INTRODUCTION
The advent of severe acute respiratory syndrome (SARS) in February 2003 raised awareness of the continued threat of previously undescribed infective agents capable of human-to-human transmission with potential fatal consequences, and highlighted the need to further investigate clusters of severe respiratory infections of unknown aetiology. Against this background, in March 2003 two family clusters of severe unexplained pneumonia were brought to the attention of a consultant in communicable disease in a UK city. All six cases within these two family clusters were aged <50 years and were resident in the same area of the city. There was one fatality. The other five cases, all of whom were hospitalized, subsequently made a good recovery on empirical therapy.
Initial routine microbiological investigations on these cases failed to identify an aetiological agent and no epidemiological link to a SARS-affected area or individual was identified. However, given the close proximity of the patients' homes within a small area and the severe nature of the pneumonia, an investigation was initiated to ascertain any potential further cases and to determine the cause of these apparent clusters.
This report summarizes results of the initial microbiological investigation and describes the use of non-culture-based molecular techniques in an attempt to confirm the disease aetiology and, where possible, to further characterize the infecting organism. The non-culture molecular techniques described include antigen detection, polymerase chain reaction (PCR) and multi-locus sequence typing (MLST).
MATERIALS AND METHODS
Epidemiological investigations, including prospective and retrospective case ascertainment, were undertaken from January 2003 to August 2003. Case definitions were formulated following review of the clinical histories of the index clusters and were as follows: any patient aged <50 years with a community-acquired pneumonia (CAP) requiring intensive or high dependency care, or a patient with CAP that was felt to be unusual (e.g. in severity?exposure?contact?clinical picture). Key clinicians, pathologists, microbiologists, epidemiologists and the coroner's office covering the area were contacted and made aware of the case-finding exercise. An additional five patients who met the case definitions were identified through this exercise, two of whom were resident in the same small area of the city as the index clusters and a further three were from differing localities. Amongst the two additional cases from the same area there was one fatality.
Initial microbiological investigations on the identified cases included blood culture from all but one (the retrospectively identified fatal case) from which no clinically significant organisms were grown. Respiratory samples from 10 of the cases were sent for microbiological examination, which included examination for viruses in four of the cases. There was no evidence of viral infection and bacterial culture from the respiratory specimens was inconclusive, however, Streptococcus pneumoniae was isolated from sputum samples in two cases. Acute serum from seven cases and paired sera (separated by >1 week) was available for serological testing from four cases. From these samples, there was no evidence of infection with ‘atypical’ bacterial respiratory pathogens (Legionella pneumophila, Mycoplasma pneumoniae, Chlamydia sp. and Coxiella burnetii) and serology for influenza A, B and respiratory syncytial virus did not reveal any significant titres. However, a slight increase in antibody levels to adenovirus was detected in one of the cases. Urine samples were taken from eight of the cases and tested for L. pneumophila antigen, for which all results were negative. In addition a serum sample from one of the cases was examined for evidence of SARS and was found to be negative.
As the clinical symptoms of many of the patients were suggestive of S. pneumoniae infection, the remnants of the samples from the initial inconclusive microbiological investigation were recovered and sent to the Respiratory and Systemic Infection Laboratory (RSIL) at the Health Protection Agency, Centre for Infections, London, UK, for further testing.
Samples for non-culture testing
Sample remnants were recovered from 10 of the 11 cases matching the case definition (see Table).
