Introduction
Cryptosporidium is an opportunistic zoonotic parasite found worldwide that infects many vertebrate hosts and typically causes self-limiting diarrhoea in humans and livestock (Kotloff, Reference Kotloff2017; Hatam-Nahavandi et al., Reference Hatam-Nahavandi, Ahmadpour, Carmena, Spotin, Bangoura and Xiao2019). Cryptosporidium is commonly found in the intestines of humans and animals and is transmitted by the fecal–oral route (Bouzid et al., Reference Bouzid, Hunter, Chalmers and Tyler2013). Children, immunodeficient individuals and newborn animals are among the groups that are susceptible to Cryptosporidium infection (Checkley et al., Reference Checkley, White, Jaganath, Arrowood, Chalmers, Chen, Fayer, Griffiths, Guerrant, Hedstrom, Huston, Kotloff, Kang, Mead, Miller, Petri, Priest, Roos, Striepen, Thompson, Ward, Van Voorhis, Xiao, Zhu and Houpt2015). Among animals susceptible to Cryptosporidium, pigs are considered as one of the main reservoir hosts (Qi et al., Reference Qi, Zhang, Xu, Zhang, Xing, Tao and Zhang2020). There are no effective vaccines that can prevent cryptosporidiosis in humans or livestock (Dumaine et al., Reference Dumaine, Tandel and Striepen2020).
Globally, the first report of 3 pig cases of cryptosporidiosis was in 1977 (Kennedy et al., Reference Kennedy, Kreitner and Strafuss1977). Pigs with cryptosporidiosis are characterized by diarrhoea, vomiting, dehydration, reduced daily gain and a lower feed conversion rate (Vítovec and Koudela, Reference Vítovec and Koudela1992; Quílez et al., Reference Quílez, Sánchez-Acedo, Clavel, del Cacho and López-Bernad1996; Enemark et al., Reference Enemark, Ahrens, Bille-Hansen, Heegaard, Vigre, Thamsborg and Lind2003), and the parasites mainly live in the intestinal tract and gallbladder (Fleta et al., Reference Fleta, Sánchez-Acedo, Clavel and Quílez1995). There is considerable genetic variation in the genus Cryptosporidium; there are 44 known species, and more than 120 genotypes of Cryptosporidium have been identified (Ryan et al., Reference Ryan, Feng, Fayer and Xiao2021). Thirteen different Cryptosporidium species/genotypes have been isolated in pigs, namely Cryptosporidium scrofarum (previously Cryptosporidium pig genotype II), Cryptosporidium suis (previously Cryptosporidium pig genotype I), Cryptosporidium muris, Cryptosporidium parvum, Cryptosporidium tyzzeri (previously Cryptosporidium mouse genotype I), Cryptosporidium hominis, Cryptosporidium meleagridis, Cryptosporidium felis, Cryptosporidium andersoni, Cryptosporidium struthioni, Cryptosporidium rat genotype, Cryptosporidium sp. Eire w65.5 and unknown Cryptosporidium genotype from pig slurry (Němejc et al., Reference Němejc, Sak, Květoňová, Hanzal, Janiszewski, Forejtek and Kváč2013b; Wang et al., Reference Wang, Gong, Zeng, Li, Zhao and Ni2021, Reference Wang, Li, Zou, Du, Song, Wang and Chen2022). Cryptosporidium scrofarum and C. suis infections account for more than 90% of cryptosporidiosis in pigs (Feng et al., Reference Feng, Ryan and Xiao2018). Cryptosporidiosis in pigs does not always cause clinical symptoms, and cases of human infection with C. scrofarum and C. suis suggest that these 2 Cryptosporidium species may be zoonotic (Kvác et al., Reference Kvác, Kvetonová, Sak and Ditrich2009c; Moore et al., Reference Moore, Elwin, Phot, Seng, Mao, Suy, Kumar, Nader, Bousfield, Perera, Bailey, Beeching, Day, Parry and Chalmers2016; Sannella et al., Reference Sannella, Suputtamongkol, Wongsawat and Cacciò2019). However, their pathogenicity and infectivity to humans are not well understood; so, they remain a potential threat to human health.
