for analysis to determine the likely strains represented in the sample, however sequences with double nucleotide peaks were excluded from phylogenetic analysis. Sequences generated in this study were compared with known Haemosporida sequences using the BLAST tool in GenBank and were aligned with the closest matches and additional publicly available avian Haemosporida species sequences representative of unique clades in previous studies (Perkins & Schall, Reference Perkins and Schall2002; Martinsen et al. Reference Martinsen, Perkins and Schall2008; Outlaw & Ricklefs, Reference Outlaw and Ricklefs2010). Samples were considered positive for prevalence estimates if a DNA sequence was obtained for which the identity matched most closely to a Haemosporida species in GenBank. Due to the sequence homology in the cytb gene across Leucocytozoon, Plasmodium and Haemoproteus species, the PCR assay used to detect Leucocytozoon has been shown to produce false positive results for Leucocytozoon due to the presence of Plasmodium or Haemoproteus (Szollosi et al. Reference Szollosi, Hellgren and Hasselquist2008). Accordingly, we considered a sample positive for Leucocytozoon infection only if a DNA sequence was obtained for which the identity matched most closely to a Leucocytozoon species in GenBank.
Phylogenetic relationships were analysed in Mega 6·0 using the maximum-likelihood method based on a general time reversible with gamma distribution (GTR+G) model of evolution using the bootstrap method with 1000 replicates (Hall, Reference Hall2011). The model was selected based on fit estimated by the Akaike information criterion (AICc) and Bayesian information criterion (BIC). Each unique sequence produced during this project and utilized in the phylogenetic analysis was deposited in GenBank (Accession no. KX223847–KX223877); when sequences occurred more than once in the dataset, the accompanying notes in GenBank identify all samples that shared the same sequence.
Statistical analysis
Statistical analysis was performed using SAS software version 9.4 (Cary, NC). Overall prevalence and confidence intervals were calculated accounting for clustering at the population level. The chi-squared (χ 2) test and logistic regression were used to investigate the relationships between haemoparasite infection and population, age and sex. Fisher's exact test was used to compare Leucocytozoon prevalence among populations due to small numbers of positive samples.
RESULTS
Molecular screening
We collected blood samples from 163 individual cranes, including 61 AWBP whooping cranes, 47 sandhill cranes captured in the Texas Panhandle, 22 sandhill cranes captured along the Texas Gulf Coast and 33 sandhill cranes captured in New Mexico (Table 1). When results from all three Haemosporida assays were interpreted in parallel, we detected Haemosporida in 83·6% (95% CI 74·0, 93·2) of whooping cranes and 59·6% (95% CI 45·0, 74·1) and 63·6% (95% CI 41·8, 85·5) in the two sympatric sandhill crane populations. Infection prevalence was significantly lower in allopatric sandhill cranes (12·1%, 95% CI 0·4, 23·9). Haemoproteus antigonis was detected in a similar proportion of whooping crane samples (57·4%, 95% CI 44·6, 70·2) and samples from two sympatric sandhill crane populations (57·5%, 95% CI 42·8, 72,1; and 50%, 95% CI 27·3, 72·7, respectively). The prevalence of H. antigonis was significantly lower in allopatric sandhill cranes (6·1%, 95% CI 0·0, 14·7). Whooping crane samples had a higher prevalence of Plasmodium spp., than sandhill crane samples overall (29·5%, 95% CI 17·7, 41·3; and 6·9%, 95% CI 1·9, 11·9, respectively), and the prevalence of Plasmodium was not significantly different among the three sandhill crane populations. The prevalence of Leucocytozoon spp. was not significantly different between whooping cranes (4·9%, 95% CI 0·0, 10·5) and sandhill cranes overall (3·9%, 95% CI 0·1, 45·1) or among sandhill crane populations.
Table 1. Prevalence of Haemosporida in AWBP whooping cranes (WHCR) and three populations of sandhill cranes (SACR). Overall prevalences (%) are given for each species, and prevalences for age (juvenile or adult) and sex (male, female or unknown) are given for each population

The 95% confidence intervals are given in parentheses.