Table. Pneumonia cases, routine test results, samples received and non-culture test results
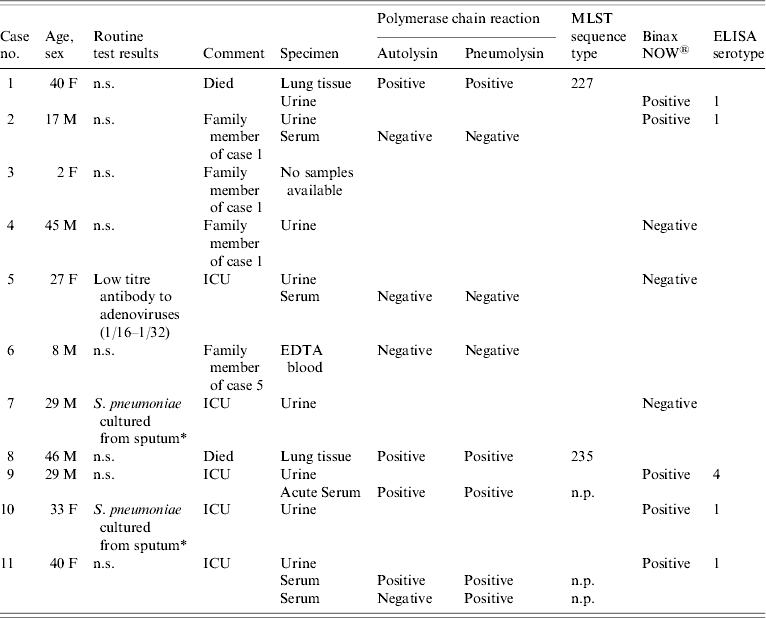
MLST, multi-locus sequence typing; ELISA, enzyme-linked immunosorbent assay; ICU, intensive care unit; n.s., nothing significant detected; n.p., not possible.
* Serotype unknown as isolates from non-sterile sites are not referred for serotyping.
Non-culture testing of samples
Urine samples were tested using the S. pneumoniae Binax NOW® immunochromatographic test kit (Launch Diagnostics, Longfield, UK) according to the manufacturer's instructions. If the sample was found positive by the Binax NOW test it was run in a serotype-specific enzyme-linked immunosorbent assay (ELISA) test using type-specific monoclonal antibodies to types 1, 3, 4, 7F, 9V, 14, 18C and 23F of S. pneumoniae (Wyeth Vaccines Research, West Henrietta, NY, USA) as previously reported [Reference Eltringham1, Reference Leeming2].
DNA was extracted from EDTA blood, serum and lung tissue samples using the Qiagen DNA mini kit (Qiagen Ltd, Crawley, UK) according to the manufacturer's instructions for the particular sample type. The samples were tested for the S. pneumoniae autolysin and pneumolysin genes using the method previously described for dual target testing of clinical samples [Reference Sheppard3]. Non-culture MLST was attempted on any dual-target PCR-positive samples following the method recently described by Birtles et al. [Reference Birtles4].
Estimation of the local prevalence of serotype 1 at the time of the clusters
To estimate the prevalence of serotype 1 pneumococci in the region at the time of the incident, pneumococcal serotyping data was analysed from invasive disease isolates that were referred to the Streptococcus and Diphtheria Unit at the Health Protection Agency (HPA) Centre for Infections (formerly Central Public Health Laboratory).
RESULTS
Urine antigen test results
Of the eight urine samples received, five were positive in the Binax NOW urine antigen test and serotypes were obtained using the serotype-specific ELISA. Four gave positive results for serotype 1 and one was positive for serotype 4 (see Table).
PCR and MLST test results
Both lung tissue samples were positive in both of the PCR assays, as were serum samples from two cases (see Table). The other samples tested by PCR gave negative results. Non-culture MLST was attempted on all samples from which a lytA and ply PCR-positive result was obtained. A full MLST allele profile and sequence type was obtained from both DNA extracts from the lung tissue samples. Due to the very small amount of pneumococcal DNA present in the serum samples (indicated by late crossing points in the real-time PCR assays), it was not possible to obtain a sequence type for those samples (see Table).
Case breakdown
The index case (case 1; Table) was part of the first family cluster and was found to be positive for S. pneumoniae by PCR of the lung extract. Urine antigen testing was also positive for S. pneumoniae antigen, and a positive result for serotype 1 was obtained in the serotype-specific urine ELISA. The MLST result from the lung sample corroborates the serotype data, as the sequence type (ST) obtained (ST 227) matched with 35 isolates of serotype 1 deposited in the MLST.net database (www.mlst.net, accessed May 2007). At the time this paper was written no other serotypes in the database had this ST.
