Introduction
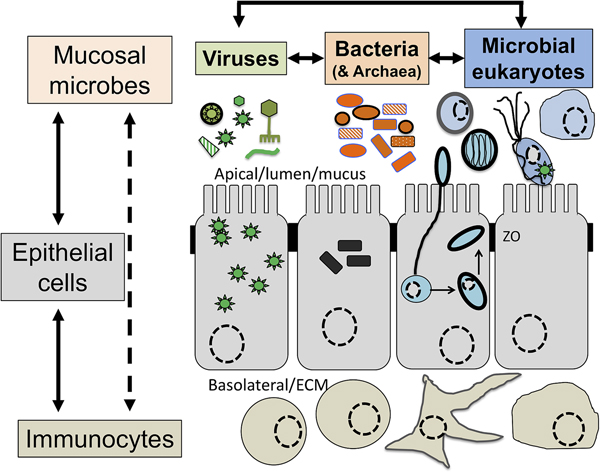
Intimately associated with the human mucosa functions and defences are the complex microbial communities, called the microbiota, which are increasingly understood to play key roles in myriads of aspects in human health and disease (Clemente et al., Reference Clemente, Ursell, Parfrey and Knight2012; Belkaid and Hand, Reference Belkaid and Hand2014). Millions of years of refinements have ensured that mammalian mucosa in a state of homoeostasis are effective at mediating simultaneously two conflicting and essential functions: (i) facilitate exchanges between the outside and the inside of the body to allow optimal breathing, nutrient and water uptake and reproduction and (ii) mediate protection against physical, chemical and biological insults, with the latter being mainly microbial in nature. Many of the molecules of the mucosal innate defence system are key for mediating interactions with microbes, including receptors sensing microbes and the central components of mucus (mucins and antimicrobial peptides) can be traced back to early phases of metazoans evolution (Schroder and Bosch, Reference Schroder and Bosch2016; Bakshani et al., Reference Bakshani, Morales-Garcia, Althaus, Wilcox, Pearson, Bythell and Burgess2018). In contrast, some of the effector molecules and cells characteristic of the human adaptive mucosal immune system represent more recent additions to the mucosal armoury against microbes with secretory IgAs, the archetypal antibody in human mucosal secretions, being only shared with reptiles and birds (Smith et al., Reference Smith, MacDonald, Blumberg, Smith, MacDonald and Blumberg2013). The mucosal microbiota form extraordinarily complex microbial ecosystems where bacteria, archaea, microbial eukaryotes and viruses form an intricate network of microbe–microbe and host–microbe interactions that can be broadly defined as eubiotic, associated with health, or dysbiotic, associated with disease (Petersen and Round, Reference Petersen and Round2014; Levy et al., Reference Levy, Kolodziejczyk, Thaiss and Elinav2017) (Fig. 1). Eubiotic relationships at mucosal surfaces are dependent on the functional characteristics of the microbiota community and corresponding finely tuned mucosal innate and adaptive immune responses to microbes, that together are required for harmonious, highly dynamic and continuous host–microbes interactions at mucosal surfaces (Clemente et al., Reference Clemente, Ursell, Parfrey and Knight2012; Belkaid and Hand, Reference Belkaid and Hand2014; Levy et al., Reference Levy, Kolodziejczyk, Thaiss and Elinav2017). Finely choreographed host–microbiota interactions are essential to maintain mucosal homoeostasis in the broadest possible range of conditions experienced by humans, including variations in diet, exposures to various environmental microbes including pathogens and an increasing range of xenobiotics (Levy et al., Reference Levy, Kolodziejczyk, Thaiss and Elinav2017; Ferreiro et al., Reference Ferreiro, Crook, Gasparrini and Dantas2018).
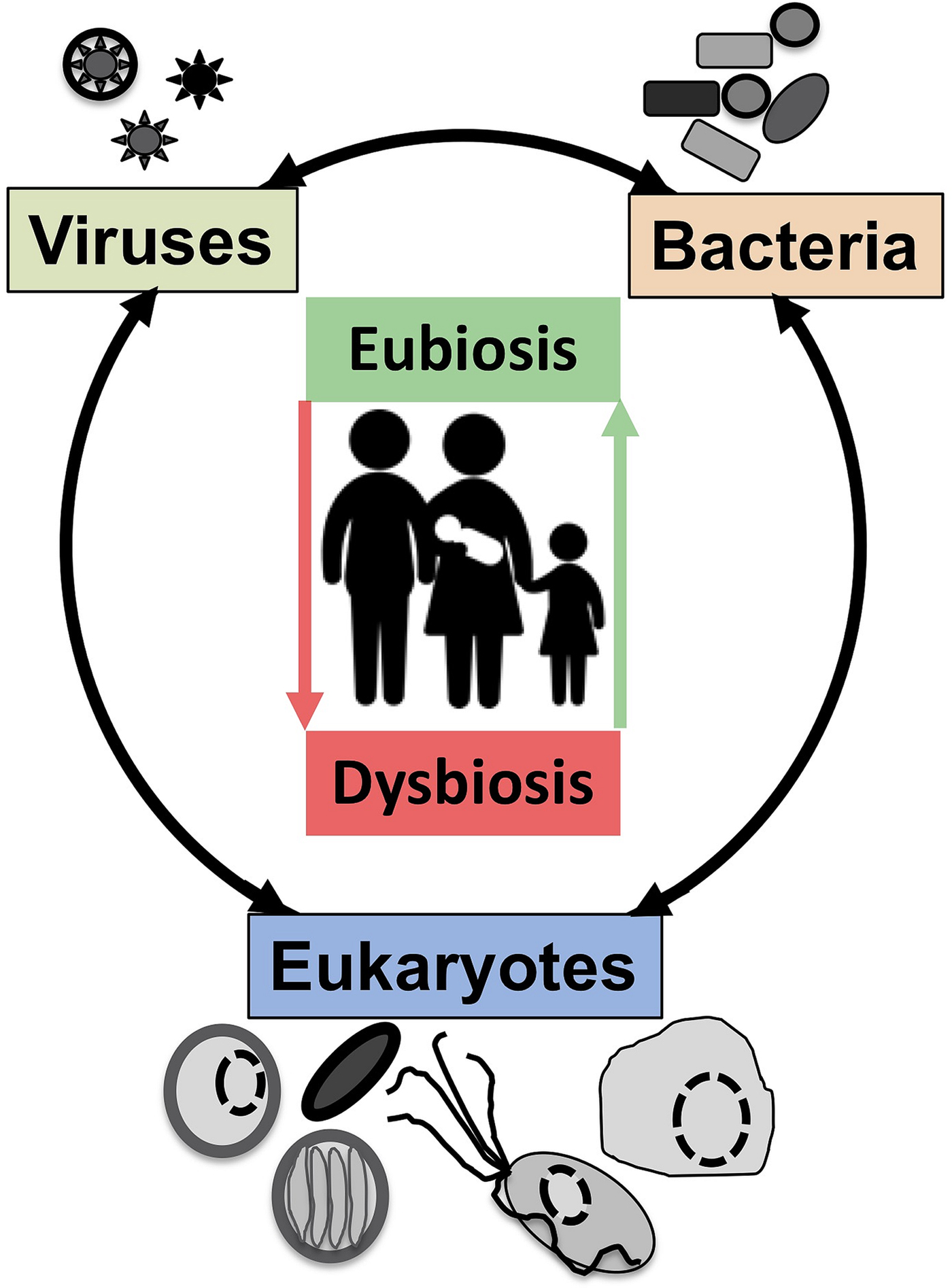
Fig. 1. The now generally accepted new paradigm of microbe–microbe/host–microbe complex network of interactions that can contribute to health (maintaining homoeostasis) or disease (inducing excessive inflammation through time and space) status of the animal/human host. The terms eubiosis and dysbiosis relate to the microbiota functional activities associated with respectively health through maintaining mucosal homoeostasis (ensuring optimal mucosal functionality) or pathologies due to excess inflammation leading to damage mucosal surfaces and that can also contribute to disrupt systemic physiology and sub-optimal cognitive capacities. See main text and cited references for the conceptual limitations on the use of eubiosis and dysbiosis.