The global pig population was estimated at 952.6 million in 2020 (https://www.fao.org/). In animal husbandry, cryptosporidiosis causes huge economic losses due to weight loss in young animals, stunted growth and reduced production in adult animals (Pumipuntu and Piratae, Reference Pumipuntu and Piratae2018). Pigs are also animals that humans often contact directly or indirectly. Therefore, we performed a systematic review and meta-analysis to assess the global prevalence of Cryptosporidium in pigs. The potential risk factors including region, age and geographical and climatic factors were also analysed. The results describe the distribution characteristics of Cryptosporidium species in different age groups of pigs, and provide a basis for the prevention and control of Cryptosporidium infections.
Materials and methods
Search strategy and selection criteria
We used 5 literature databases (PubMed, Web of Science, the China National Knowledge Infrastructure, VIP Chinese Journals Database and Wanfang Data) to search for studies on the global prevalence of Cryptosporidium in pigs. All published studies on Cryptosporidium in pigs from 31 September 2022 onwards were included. We searched the 2 English databases with the term ‘Cryptosporidium’, ‘Cryptosporidiosis’ cross-referenced with ‘pig’, ‘swine’, ‘hog’, ‘wart’, ‘warthog’, ‘Phacochoerus’, ‘Suidae’, ‘boar’ or ‘piglet’. In the 3 Chinese databases, ‘Cryptosporidium’ (Chinese) and ‘pig’ (Chinese) were used as keywords. We conducted analyses in accordance with the preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and the PRISMA 2009 checklist (Table S1). The articles for which full text was not available, the first author was not contacted for more research information and/or statistics.
The following clauses were used as the criteria for article exclusion:
1) the purpose of the study was not the prevalence of Cryptosporidium in pigs;
2) the total number of pigs tested and the number of pigs that tested positive were not provided;
3) the testing method was not clearly described;
4) the sample was a mixture of specimens from multiple pig feces;
5) the study sample size was less than 20;
6) the study was a review or a case report.
Quality assessment
We used established methods to evaluate the quality of the studies (Guyatt et al., Reference Guyatt, Oxman, Vist, Kunz, Falck-Ytter, Alonso-Coello and Group2008). Studies with scores of 0 or 1 point were classified as low quality, studies with scores of 2 or 3 points were classified as medium quality, and studies with scores of 4 or 5 points were classified as high quality. A study scored 1 point if it included one of the following items:
1) a clear research goal;
2) a clearly defined research period;
3) a sample size of greater than 200;
4) a clear detection method;
5) analysis involving 3 or more influencing factors.
Data extraction
Two authors (Y. C. and H. Q.) separately screened all titles, abstracts and full texts and independently extracted the data. Disagreements were resolved by discussion with Y. W. Y. C. and H. Q. extracted information, including the first author, publication date, country, sampling time, detection method, total samples, positive samples, prevalence, study quality and Cryptosporidium species (Table S2).
Statistical analysis
All data were analysed using Stata version 14.0 (https://www.stata.com). Due to high heterogeneity (I 2 > 50%, P < 0.1) of the data, the random-effects model was used for the meta-analysis. To investigate the potential sources of heterogeneity, sensitivity analysis, subgroup analysis and meta-regression analysis were performed on the extracted data. If a study involved multiple detection methods for Cryptosporidium, the molecular results in the analysis were the first choice. We used sensitivity analysis to test the stability of the data, and the overall study was evaluated using forest plots. We evaluated the effect of selected studies on the pooled prevalence by excluding single studies sequentially (Wang et al., Reference Wang, Liu, Liu, Li, Zhang, Zhao and Zhu2018b). Publication bias of the study was evaluated using a funnel plot and Egger's tests (Egger et al., Reference Egger, Davey Smith, Schneider and Minder1997). The following potential sources of heterogeneity were examined: region (Asia compared to other regions), age (post-weaned compared to the other age groups), presence or absence of diarrhoea (diarrhoea compared to non-diarrhoea) and Cryptosporidium species (C. scrofarum compared to the other species).