Case 2 (family member of case 1) also gave a positive result in the Binax urinary antigen test and was positive for serotype 1 pneumococcal capsular type in the urine ELISA. This provided strong evidence that the cases were probably infected by the same pneumococcal strain. A serum sample taken 12 days after admission was negative for pneumococcus by PCR. Cases 3 and 4 were also from the same family as the index case, there were no samples available for testing from case 3 and case 4 gave a negative result in the Binax NOW test. Thus the aetiology of the disease in cases 3 and 4 was undetermined by this investigation.
Cases 5 and 6 were a second family and were residents of the same postcode district as the other family cluster. None of the culture or non-culture tests performed on the samples taken from these patients were positive for any clinically significant organism other than a low antibody titre to adenovirus from one of the cases.
Case 7 was a prospectively identified case that fitted the case definition but was a resident of a different area of the city. Although the urine sample was negative for S. pneumoniae antigen using the Binax NOW test. S. pneumoniae was also cultured from a respiratory sample from this patient during the initial clinical investigations, but the isolate that was cultured was not stored or sent to RSIL for serotyping.
Case 8 was a resident of the same postcode district of the city as cases 1–6 and was found dead at home. This patient had a medical history of splenectomy and was positive for S. pneumoniae by PCR from the lung sample taken at post-mortem. Non-culture MLST yielded a profile (ST 235), which was an exact match to one serotype 20 isolate from Spain and two from Poland, and one serotype 7C isolate from Austria. Because the sequence type did not match that from case 1, there was no evidence of a microbiological link between these cases.
Case 9 was a resident of the same postcode district as the index case. The urine sample from this patient gave a positive result for serotype 4 pneumococcal antigen. The serum sample was positive by PCR but no MLST profile was obtained from this sample. The presence of serotype 4 antigen in the urine suggested that this case was not linked to the index cluster. There were no isolates of serotype 4 on the MLST database that had ST 227 or 235 and none with single locus variants of these.
Case 10 was retrospectively identified and lived in a different area of the city to any of the other cases. Urine from this patient was positive for pneumococcal antigen of serotype 1. Based on the serotype data alone the possibility of a link with the index case cannot be ruled out. S. pneumoniae was also cultured from a respiratory sample but the isolate was not stored or sent to RSIL for serotyping.
Case 11 was a prospectively identified case from a different area of the city from the other identified cases. PCR testing of serum was positive and the urine sample was positive for pneumococcal serotype 1 antigen. MLST was attempted on the serum extract but failed due to insufficient DNA target concentration. Again, based on the serotype data alone it is not possible to rule out a link to the index case.
Estimation of the local prevalence of serotype 1 at the time of the clusters
The serotypes of invasive isolates obtained from the area of study in the seasons before and during the study incident showed that the prevalence of serotype 1 isolates referred to the reference laboratory increased from being an unusual serotype (representing below 5% of referred isolates in the season September 2000 to August 2001) to the most prevalent (up to 28%) in the season September 2002 to August 2003, the period during which the cases were seen.
DISCUSSION
S. pneumoniae is the most common cause of CAP, but diagnosis of the infection is hampered by the insensitivity of culture techniques [Reference Musher5]. The use of non-culture methods is therefore of considerable interest as a means of increasing diagnostic yield. This study describes the investigation of a putative cluster of unexplained pneumonia cases and involved the use of non-culture techniques on sample remnants following initial inconclusive routine laboratory investigations.
A recent study of the clonal diversity and geographical distribution of serotype 1 isolates showed that there was a striking geographical structure in sequence type, possibly due to its rare occurrence in carriage in healthy individuals. Nine of ten isolates submitted to the study from England were of ST 227 (the sequence type of the isolate from case 1), and it was part of a group of related sequence types recovered from Europe the United States and Canada, which were different to those recovered from Africa and Israel [Reference Brueggemann and Spratt6].