Mucosal microbial parasites (also referred to as parasitic protozoa or parasitic protists) are phylogenetically highly diverse and heterogenous that can be broadly distributed across human populations and can contribute to important pathologies but that are also often associated with asymptomatic interactions (Lukes et al., Reference Lukes, Stensvold, Jirku-Pomajbikova and Wegener Parfrey2015; Chabe et al., Reference Chabe, Lokmer and Segurel2017). Thus human–microbial parasite symbiotic relationships can range from commensalism to parasitism and more recently some host–parasites interactions are suggested to have evolved into mutualistic associations too (Lukes et al., Reference Lukes, Stensvold, Jirku-Pomajbikova and Wegener Parfrey2015; Loke and Lim, Reference Loke and Lim2016; Chabe et al., Reference Chabe, Lokmer and Segurel2017; Stensvold, Reference Stensvold2019). Hence these mucosa residents will be referred to here as ‘microbial eukaryote symbionts’ to better capture the diversity of symbiotic interactions mediated by organisms historically typically referred to as parasites (Lukes et al., Reference Lukes, Stensvold, Jirku-Pomajbikova and Wegener Parfrey2015; Stensvold, Reference Stensvold2019). Notably it is increasingly appreciated that this diversity of symbiotic outcomes is the product of parasites–microbiota–host complex network of interactions (Clemente et al., Reference Clemente, Ursell, Parfrey and Knight2012; Burgess et al., Reference Burgess, Gilchrist, Lynn and Petri2017; Rowan-Nash et al., Reference Rowan-Nash, Korry, Mylonakis and Belenky2019), further highlighting the importance of contextuality for the phenotypic outcome of human–microbe interactions (Clemente et al., Reference Clemente, Ursell, Parfrey and Knight2012; Belkaid and Hand, Reference Belkaid and Hand2014; Levy et al., Reference Levy, Kolodziejczyk, Thaiss and Elinav2017). In this editorial, a selection of examples will illustrate how mucosal microbial parasites/symbionts (MMPS) can represent disruptive nodes of the host–microbes complex network of interactions underlying mucosal homoeostasis and thus contribute directly or indirectly to mucosal dysbiosis. In contrast, other examples will illustrate the potential of microbial eukaryote symbionts to contribute to eubiosis (Stensvold and van der Giezen, Reference Stensvold and van der Giezen2018). With these seemingly contradictory considerations in mind, it will be argued that MMPS diversity will represent an important resource to help researchers to dissect the potential causal link between eubiosis and health and dysbiosis and disease through comparative studies. This is a research topic not without controversies and important difficulties and that will require a dramatic increase in the physiological functional characterization of members of the mucosal microbial communities (Hooks and O'Malley, Reference Hooks and O'Malley2017) including microbial eukaryotes (Chabe et al., Reference Chabe, Lokmer and Segurel2017; Stensvold and van der Giezen, Reference Stensvold and van der Giezen2018).
Several papers associated with this Special Issue are derived from talks that were delivered at the EMBO Conference ‘Anaerobic protists: Integrating parasitology with mucosal microbiota and immunology’ (http://meetings.embo.org/event/17-anaerobic-protists) (Labruyere et al., Reference Labruyere, Thibeaux, Olivo-Marin and Guillen2017; Leitsch, Reference Leitsch2017; Dessi et al., Reference Dessi, Margarita, Cocco, Marongiu, Fiori and Rappelli2019; Miranda-Ozuna et al., Reference Miranda-Ozuna, Rivera-Rivas, Cardenas-Guerra, Hernandez-Garcia, Rodriguez-Cruz, Gonzalez-Robles, Chavez-Munguia and Arroyo2019; Stensvold, Reference Stensvold2019). These are complemented by articles providing broader perspectives on the study of the MMPS (Chabra et al., Reference Chabra, Rahimi-Esboei, Habibi, Monadi, Azadbakht, Elmi, Keshavarz, Akhtari, Fakhar and Naghshvar2019; Liu et al., Reference Liu, Xu, Yin, Yuan, Shen and Cao2019; Midlej et al., Reference Victor, Felipe, Wilmer, Érica, Maribel, Wanderley de and Marlene2019; Bartley et al., Reference Bartley, Roehe, Thomson, Shaw, Peto, Innes and Katzer2018; Deere et al., Reference Deere, Parsons, Lonsdorf, Lipende, Kamenya, Collins, Travis and Gillespie2018; van Gestel et al., Reference van Gestel, Kusters and Monkelbaan2018; Vargas Rigo et al., Reference Vargas Rigo, Petro-Silveira, Devereux, McCann, Souza Dos Santos and Tasca2018; Chihi et al., Reference Chihi, Stensvold, Ben-Abda, Ben-Romdhane, Aoun, Siala and Bouratbine2019; Rush et al., Reference Rush, Reynolds, Calvani and Slapeta2019). For more in-depth coverage of MMPS biology including broader taxonomic coverage (e.g. Fungi and Helminths), mucosal sites and biology (e.g. lungs, mucus) and topics including parasite genomics, parasite diagnostics and mucosal vaccine, the reader is directed to the following reviews or original papers (Hupalo et al., Reference Hupalo, Bradic and Carlton2015; Serradell et al., Reference Serradell, Saura, Rupil, Gargantini, Faya, Furlan and Lujan2016; Baker et al., Reference Baker, Bor, Agnello, Shi and He2017; Lemieux et al., Reference Lemieux, Sonzogni-Desautels and Ndao2017; Ryan et al., Reference Ryan, Paparini and Oskam2017; Collins and Belkaid, Reference Collins and Belkaid2018; Corfield, Reference Corfield2018; Rowan-Nash et al., Reference Rowan-Nash, Korry, Mylonakis and Belenky2019).