The global longitude and latitude span was large, and there were significant geographical differences. The data related to geographic factors were obtained from the National Oceanic and Atmospheric Administration (NOAA, https://gis.ncdc.noaa.gov/maps/ncei/cdo/monthly). We also used subgroup analysis and meta-regression analysis to evaluate the impact of geographical risk factors, including latitude (30°–60° vs others), longitude (<−60° vs others), mean yearly temperature (5–10 °C vs others), mean yearly relative humidity (<60% vs others), mean yearly precipitation (0–400 mm vs others).
Results
Characteristics of studies
A total of 833 publications were initially identified. After screening of the title and abstract, 162 potentially relevant articles were selected for full text search. Of these, 6 were review studies, 9 had incomplete information or only provided prevalence, 6 had sample sizes less than 20, 4 were case reports and 9 lacked full text. In total, 128 publications (including 131 datasets) were of sufficient quality and were considered suitable for meta-analysis (Fig. 1).

Fig. 1. Flow diagram of the selection of eligible studies.
The selected studies came from 36 countries (Fig. 2, Table 1). A total of 71 datasets originated from Asia [China (n = 54), India (n = 2), Indonesia (n = 1), Japan (n = 6), Korea (n = 3), Thailand (n = 1), Turkey (n = 1), Vietnam (n = 3)]. A total of 30 datasets were from countries in Europe [Austria (n = 1), Czech Republic (n = 6), Denmark (n = 2), Germany (n = 2), Ireland (n = 1), Norway (n = 1), Poland (n = 2), Serbia (n = 1), Slovak Republic (n = 2), Spain (n = 8), Sweden (n = 1), Switzerland (n = 1) and the UK (n = 1)]. Eight datasets were from countries in Africa [Ghana (n = 1), Madagascar (n = 1), Malawi (n = 1), Nigeria (n = 3), South Africa (n = 1), Zambia (n = 1)]. A total of 10 datasets were from countries in North America [Canada (n = 4), Trinidad (n = 1), the USA (n = 4), Cuba (n = 1)]. Eight datasets were from South America [Argentina (n = 