Using the techniques described, a considerable amount of information was gained about a cluster of cases of initially unexplained pneumonia occurring within a small region of a UK city. In addition to confirming the aetiology of the disease in six of the cases, valuable epidemiological data was obtained which would not otherwise have been available. Two of the possible cases have been shown to be unrelated to the index case (cases 8 and 9), two more cases have shown a possible link both being due to serotype 1 (cases 10 and 11).
A transmission link between all the cases shown to have been caused by pneumococci of serotype 1 cannot be confirmed, as this was shown to be the most commonly isolated serotype in the region during the time of the study. Given the clonal nature of serotype 1 pneumococci in a given geographical region, it is probable that all the serotype 1 pneumococci in this study are of the same sequence type, whether or not they were the result of direct transmission between the patients in the study. The public health interpretation of these results, therefore, is that in the area at the time there was a high background rate of pneumonia caused by pneumococci of serotype 1 which was capable of infecting an age group not normally associated with severe pneumococcal pneumonia [Reference Laurichesse7]. However, within the cases of pneumonia caused by pneumococcus serotype 1 there was a small family cluster of two linked cases.
The other family involved in the initially recognized cluster (cases 5 and 6), gave negative results by all methods of testing. Only EDTA blood was received from patient no. 6 and serum and urine was available from patient no. 5. The sensitivity of PCR on stored EDTA blood samples has been shown to be poor [Reference Sheppard8] and is probably even worse on serum samples. However, given the severity of the disease and the good sensitivity of the Binax NOW urine antigen test, a positive result would have been expected if pneumococci were the cause of the disease in case 5, from which a urine sample was available for testing. The disease cause for case 7 was not resolved using the non-culture pneumococcal testing techniques either. Only urine was available from this patient, who was a resident of a different area of the city.
From this study, using sample remnants, the use of these non-culture techniques has proved to be a powerful tool where blood cultures are negative and sputum cultures inconclusive. The samples available were not ideal for the molecular testing performed either in terms of completeness (same sample types for all patients), or suitability for testing. For example, serum samples were tested by PCR in the absence of more suitable samples. While the PCR technique is widely applicable to many sample types it is most sensitive when used on CSF or tissue samples: PCR sensitivity on whole blood may be improved if the samples tested are freshly collected for testing or stabilized with guanidinium prior to freezing [Reference Michelow9]. MLST, like diagnostic PCR, is affected by the sample type and storage conditions. Furthermore, the necessity to amplify seven separate regions further reduces sensitivity meaning that only samples giving strong positive results by the diagnostic PCR assays have the potential to be typed using this method. The timely collection of suitable samples would improve the ability of the PCR or MLST techniques to yield useful results in future investigations.
Urine antigen testing has proven to be a valuable and sensitive technique for diagnosis of pneumococcal infections [Reference Smith10], and also has the potential for use as a serotyping method. Urine is an easy sample to obtain and its antigenicity is not much affected by storage [Reference Coonrod11], therefore samples would be widely available for the application of antigen detection/serotyping techniques. Serotyping directly from urine samples by antigen detection assays will be a valuable source of data to inform vaccine development strategies, as serotype data will become available from the many cases in which an isolate is never obtained for standard serotyping methods. This study highlights the usefulness of non-culture methods (particularly in combination) in the investigation of clusters of pneumonia where there is an absence of conclusive culture results.
ACKNOWLEDGEMENTS
The authors acknowledge the intellectual input of Dr Andrew Birtles for the non-culture MLST method. We also acknowledge the hard work of the clinical teams and others at NPHS and the Coroner's office.
DECLARATION OF INTEREST
Dr R. C. George's laboratory has received financial support from Wyeth Vaccines Research, for assay development and attendance at international conferences.