Mucosal microbial eukaryote diversity, host range and zoonoses
What range of microbial eukaryote symbionts can thrive at our different mucosa, how broadly are they distributed across human populations and what are their host range beyond humans, how genetic diverse are they, what species/genetic lineage are associated with disease and how do these influence the mucosal microbiota and vice versa? These are some of the most basic questions for which we still have relatively limited knowledge for most species. This important knowledge gap currently limits us to properly assess the role in health and disease of microbial eukaryote symbionts and reflects the difficulty of studying mucosal associated organisms and viruses more generally through reductive approaches. A few examples will illustrate the importance of new perspectives one can gain from working on answering these basic questions in humans and animal models. New diagnostic technologies (Ryan et al., Reference Ryan, Paparini and Oskam2017) and the increasing number of microbial eukaryote symbionts genome sequence data (Hupalo et al., Reference Hupalo, Bradic and Carlton2015) are all contributing at providing a better picture of the natural history of MMPS, including non-pathogenic species (Chihi et al., Reference Chihi, Stensvold, Ben-Abda, Ben-Romdhane, Aoun, Siala and Bouratbine2019). These in combination with metagenomics surveys (Lokmer et al., Reference Lokmer, Cian, Froment, Gantois, Viscogliosi, Chabe and Segurel2019) will generate a more comprehensive knowledge on MMPS diversity and host range and their link with health and disease.
The relatively common gut MMPS Blastocystis spp., Dientemoeba fragilis are reviewed by Stensvold (Reference Stensvold2019) (both species) and van Gestel et al. (Reference van Gestel, Kusters and Monkelbaan2018) (D. fragilis). These species are thought to be common in some populations but there are a number of contradictory datasets in relation to their potential role in both disease and health and issues with the apparent important prevalence variations between populations (van Gestel et al., Reference van Gestel, Kusters and Monkelbaan2018). Although potentially misleading detection tools can explain some variation between studies (van Gestel et al., Reference van Gestel, Kusters and Monkelbaan2018; Gough et al., Reference Gough, Ellis and Stark2019), a combination of environmental and biological explanations are also likely to play a role. An intriguing possibility suggested for D. fragilis higher prevalence in some countries is pig farming, which could potentially play a role in its higher prevalence in Denmark and the Netherlands where both humans and pigs cohabit in relatively higher densities (van Gestel et al., Reference van Gestel, Kusters and Monkelbaan2018). This highlights the importance of considering both human and animal prevalence and study in detail the genetic diversity and phylogeny of the microbial eukaryote symbionts to establish their origins among humans and their potential association with animal reservoirs. This is also relevant for the relatively better known species such as Giardia, including in developed countries such as the UK (Horton et al., Reference Horton, Bridle, Alexander and Katzer2019). A recent survey for Giardia duodenalis among cattle in Scotland further illustrates the importance of studying animal populations, where this species was shown to be common across surveyed beef and dairy cattle (~32%) and included genetic lineages associated with human symptomatic infections (Bartley et al., Reference Bartley, Roehe, Thomson, Shaw, Peto, Innes and Katzer2018). In another example, vaccination to protect dogs and cats from G. duodenalis infections (100% prevalence) in a peri-urban disadvantaged community in Argentina, using an elegant vaccination strategy (Rivero et al., Reference Rivero, Saura, Prucca, Carranza, Torri and Lujan2010), was shown to reduce dog and cat infections with the concomitant reduction of children infections in the community associated with the vaccinated pets (Serradell et al., Reference Serradell, Saura, Rupil, Gargantini, Faya, Furlan and Lujan2016). This example illustrates the importance of both the knowledge of the epidemiology of a potential pathogen and the molecular mechanisms underlying surface antigen variation to develop an effective vaccine for relevant hosts to eventually also control infections among humans. Similarly, the prevalence of Entamoeba spp., including Entamoeba histolytica, among humans, chimpanzees and baboon in the Greater Gombe Ecosystem in Tanzania, where the human and nonhuman primate populations overlap, demonstrated a high level of prevalence (~60% for all Entamoeba spp. and ~10% of E. histolytica) among all three species highlighting the potential for zoonotic transmission of Entamoeba species (Deere et al., Reference Deere, Parsons, Lonsdorf, Lipende, Kamenya, Collins, Travis and Gillespie2018). Notably the presence of E. histolytica in chimpanzees was apparently never associated with symptoms in the tested population, in contrast to human infections (Deere et al., Reference Deere, Parsons, Lonsdorf, Lipende, Kamenya, Collins, Travis and Gillespie2018).