Haemoproteus antigonis
Among the four crane populations, we detected significantly fewer infections with H. antigonis in the New Mexico population of sandhill cranes than in the other three populations (χ 2 = 26·99, df = 3, P < 0·0001) (Table 1). Age and sex were not significant predictors of infection; however, age and sex were retained in the regression model because previous studies in other avian species have found differences in infection prevalence between juveniles and adults (Bennett & Fallis, Reference Bennett and Fallis1960; Mendes et al. Reference Mendes, Piersma, Lecoq, Spaans and Ricklefs2005; Atkinson & Samuel, Reference Atkinson and Samuel2010; Hill et al. Reference Hill, Howe, Gartrell and Alley2010), and sex was not distributed equally across populations in our data. When controlling for age and sex of the bird, the odds of infection were 22·9 (95% CI 4·8, 109·1), 27·4 (95% CI 5·1, 148·2) and 17·5 (95% CI 3·3, 92·6) times higher in AWBP whooping cranes, sandhill cranes captured in the Panhandle and along the Gulf Coast than in sandhill cranes captured in New Mexico, respectively. Haemoproteus antigonis infection was present in hatch-year birds, including three of 21 hatch-year whooping cranes captured at Wood Buffalo National Park, four of five hatch-year sandhill cranes captured along the Gulf Coast, and 21 of 32 hatch-year sandhill cranes captured in the Panhandle.
Plasmodium
Among the four crane populations, we detected significantly more infections with Plasmodium in the AWBP whooping cranes than in the three sandhill crane populations (χ 2 = 17·87, df = 3, P = 0·001), and hatch-year birds were significantly more likely to be parasitaemic than adults (χ 2 = 10·73, df = 1, P = 0·001). When controlling for population and sex of the bird, the odds of infection were 6·62 (95% CI 2·16, 20·32) times higher in hatch-year than in adult birds, and when controlling for age and sex the odds of infection were 34·4 (95% CI 4·0, 333), 1·35 (95% CI 0·3, 5·6) and 8·26 (95% CI 0·9, 71·4) times higher in the AWBP whooping cranes than in the Panhandle, Gulf Coast and New Mexico populations, respectively. Plasmodium infection was present in hatch-year birds, including 14 of 21 hatch-year whooping cranes captured at Wood Buffalo National Park, one of five hatch-year sandhill cranes captured along the Gulf Coast, and two of 32 hatch-year sandhill cranes captured in the Panhandle.
Leucocytozoon
We found an overall prevalence of 4·29% (95% CI 0·75, 7·84) of Leucocytozoon infection in cranes, and we did not detect a significant difference in prevalence among the four crane populations (Fisher's exact test, P = 0·601). Leucocytozoon infection was present in hatch-year birds, including two of 21 hatch-year whooping cranes captured at Wood Buffalo National Park, and one of 32 hatch-year sandhill cranes captured in the Panhandle.
Comparison of PCR assays and phylogenetic analysis
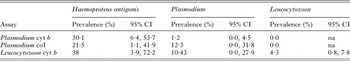
The three primer sets produced different results for some samples with respect to parasitaemia (Table 2). Results agreed between the Haemosporida cyt b and coI assays for 117 (72%) samples, between the Haemosporida cyt b and Leucocytozoon cyt b assays for 116 (71%) samples, and between the Haemosporida coI and Leucocytozoon cyt b assays for 121 (74%) samples.
Table 2. Prevalence of Haemosporida detected by each PCR assay in 163 whooping crane and sandhill crane blood samples
Haemosporida cyt b
We obtained DNA sequences from the cyt b gene for 37 samples, which were included in the phylogenetic analysis. Upon manual examination of the chromatograph traces, an additional three samples had double nucleotide peaks, indicating mixed infections with two strains of the parasite. The three samples were from two whooping cranes and one sandhill crane from the Gulf Coast and had the same double nucleotide peak at the same base pair. Sequences for these samples were separated using phasing. We determined that these three samples were infected with both H. antigonis strains (KX223856 and KX223860).
Phylogenetic analysis revealed 33 sequences, identified as H. antigonis, were identical to each other (KX223856–KX223859) and formed a novel clade, along with an additional three sequences which each differed from the sequence of the majority clade at the same single base pair (KX223860–KX223861) (Fig. 2). Haemoproteus antigonis was detected in all four crane populations, and sequences from all four crane populations were identical to each other. Additionally, one sequence from a whooping crane sample aligned in a clade with a previously published Plasmodium sequence.

Fig. 2. Cladogram using avian Haemosporida cyt b sequences (399 bp). The tree was created using the maximum-likelihood method with a GTR + G model of evolution. Bootstrap values are based on 1000 replicates, and nodes with <50% support are collapsed. Sequences in bold were generated in this study. NOCA – positive control sample from a northern cardinal. W – AWBP whooping crane, STP – sandhill crane harvested in the Texas, SGC – sandhill crane harvested on the Texas Gulf Coast, SNM – sandhill crane harvested in New Mexico.