1), Brazil (n = 4), Colombia (n = 2), Ecuador (n = 1)]. Four datasets were from countries in Oceania [Australia (n = 4)] (Tables 1 and 2). Pre-weaned pigs were described in 48 datasets, post-weaned pigs were described in 63 datasets, fattening pigs were described in 48 datasets and adult pigs were described in 53 datasets. Most datasets lacked information on pig health status. Diarrhoea in pigs was reported in 14 datasets, and no diarrhoea in pigs was reported in 10 datasets (Table 2).
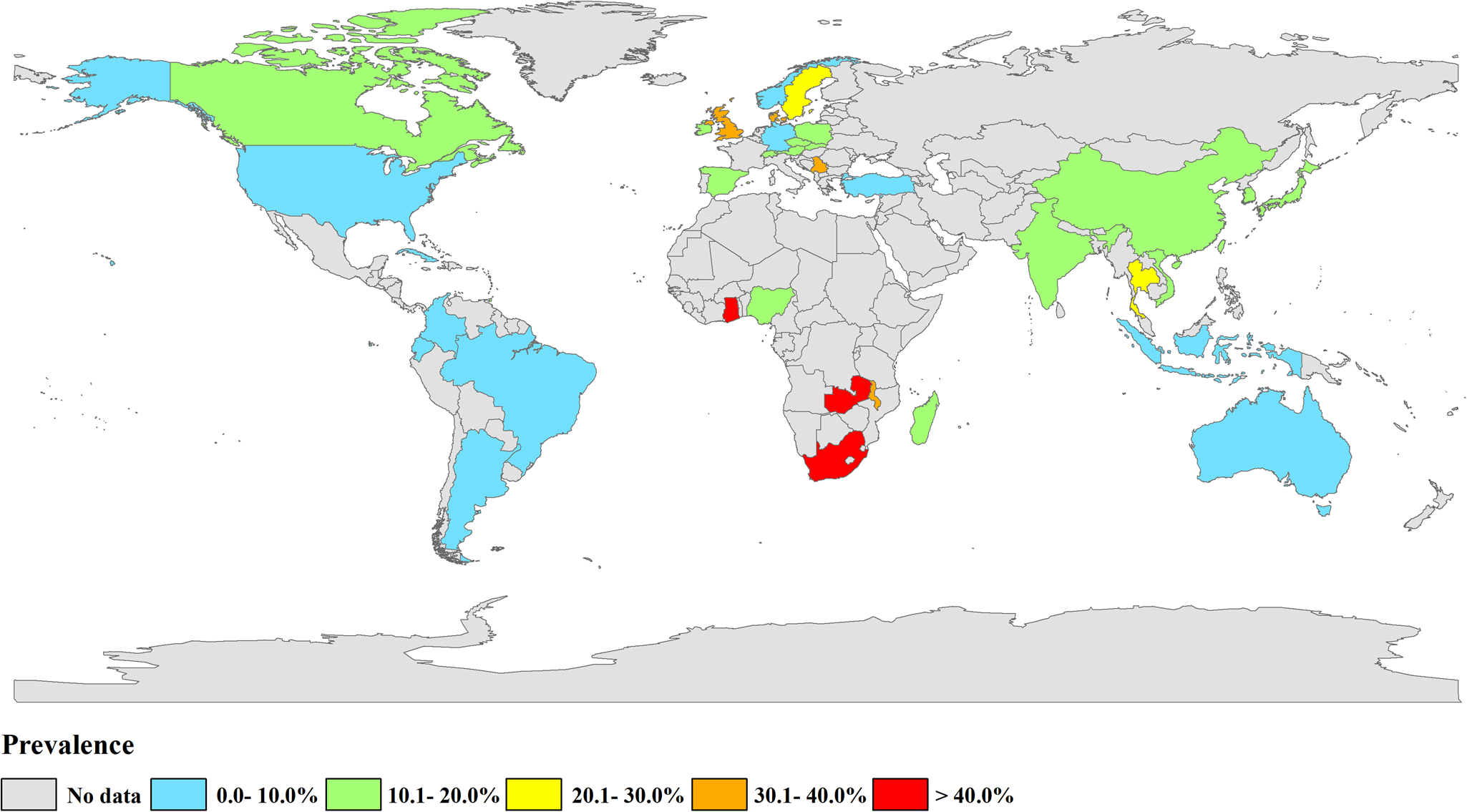
Fig. 2. Map of Cryptosporidium infection in pigs across the world. Prevalence ranges are shown in different colours. [The figure was designed using Arcgis 10.2, and the original vector diagram imported in Arcgis was adapted from Natural Earth (http://www.naturalearthdata.com).]
Table 1. Estimated pooled prevalence of Cryptosporidium infection by country/region

Table 2. Pooled prevalence of Cryptosporidium infection in pigs across the world

a Including C. parvum, C. muris, C. tyzzeri, C. andersoni, C. struthioni, Cryptosporidium spp.
Table 3. Extracted data from included studies for molecular methods of Cryptosporidium species

a Mixed infection
Cryptosporidium infection in pigs by region
The estimated Cryptosporidium prevalence in pigs ranged from 7.1% [95% confidence interval (CI) 3.6–10.5%] to 40.8% (95% CI 20.6–61.0%), with substantial heterogeneity (I 2 = 98.8%, P < 0.001). On a global scale, pooled estimated prevalence of Cryptosporidium infection in pigs was 16.3% (95% CI 15.0–17.6%, 8560/64 809) (Table 2). On 6 continents (Table 2, Figs 3–8), the infection rates of Cryptosporidium in pigs were 14.8% in Asia, 18.3% in Europe, 40.8% in Africa, 13.6% in North America, 7.1% in South America and 9.3% in Oceania. The highest number of studies on Cryptosporidium infections in pigs originated from Asia (n = 71). The highest prevalence rate was reported in South Africa [80.0% (95% CI 71.6–88.4%)], and the lowest prevalence rate was in Germany [0.4% (95% CI 0.1–0.6%)] (Table 1).