Beyond the gut, an interesting set of data for Trichomonas vaginalis and Trichomonas tenax, infecting respectively the urogenital tract (Hirt and Sherrard, Reference Hirt and Sherrard2015) and oral cavities (Marty et al., Reference Marty, Lemaitre, Kemoun, Morrier and Monsarrat2017) also highlight the importance of specific and sensitive diagnostics and the knowledge of their distributions beyond humans (Maritz et al., Reference Maritz, Land, Carlton and Hirt2014). Through carefully testing the specificity of a molecular diagnostic tool used for T. vaginalis it was discovered that some infections of the urogenital tract (three male urine samples) could be due to T. tenax rather than T. vaginalis (Brosh-Nissimov et al., Reference Brosh-Nissimov, Hindiyeh, Azar, Smollan, Belausov, Mandelboim, Rahav, Keller and Gefen-Halevi2019). A screening across dogs and cats for oral trichomonads also indicated a potential zoonotic source for T. tenax from pets (Kellerova and Tachezy, Reference Kellerova and Tachezy2017). Genotyping T. tenax clinical isolates from humans also established that a subset of genetic lineages are significantly associated with periodontal patients, in addition of being common among the tested population in an affluent setting (35% among patients with periodontitis and 19% among healthy controls in the studied French cohort) (Benabdelkader et al., Reference Benabdelkader, Andreani, Gillet, Terrer, Pignoly, Chaudet, Aboudharam and La Scola2019). Notably both T. vaginalis and T. tenax are likely derived from species infecting birds (Maritz et al., Reference Maritz, Land, Carlton and Hirt2014) as these two species are respectively more closely related phylogenetically to a distinct set of species infecting birds including Trichomonas gallinae, common among pigeons, and Trichomonas gypaetinii isolated from vultures among other Trichomonas spp. isolated from various bird species (Martinez-Diaz et al., Reference Martinez-Diaz, Ponce-Gordo, Rodriguez-Arce, del Martinez-Herrero, Gonzalez, Molina-Lopez and Gomez-Munoz2015). Transfer of T. gallinae from columbiform to passerines has led to important mortality rates for some passerine species dramatically illustrating the potential for a Trichomonas species to jump host and spread rapidly through populations and to become a virulent parasite in some contexts (wild finches such as the common chaffinch) whereas it is often a commensal in others (the columbiform rock pigeon) (Amin et al., Reference Amin, Bilic, Liebhart and Hess2014). The comparative study of the molecular basis of the interactions between these various Trichomonas species and mucosal landmarks required to initiate and sustain the colonization of various hosts and mucosa will be of great interest and represent a fascinating model system to study MMPS transfers between birds and from birds to mammals, including humans (Maritz et al., Reference Maritz, Land, Carlton and Hirt2014).
Symbiosis: from parasitism to commensalism to mutualism
Although a number of MMPS are known to be associated with pathologies, leading to important morbidities and mortality rates in some contexts (Bar et al., Reference Bar, Phukan, Pinheiro and Simoes-Barbosa2015; Burgess et al., Reference Burgess, Gilchrist, Lynn and Petri2017), many infections by the same species are asymptomatic (Lukes et al., Reference Lukes, Stensvold, Jirku-Pomajbikova and Wegener Parfrey2015; Chabe et al., Reference Chabe, Lokmer and Segurel2017; Stensvold, Reference Stensvold2019). The outcome of host–microbial eukaryote symbiont interactions is dependent on the combination of the characteristics of the host, the microbial eukaryote and the mucosa microbiota, with increasing evidence for an important role played by cross kingdoms interactions (Fig. 1) (Burgess et al., Reference Burgess, Gilchrist, Lynn and Petri2017; Rowan-Nash et al., Reference Rowan-Nash, Korry, Mylonakis and Belenky2019). Inter-kingdom interactions can modulate the inflammatory tone of the mucosa through multiple possible direct and indirect interactions between mucosal microbes, microbes and epithelial cells and microbes and immunocytes (Fig. 2). Notably the epithelial cells play key roles in both sensing microbes and orchestrating the mucosal immunological innate and adaptive responses mediated by the combination of epithelial cells and immunocytes (Fig. 2) (Petersen and Round, Reference Petersen and Round2014; Levy et al., Reference Levy, Kolodziejczyk, Thaiss and Elinav2017). Primary immunodeficiencies, due to specific genetic background interfering with epithelial cells and/or immunocytes–microbes interactions, or secondary immunodeficiencies due to infections (e.g. HIV/AIDS) or malnutrition, can dramatically increase the susceptibility of the host to numerous infections including by those of MMPS. This is particularly marked for intracellular parasites such as Cryptosporidium and Microsporidia, with the HIV/AIDS pandemic highlighting both the importance of the adaptive immune response in controlling these parasites and the high level of human exposure to these opportunistic intracellular pathogens from diverse zoonotic reservoirs (Stentiford et al., Reference Stentiford, Becnel, Weiss, Keeling, Didier, Williams, Bjornson, Kent, Freeman, Brown, Troemel, Roesel, Sokolova, Snowden and Solter2016; Khan et al., Reference Khan, Shaik and Grigg2018).