Haemosporida coI
We obtained sequences from the coI gene for 31 samples, which were included in the phylogenetic analysis. One additional sample had double nucleotide peaks, and phasing determined this sample was infected with two H. antigonis strains (KX223853 and KX223855).
Phylogenetic analysis revealed 13 identical H. antigonis sequences (KX223854–KX223855) that formed a novel clade along with seven additional sequences (KX223853), as was seen on the Haemosporida cyt b phylogenetic analysis (Fig. 3). One sequence aligned in a clade with a previously published Plasmodium sp. (Martinsen et al. Reference Martinsen, Perkins and Schall2008) sequence, two identical sequences aligned with a previously published Plasmodium circumflexum sequence, and an additional eight identical sequences formed a separate, closely related clade. Three sequences in this clade (W02, W14 and SGC20) aligned with H. antigonis in the Haemosporida cyt b analysis. AWBP whooping cranes and sandhill cranes captured in the Panhandle and along the Gulf Coast were represented in each of the unique clades, whereas sandhill cranes captured in New Mexico were not represented in the phylogenetic analysis for the coI assay.

Fig. 3. Cladogram using avian Haemosporida coI sequences (370 bp). The tree was created using the maximum-likelihood method with a GTR + G model of evolution. Bootstrap values are based on 1000 replicates, and nodes with <50% support are collapsed. Sequences in bold were generated in this study. NOCA – positive control sample from a northern cardinal. W – AWBP whooping crane, STP – sandhill crane harvested in the Texas, SGC – sandhill crane harvested on the Texas Gulf Coast, SNM – sandhill crane harvested in New Mexico.
Leucocytozoon cyt b
Although 74 of 163 samples produced a sequence from the Leucocytozoon PCR assay, a majority of the sequences (94%) revealed the presence of other Haemosporida species, which was not unexpected given the sequence homology across taxa (Szollosi et al. Reference Szollosi, Hellgren and Hasselquist2008). We obtained DNA sequences from the cyt b gene for 62 samples, which were included in the phylogenetic analysis. An additional 12 samples had double nucleotide peaks in the chromatographs. Ten samples, from whooping cranes and sandhill cranes captured along the Gulf Coast, had double nucleotide peaks at the same base pairs and were considered mixed infections with the two H. antigonis strains (KX223875 and KX223876). One sample had a mixed infection with two Leucocytozoon strains (KX223864 and KX223866), and one sample had a mixed infection with Plasmodium and H. antigonis (KX223867 and KX223873).
Phylogenetic analysis revealed four sequences aligned with a clade containing previously published Leucocytozoon sequences; however, the crane sequences formed a distinct group within this clade (Fig. 4). All four crane populations were represented in this group. One sample in this group (SGC05) was also represented in the coI analysis, in which it grouped with Plasmodium sequences.

Fig. 4. Cladogram using Leucocytozoon cyt b sequences (617 bp). A) Tree showing samples that do not fall in the novel crane Haemosporida clade. B) Tree showing samples that are included in the novel clade. The tree was created using the maximum-likelihood method with a GTR + G model of evolution. Bootstrap values are based on 1000 replicates, and nodes with <50% support are collapsed. Leucocytozoon A and B are positive control samples. Sequences in bold were generated in this study, and Leucocytozoon sequences are underlined. W – AWBP whooping crane, STP – sandhill crane harvested in the Texas, SGC – sandhill crane harvested on the Texas Gulf Coast, SNM – sandhill crane harvested in New Mexico.
Of the 58 non-Leucocytozoon Haemosporida sequences obtained from this assay, 46 sequences, representing all four crane populations, were H. antigonis and formed a novel clade comprised two strains (KX223873–KX223875, KX223876–KX223877), consistent with results from our analyses of Haemosporida cyt b and coI. Seven were identical sequences (KX223867) and grouped with a previously published Plasmodium polare sequence. An additional five sequences (KX223869–KX223872) grouped with P. circumflexum. One sample in this clade (SGC04) also aligned with Plasmodium in the coI analysis, but aligned with H. antigonis in the Haemosporida cyt b analysis. The P. polare group consisted of whooping crane samples exclusively, whereas all four crane populations were represented in the other Plasmodium group.
Nine samples were represented in the H. antigonis clade in all three analyses, and an additional 21 samples were represented in the H. antigonis clade in two of the three analyses. One sample (W55) aligned with Plasmodium in all three analyses, and an additional two samples aligned with Plasmodium in two of the three analyses. All sequences represented in multiple analyses grouped with the same strain in each analysis, with the exception of the samples mentioned above, which aligned with different species in different analyses.