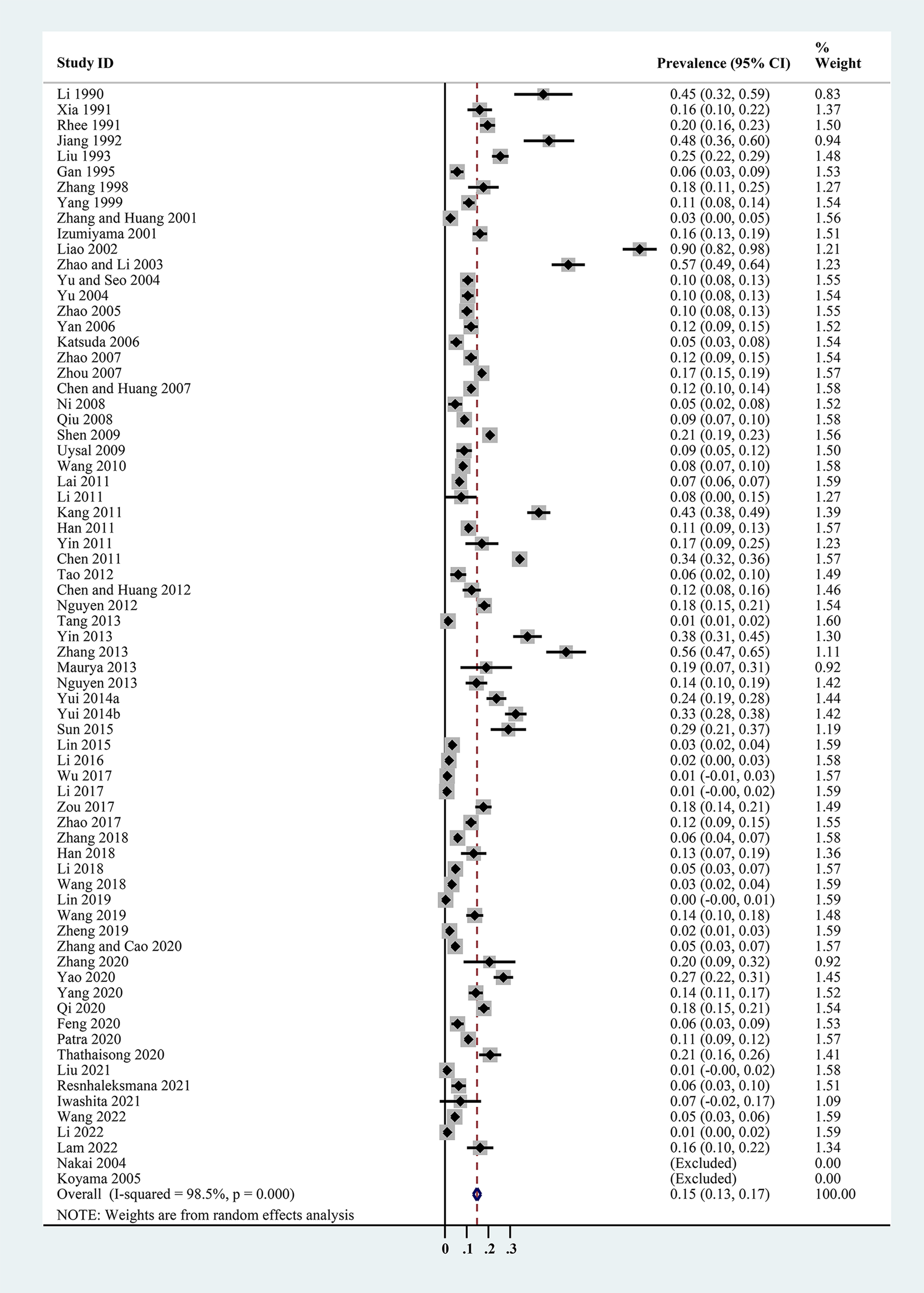
Fig. 3. Forest plot of the prevalence estimates of Cryptosporidium infection in pigs in Asia.