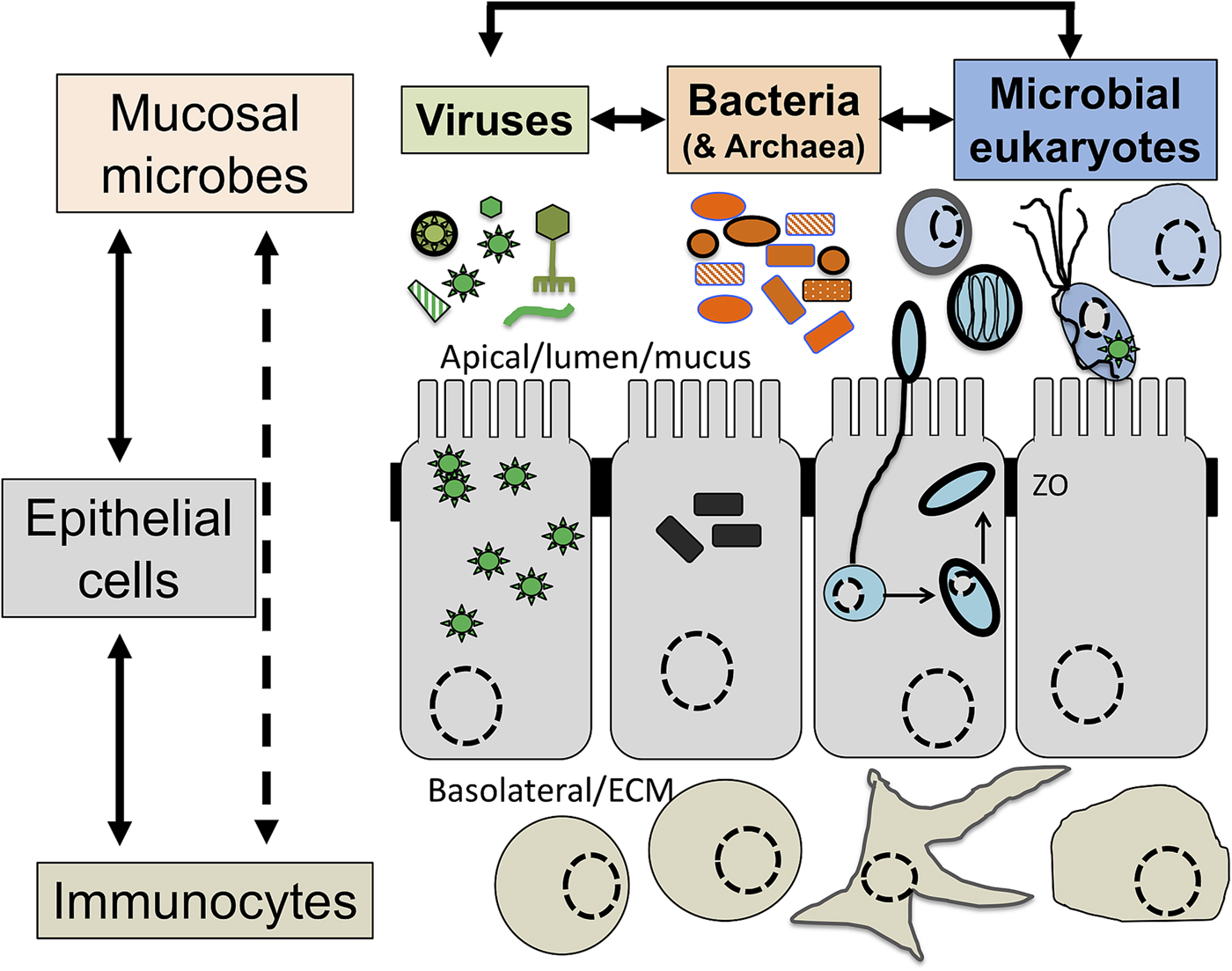
Fig. 2. The complex network of interactions at mucosal surfaces between microbes, epithelial cells and immunocytes modulating the immunological and inflammatory status of the mucosal surfaces. Optimal interactions ensure adequate responses to the presence of members of the microbiota and robust challenges to pathogens and at the same time tolerance to innocuous antigens required to maintain long term functionality of the mucosal surface underlying optimal digestion and nutrient uptake, breathing, or reproduction. Arrows indicate direct (e.g. physical contact) and indirect (e.g. metabolites or signalling molecules) interactions such as infection of epithelial cells by intracellular pathogens (viruses or Microsporidia, both illustrated) and doted arrows indicate indirect (e.g. though metabolites) interactions between illustrated cells. Note in particular the central node/role of epithelial cells that integrate, and in effect coordinate/orchestrate the complex network of interactions between microbes and immunocytes. A virus (several green ‘stars’) infected epithelial cell is illustrated as is a virus infected trichomonad (one green ‘star’, see example in the text). In addition, some viruses/phages infect bacteria are also contributing to the overall functional properties of the mucosa microbial ecology. Intracellular bacteria (black rectangles) and Microsporidia (blue cell and spores) are also illustrated within epithelial cells. ZO, Zonula occludens – tight junction; ECM, extra cellular matrix. For simplicity, the presence of mucus and the glycocalyx interacting with luminal microbes are not shown and only a monolayer of epithelial cells (e.g. as in the intestine) is illustrated.
In other contexts, MMPS could provide benefit to their mammalian carrier. Mice carrying the recently described gut trichomonad Tritrichomonas musculis were shown to be more resistant to challenges by the bacterial pathogen Salmonella typhimurium through enhancing mucosal defences by increasing intestinal inflammation via inflammosome activation and increase of the proinflammatory IL-18 production leading to a TH1/TH17 immune response (Chudnovskiy et al., Reference Chudnovskiy, Mortha, Kana, Kennard, Ramirez, Rahman, Remark, Mogno, Ng, Gnjatic, Amir, Solovyov, Greenbaum, Clemente, Faith, Belkaid, Grigg and Merad2016). This higher protection level to Salmonella was however associated with a cost as T. musculis colonization was also associated with a higher rate of colorectal cancer (Chudnovskiy et al., Reference Chudnovskiy, Mortha, Kana, Kennard, Ramirez, Rahman, Remark, Mogno, Ng, Gnjatic, Amir, Solovyov, Greenbaum, Clemente, Faith, Belkaid, Grigg and Merad2016). This contrasts with helminths infections that tend to inhibit gut inflammation through stimulating TH2/Treg responses (Cortes et al., Reference Cortes, Toledo and Cantacessi2018).
These contrasting examples illustrate the importance, and potential great value, of increasing our knowledge of the natural history of mammal–MMPS interactions and the importance of studying various microbial eukaryote symbiont species in humans and animal models to dissect the complex host–MMPS–microbiota interactions to illuminate their influence in both health and disease (Loke and Lim, Reference Loke and Lim2016). Additional examples of potentially beneficial microbial eukaryote symbionts, including Entamoeba spp. and Blastocystis, are discussed in this Special Issue (Stensvold, Reference Stensvold2019) and in other contexts (Lukes et al., Reference Lukes, Stensvold, Jirku-Pomajbikova and Wegener Parfrey2015; Chabe et al., Reference Chabe, Lokmer and Segurel2017; Stensvold and van der Giezen, Reference Stensvold and van der Giezen2018) and in the next section.
Microbial eukaryote symbionts/parasite–bacteria–virus interactions
The complex interplay between MMPS, bacteria, archaea and viruses and mammalian host health and disease status is increasingly being uncovered through the study of various microbial cellular species, bacteriophages and eukaryote infecting viruses, different mucosal surfaces and mammalian species, including humans (Clemente et al., Reference Clemente, Ursell, Parfrey and Knight2012; Burgess et al., Reference Burgess, Gilchrist, Lynn and Petri2017; Chabe et al., Reference Chabe, Lokmer and Segurel2017; Rowan-Nash et al., Reference Rowan-Nash, Korry, Mylonakis and Belenky2019). Here a few examples illustrating the link between these interactions and health and disease are covered with MMPS potentially contributing to either eubiosis or dysbiosis depending on the context of the hosts and their associated microbiota and environmental factors such as diet and xenobiotics (e.g. antibiotics) (Fig. 3).