DISCUSSION
A surrogate species is one that is expected to respond to environmental conditions in a manner similar to the species of interest, usually a species of conservation concern (Caro et al. Reference Caro, Eadie and Sih2005; Murphy et al. Reference Murphy, Weiland and Cummins2011).Surrogate species are often used to study endangered species when the endangered species is particularly susceptible to disturbance, individuals are difficult to locate, the population is too small to afford an adequate sample size, or when use of the endangered species would be impossible (e.g. experimental studies). We applied the surrogate species approach in a parasitological context to describe infections with Haemosporida in the genera Haemoproteus, Plasmodium and Leucocytozoon in endangered wild whooping cranes and three wintering populations of sandhill cranes. We found a similar prevalence and diversity of Haemosporida in whooping cranes and sympatric sandhill cranes, and markedly lower infection in allopatric sandhill cranes, supporting our hypothesis that sandhill cranes that co-occur with whooping cranes are appropriate surrogates.
We found a high prevalence (50–57%) of H. antigonis in AWBP whooping cranes and the two sympatric sandhill crane populations that winter in Texas. In contrast, the sandhill crane population wintering in New Mexico, which does not overlap with whooping cranes, had a significantly lower prevalence (6%) of H. antigonis. The lower prevalence in sandhill cranes harvested in New Mexico may reflect that the majority of sandhill cranes wintering in New Mexico are part of the Rocky Mountain population, which breeds in and around Southeastern Idaho, whereas the two Texas wintering sandhill crane populations are part of the Mid-continent population which breeds in Northern Canada.
Our observations include Haemosporida (H. antigonis, Plasmodium and Leucocytozoon) infection in juvenile whooping cranes sampled on their breeding grounds at Wood Buffalo National Park. This finding indicates that vector-borne transmission is occurring on the breeding grounds. We also noted infections in sympatric juvenile sandhill cranes harvested on the wintering grounds. The prepatent period for most Haemosporida is 11 days to three weeks (Valkiunas, Reference Valkiunas2005), and the sandhill cranes were harvested shortly after arrival to the wintering grounds, which suggests that transmission occurred prior to the arrival of the birds to the wintering grounds. These observations support previous studies, including one on sandhill cranes, which indicate birds usually acquire haemoparasite infections from biting Diptera on their breeding grounds (Lee et al. Reference Lee, Pence and Gaines1985).
The specific vector species are unknown for most avian Haemosporida, including those infecting cranes. Previous studies have suggested Culicoides midges (Bennett & Fallis, Reference Bennett and Fallis1960) and Hippoboscid flies (Lee et al. Reference Lee, Pence and Gaines1985) are the primary vectors for Parahaemoproteus and Haemoproteus, respectively, and mosquitoes are the primary vectors for Plasmodium [reviewed by Valkiunas (Reference Valkiunas2005)]. Interestingly, the most common haemosporidian parasite (H. antigonis) in this study infected all four crane populations, suggesting its vector has a broad geographic range. Despite the uncertainty of vector identity, whooping cranes and Mid-continent sandhill cranes are likely exposed to the same vector communities on their breeding grounds in Northern Canada, resulting in a similar prevalence and diversity of Haemosporida among whooping cranes and sympatric sandhill cranes and further supporting sandhill cranes as a surrogate species.
This is the first study to use molecular techniques to detect haemoparasite infection in North American cranes, and the prevalence reported here is higher than previous studies of sandhill cranes that relied on microscopy. Morphologic studies of Haemosporida are dependent on the quality of the blood smear and the search effort (Valkiunas et al. Reference Valkiunas, Lezhova, Krizanauskiene, Palinauskas, Sehgal and Bensch2008) and may underestimate infection prevalence, especially in chronic infections with low parasitaemia (Garamszegi, Reference Garamszegi2010). Further, many Haemosporida are difficult to identify to species based on morphology, especially when infection intensity is low and not all stages of the parasite are represented in the sample. We used three different PCR assays in parallel to detect infection and mitigate differences in sensitivity and specificity among individual assays, which impact prevalence estimates determined by single assays (Garamszegi, Reference Garamszegi2010). Nonetheless, 46 of 97 positive samples were represented in at least two of the three phylogenetic analyses. Different primers may have differing affinities for different haemosporidian lineages, which can result in bias, especially when mixed infections are present. Additionally, when there are large differences in parasitaemia between co-infecting lineages, the consensus sequence may be biased towards the lineage with the higher concentration of DNA in the sample. In our study, the coI and Leucocytozoon cyt b primers resulted in a higher prevalence of Plasmodium than the Haemosporida cyt b primers. Further, our approach revealed mixed infections in seven samples with H. antigonis and Plasmodium, which may be missed by a single assay (Valkiunas et al. Reference Valkiunas, Bensch, Iezhova, Krizanauskiene, Hellgren and Bolshakov2006). Our results underscore the importance of using multiple assays to increase probability of detection of avian Haemosporida and to help uncover mixed infections.