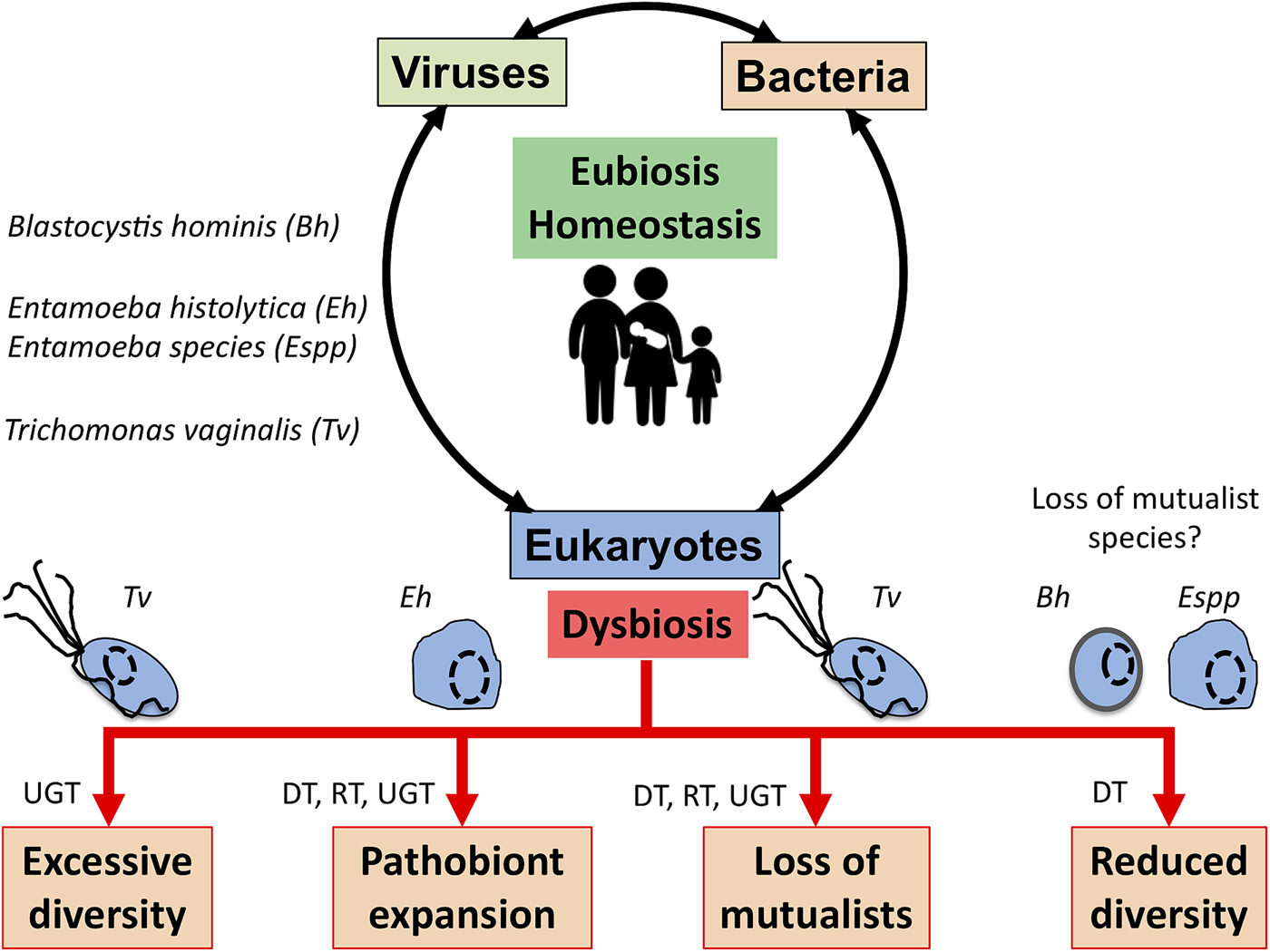
Fig. 3. Potential role of MMPS in inducing dysbiosis or eubiosis at mucosal surfaces. In the context of dysbiosis this would contribute to the loss of mucosal homoeostasis, and by doing so to a number of potential pathologies that eventually will translate in dysfunctional mucosa leading to disease both locally, e.g. mucosa inflammation, or more distal impacts. The illustrated examples include Trichomonas vaginalis (Tv) contributing to increasing the vaginal bacterial diversity associated with bacterial vaginosis, a form of pro-inflammatory dysbiosis of the urogenital tract (UGT). Trichomonas vaginalis infections are also associated with the loss of mutualists in the UGT. Entamoeba histolytica (Eh) can contribute to colitis, mucosa perforation and translocation of both parasites and some member of the gut microbiota into the portal vein and systemic tissues that can contribute to highly damaging systemic and local inflammations in the digestive tract (DT) and beyond. In contrast, the loss of some microbial eukaryotes, including potentially Blastocystis hominis (Bh) and some Entamoeba spp. (especially, non-histolytica species), could contribute to the gut microbiota reduced bacterial diversity associated with a dysbiotic state. Similarly, metronidazole treatments aiming at eradicating anaerobic mucosal microbial parasites such as Giardia and Trichomonas species will also contribute at disrupting the mucosal microbiota by killing important bacterial anaerobes and can favour the expansion of bacterial pathobionts in the DT and the respiratory tract (RT). See main text for examples and citations. MMPS can also influence the RT – e.g. (Maritz et al., Reference Maritz, Land, Carlton and Hirt2014) – but this is not covered here.
Arguably one of the most fascinating and complex examples includes Trichomonas vaginalis that infect the urogenital tracts (UGT) of humans (Hirt and Sherrard, Reference Hirt and Sherrard2015). A complex set of interactions between T. vaginalis, RNA viruses infecting T. vaginalis (TVV), the bacteria Mycoplasma hominis forming symbiosis with T. vaginalis and other bacteria associated with bacterial vaginosis, are all thought to contribute in concert to symptomatic infections, adverse pregnancy outcomes and increase transmission and acquisition of human infecting viruses, including HIV, HPV and HSV-2 (Hirt and Sherrard, Reference Hirt and Sherrard2015; Kissinger, Reference Kissinger2015). This is thought to be mediated through several mechanisms including, boosting the inflammatory tone of the UGT, increasing the population of target immunocytes for HIV and induction of microlesions disrupting the mucosal barrier (Kissinger, Reference Kissinger2015). Furthermore, although human viruses including HIV and HSV are not known to infect T. vaginalis, HIV and HSV viral particles can be internalized by the parasite and potentially be transferred to, and infect, human cells in a new host (Pindak et al., Reference Pindak, Mora de Pindak, Hyde and Gardner1989; Rendon-Maldonado et al., Reference Rendon-Maldonado, Espinosa-Cantellano, Soler, Torres and Martinez-Palomo2003). Although T. vaginalis can induce tissue damage and inflammation on its own, TVV and M. hominis can act synergistically to dramatically boost inflammations associated with T. vaginalis infections as reviewed by Dessi et al. (Reference Dessi, Margarita, Cocco, Marongiu, Fiori and Rappelli2019). Dysbiosis associated with infections by T. vaginalis is also thought to contribute to the pathobiology of T. vaginalis (Fichorova et al., Reference Fichorova, Fraga, Rappelli and Fiori2017; Mercer and Johnson, Reference Mercer and Johnson2018). Direct targeting of bacteria peptidoglycans by the parasite through enzymes of bacterial origins (Pinheiro et al., Reference Pinheiro, Biboy, Vollmer, Hirt, Keown, Artuyants, Black, Goldstone and Simoes-Barbosa2018) could potentially contribute to modulate the microbiota bacterial taxonomic composition in addition to contributing to T. vaginalis capacity to colonize the mucosal surface. The combination of the parasite and several bacterial species characteristic of dysbiotic vaginal microbiota associated with trichomoniasis, were also recently shown to synergistically affect the integrity of the tight junction complex of the cervicovaginal epithelial cells (Hinderfeld et al., Reference Hinderfeld, Phukan, Bar, Roberton and Simoes-Barbosa2019). Notably, treating T. vaginalis infections with metronidazole can liberate from the killed parasite TVV particles and/or M. hominis cells leading to the boosting of inflammation and to infection of human cells by M. hominis (Thi Trung Thu et al., Reference Thi Trung Thu, Margarita, Cocco, Marongiu, Dessi, Rappelli and Fiori2018; Dessi et al., Reference Dessi, Margarita, Cocco, Marongiu, Fiori and Rappelli2019). These different aspects associated with T. vaginalis infections illustrates the intricate associations of the parasite with bacterial (Mycoplasma) and viral (TVV) endosymbionts, the bacterial members of the UGT microbiota and how these interactions can influence the parasite pathobiology including increasing human infecting virus transmission rates. These considerations will be important to complement more traditional investigations focusing on the study of specific aspects of host–parasite interactions, such as the potential role of environmental glucose concentration variation (Miranda-Ozuna et al., Reference Miranda-Ozuna, Rivera-Rivas, Cardenas-Guerra, Hernandez-Garcia, Rodriguez-Cruz, Gonzalez-Robles, Chavez-Munguia and Arroyo2019) and cell surface and secreted factors such as exosomes (Mercer and Johnson, Reference Mercer and Johnson2018), in modulating the virulence of the parasite. These examples illustrate dramatically the importance to investigate host–MMPS–microbiota–virus interactions in an integrative manner to develop more refined diagnostics and novel prophylactic and therapeutic strategies to eventually promote reproductive and sexual health more efficiently. It will also be of interest to investigate the possibility that related endosymbionts (to TVV and Mycoplasma) are also present in other Trichomonas species including bird infecting species and T. tenax associated with periodontitis (described in the previous section).
The MMPS Giardia, Entamoeba and Cryptosporidium are also known to be infected by RNA viruses (Gomez-Arreaza et al., Reference Gomez-Arreaza, Haenni, Dunia and Avilan2017). Cryptosporidium-infected virus is associated with a higher rate of the parasite propagation capacity; however, it is not clear if this increases the virulence of such infections. Similarly there is currently no evidence for Giardia and Entamoeba that their RNA viruses can contribute to boosting the pathobiology of these parasites (Gomez-Arreaza et al., Reference Gomez-Arreaza, Haenni, Dunia and Avilan2017). Complex interplay between Giardia, Entamoeba and Cryptosporidium with bacteria members of the microbiota have also been shown to influence the virulence of these parasites in both negative (e.g. inhibiting infections) and positive ways (e.g. promoting virulence) (Burgess et al., Reference Burgess, Gilchrist, Lynn and Petri2017; Rowan-Nash et al., Reference Rowan-Nash, Korry, Mylonakis and Belenky2019). A remarkable example illustrating the importance of the microbiota in playing a role in reducing the impact of Cryptosporidium infection was uncovered when investigating two candidate drugs to treat the parasite. Two novel drugs that had promising properties in initial in vitro tests had an opposite effect on Cryptosporidium infections in a mouse model (Gorla et al., Reference Gorla, McNair, Yang, Gao, Hu, Jala, Haribabu, Striepen, Cuny, Mead and Hedstrom2014). Although one of the drugs was potent in controlling the parasite, the other drug was shown to actually boost infection levels, which was associated with a significant change in the bacterial taxonomic composition of the gut microbiota, with in particular a dramatic increase of the population of the mucin loving gut bacteria Akkermansia muciniphila (2800-fold increase compared to the pre-treatment state), suggesting a dysbiotic microbiota (Gorla et al., Reference Gorla, McNair, Yang, Gao, Hu, Jala, Haribabu, Striepen, Cuny, Mead and Hedstrom2014). This was rationalized as an off-target impact of the drug on members of the gut microbiota. Although A. muciniphila is considered to be an important mutualist associated with human health (Cani and de Vos, Reference Cani and de Vos2017), the significant boost in Cryptosporidium infection level could be explained by an excessive degradation by A. muciniphila of the mucus protective layer in the gut facilitating access to, and eventual infection of, epithelial cells by Cryptosporidium. An apparently similar outcome was observed in a mouse model with a humanized gut microbiota fed with a diet depleted from plant fibbers, which led to the depletion of the mucus protective layers by the microbiota and higher susceptibility to pathogens (Desai et al., Reference Desai, Seekatz, Koropatkin, Kamada, Hickey, Wolter, Pudlo, Kitamoto, Terrapon, Muller, Young, Henrissat, Wilmes, Stappenbeck, Nunez and Martens2016). These examples illustrate how environmental factors, including xenobiotics (an antibiotic in the example above) and diet, can influence the mucosal microbial ecology and by doing so modulate the host susceptible to infections by potential pathogens, including MMPS.