We detected a low prevalence of Leucocytozoon infection (2–9%) in the four crane populations, which constitutes the first report of Leucytozoon infection in whooping cranes and in sandhill cranes wintering in Texas and New Mexico. This infection prevalence is conservative because we required sequence confirmation of Leucocytozoon to consider a sample positive. Although we screened for coinfections as evidenced by double nucleotide peaks in the sequence chromatographs, we may have failed to detect coinfections, especially if another Haemosporida was present at higher levels than Leucocytozoon. A single species of Leucocytozoon, L. grusi, has been reported in sandhill cranes (Bennett et al. Reference Bennett, Khan and Campbell1974). Previous studies, based on microscopy, have reported a prevalence of 8–18% in the non-migratory Florida sandhill cranes (Bennett et al. Reference Bennett, Khan and Campbell1974; Forrester et al. Reference Forrester, Bush and Williams1975). In contrast, infection prevalence was 50% in one study examining Florida sandhill crane chicks only (Dusek et al. Reference Dusek, Spalding, Forrester and Greiner2004). Forrester et al. (Reference Forrester, Bush, Williams and Weiner1974) did not detect L. grusi in any of 51 Greater (migratory) sandhill cranes wintering in Florida, and Leucocytozoon infection has not previously been reported in whooping cranes (Forrester & Spalding, Reference Forrester and Spalding2003). The Florida sandhill cranes were likely infected during the summer, when Simuliid flies, the vector for Leucocytozoon, were abundant (Forrester & Spalding, Reference Forrester and Spalding2003). Our observations of infections of Leucocytozoon in two juvenile whooping cranes sampled on the breeding grounds in Canada and one sandhill crane harvested in the Texas Panhandle provides further evidence that exposure is occurring during the summer on breeding grounds for both species.
We found many identical parasite sequences in samples from whooping and sandhill cranes, suggesting these parasites are shared between the two crane species. This is likely the result of generalist haematophagous arthropod vectors that feed opportunistically on either crane species as well as the parasite's propensity to infect closely related hosts (Medeiros et al. Reference Medeiros, Hamer and Ricklefs2013; Ellis et al. Reference Ellis, Collins, Medeiros, Sari, Coffey, Dickerson, Lugarini, Stratford, Henry, Merrill, Matthews, Hanson, Roberts, Joyce, Kunkel and Ricklefs2015). This highlights the utility of sympatric sandhill cranes as a surrogate for the whooping crane. Given the establishment of baseline population-level infection prevalence data and validation of the surrogate species, future studies investigating the individual clinical impacts as well as population-level effects (e.g. altered survivorship or recruitment) of avian Haemosporida will aid in the management of both sandhill cranes and whooping cranes.
ACKNOWLEDGEMENTS
We thank Lisa Auckland, Lee Vang, Angela Atkins, Chris Beck, Martha Hensel, and Andrew Golnar for assistance with sample collection, preparation and molecular work. Ravinder Sehgal provided Leucocytozoon-positive control material. We thank Kevin Kraai with the Texas Parks and Wildlife Department; Dan Collins with the United States Fish and Wildlife Service, Region 2, Division of Migratory Birds; Kristin Madden and Aaron Roberts with the New Mexico Department of Game and Fish; Straightline Outfitters; and Thunderbird Hunt Club for supporting the collection of sandhill cranes used in this study. We also thank the Whooping Crane Tracking Partnership, Parks Canada, and Gulf Coast Bird Observatory for facilitating permitting, logistics, and providing personnel to assist in the collection of whooping crane blood samples used in this study.
FINANCIAL SUPPORT
This work was supported by the United States Fish and Wildlife Service, Region 2, Division of Migratory Birds, Avian Health and Disease Program (SAH, GLH, Award NO. F12AC00423); Cooper Ornithological Society, Mewaldt-King Award (MRB); and the College of Veterinary Medicine and Biomedical Sciences at Texas A&M University (Graduate Merit Fellowship to MRB). Additional funding was provided by the University of Wisconsin Companion Animal Fund (BKH) and a USGS Cooperative Agreement (BKH, Award No. G13AC00321).