Antibiotics and vaccines for mucosal parasites/symbionts
In contrast to the availability of a broad range of antibiotic treatment regiments for bacteria, there are far less efficient options to treat with drugs symptomatic infections due to microbial parasites (Farthing, Reference Farthing2006; Leitsch, Reference Leitsch2017). As for bacteria, there is also the issue of microbial parasites developing resistance to existing drugs regiments and for off-target effects on the microbiota (Wypych and Marsland, Reference Wypych and Marsland2018). Furthermore, some patients can develop strong reactions to some drugs including to the commonly used metronidazole targeting anaerobic parasites (Leitsch, Reference Leitsch2017). These considerations stimulate continuous research efforts to identify new drugs to treat microbial parasites, either based on modifying existing well established drugs such as 5-nitroimidazole (Leitsch, Reference Leitsch2017), or new drugs such as plant derived phenanthrenes (Vargas Rigo et al., Reference Vargas Rigo, Petro-Silveira, Devereux, McCann, Souza Dos Santos and Tasca2018). Irrespective of the drug, it is increasingly appreciated necessary to consider their broad impact on the host microbiota, with increasing evidence that antibiotic treatments are being associated with dysbiosis favouring opportunistic pathogens, including pathobionts, and/or leading to a difunctional immune response to microbial and other antigens that can lead to debilitating conditions such as allergies and asthma (Wypych and Marsland, Reference Wypych and Marsland2018). In the case of the treatment of anaerobic mucosal parasites (such as Trichomonas and Giardia) with metronidazole/imidazole, the anaerobic members of the microbiota will also be affected (Leitsch, Reference Leitsch2017). This can contribute to dysbiosis in the gut microbiota in particular where anaerobes are known to play important roles (Wypych and Marsland, Reference Wypych and Marsland2018) (Fig. 3).
In comparison to drug treatments options, vaccines for MMPS are even less well developed. This is due to the combination of the inherent difficulties in developing effective mucosal vaccines (Lycke, Reference Lycke2012) and the complex biology of MMPS, including their capacity to mediate cell surface antigen variation (Deitsch et al., Reference Deitsch, Lukehart and Stringer2009; Gargantini et al., Reference Gargantini, Serradell, Rios, Tenaglia and Lujan2016) and the little knowledge we have on the nature of the host immune response to eradicate MMPS (Farthing, Reference Farthing2006; Chapwanya et al., Reference Chapwanya, Usman and Irons2016). One promising strategy that takes advantage of the properties of VSP proteins from Giardia (Gargantini et al., Reference Gargantini, Serradell, Rios, Tenaglia and Lujan2016) and viral-like particles has great potential to develop novel oral vaccines for various pathogens (Serradell et al., Reference Serradell, Rupil, Martino, Prucca, Carranza, Saura, Fernandez, Gargantini, Tenaglia, Petiti, Tonelli, Reinoso-Vizcaino, Echenique, Berod, Piaggio, Bellier, Sparwasser, Klatzmann and Lujan2019), including a broad range of MMPS in addition to Giardia (Serradell et al., Reference Serradell, Saura, Rupil, Gargantini, Faya, Furlan and Lujan2016).
Conclusion and some speculations
From the examples covered here and in cited publications one can conclude that it might be more appropriate to refer to many extracellular microbial eukaryotic symbionts with various pathogenic potential as pathobionts that is, they are members of the mucosal microbial ecosystems that can become pathogenic in some contexts where host genetic, environment and properties of the microbial community as a whole all play a role (Chow et al., Reference Chow, Tang and Mazmanian2011). Acquired immunodeficiencies or transfer of MMPS between different hosts species can lead to sub-optimal interactions with some species becoming pathogenic (Farthing, Reference Farthing2006; Price et al., Reference Price, Hungate, Koch, Davis and Liu2017). In contrast intracellular parasites, including the Apicomplexa Cryptosporidium and the Microsporidia, are typically thought to be primarily gut pathogens (Farthing, Reference Farthing2006), as they must directly exploit their host cell energy and metabolites to proceed through their life cycle and in the process compromise the integrity of the epithelial monolayer of the gut (Farthing, Reference Farthing2006; Dean et al., Reference Dean, Hirt and Embley2016). One aim of this editorial was to illustrate specific aspects of the intricate and complex interactions taking place between MMPS, the other members of the microbiota and their animal or human hosts. These highlight the importance of collaborative research projects integrating parasitology, microbiology, virology, pharmacology and mucosal immunology in the context of both basic and medical and veterinary research on the factors influencing mucosa health and disease. Generating more comprehensive knowledge on the link between these microbial interactions and mucosal and systemic health and disease is undoubtedly one of ‘the most difficult and challenging scientific endeavour of our time’(Birchenough and Hansson, Reference Birchenough and Hansson2017), as it will need to identify and characterize key aspects of thousands of highly dynamic interactions mediated by a complex cocktail of metabolites, cell–virus and cell–cell interactions involving complex microbial communities, epithelial cells and immunocytes. The knowledge derived from the study of these complex network of interactions will be required to eventually develop much needed novel prophylactic, including mucosal vaccines for overt pathogens, and therapeutic strategies (including highly specific drugs, prebiotics, fecal transplants), to regenerate, maintain and promote human and animal health at mucosal surfaces. It is also suggested that considering microbial eukaryote symbionts/parasites will provide important opportunities for much required comparative studies to delicately dissect key nodes orchestrating mucosal–microbes interactions and how these are causally linked to the specific phenotypic outcomes in their human and animal hosts. Contextualization of the diversity of both MMPS, the microbiota at large (bacteria, archaea and viruses) and their host within an evolutionary and ecological framework will also likely be important at helping building a more predictive theoretical framework for the outcome of host–microbes interactions (Amato, Reference Amato2016; Davenport et al., Reference Davenport, Sanders, Song, Amato, Clark and Knight2017; Rook et al., Reference Rook, Backhed, Levin, McFall-Ngai and McLean2017; Ferreiro et al., Reference Ferreiro, Crook, Gasparrini and Dantas2018).
Author ORCIDs
Robert P. Hirt, 0000-0002-3760-9958.
Acknowledgments
EMBO and Newcastle University were the main sponsors of the EMBO conference: ‘Anaerobic protists: Integrating parasitology with mucosal microbiota and immunology’ and Cambridge University Press, for providing the opportunity to put together this Special Issue.
Conflict of interest
None.
Ethical standards
Not applicable.