Fig. 4. Forest plot of the prevalence estimates of Cryptosporidium infection in pigs in Europe.

Fig. 5. Forest plot of the prevalence estimates of Cryptosporidium infection in pigs in Africa.

Fig. 6. Forest plot of the prevalence estimates of Cryptosporidium infection in pigs in North America.

Fig. 7. Forest plot of the prevalence estimates of Cryptosporidium infection in pigs in South America.

Fig. 8. Forest plot of the prevalence estimates of Cryptosporidium infection in pigs in Oceania.
Prevalence related to age, presence or absence of diarrhoea and Cryptosporidium species
The Cryptosporidium infection rate in post-weaned pigs was 25.8% (95% CI 21.8–29.8%, 2739/11 824). This was significantly higher than that in pre-weaned pigs [12.0%, 95% CI 9.9–14.0%, 1061/11 370, odds ratio (OR) 2.93, P < 0.05], fattening pigs (17.4%, 95% CI 14.8–20.0%, 1186/8815, OR 1.94, P < 0.05) and adult pigs (12.7%, 95% CI 10.4–15.1%, 980/9658, OR 2.67, P < 0.05) (Table 2). The infection rate for pigs with diarrhoea was 8.0% (95% CI 5.6–10.3%, 348/4874), while the infection rate for pigs without diarrhoea was 12.2% (95% CI 8.4–15.9%, 371/3501) (Table 2). Seven Cryptosporidium species (C. scrofarum, C. suis, C. parvum, C. muris, C. tyzzeri, C. andersoni, C. struthioni) were detected in pigs globally (Table 3). The prevalence rate of C. scrofarum was 7.9% (95% CI 6.9–8.8%, 1491/23 168) and that of C. suis was 4.7% (95% CI 3.8–5.6%, 1385/25 036) (Table 2). In Europe, C. scrofarum and C. suis infection rates were the highest, at 10.3% (678/6613) and 8.0% (881/10 951), respectively (Table S2).
Prevalence according to geographic and climatic variables
We analysed geographic subgroup factors. The prevalence of Cryptosporidium in pigs in regions with a −30° to 0° latitude range (22.9%, 95% CI 8.3–37.5%, 193/872), 0°–60° longitude range (29.3%, 95% CI 17.9–40.7%, 774/5729), 5–10 °C mean yearly temperature (25.4%, 95% CI 16.3–34.6%, 603/4991), <60% mean yearly relative humidity (21.5%, 95% CI 15.0–28.0%, 627/3921), 800–1200 mm mean yearly precipitation (20.7%, 95% CI 15.5–25.9%, 2006/10 586) was higher than that in other regions (Table S3).
Sensitivity analysis and publication bias
Sensitivity analysis indicated that the analysis was reliable (Figs S1–S6). We often used a funnel plot to measure the publication bias in selected articles. Some points fell outside the funnel and the funnel plot showed obvious asymmetry (Fig. 9). The P value was less than 0.001 by Egger's test (Table S4), indicating that obvious publication bias was found.

Fig. 9. Funnel plot for examination of publication bias of the prevalence estimates of Cryptosporidium infection in pigs across the world.
Sources of heterogeneity by meta-regression analysis
Univariate meta-regression analysis was used to determine the sources of heterogeneity. Age (P < 0.001), Cryptosporidium species (P = 0.002) and latitude (P = 0.028) were the factors that fostered heterogeneity. Region (P = 0.381), presence or absence of diarrhoea (P = 0.367), longitude (P = 0.793), mean temperature (P = 0.345), mean relative humidity (P = 0.356) and mean yearly precipitation (P = 0.548) were the factors that affected heterogeneity (Tables 2 and S3).
Discussion
A meta-analysis based on selected datasets from 36 countries on 6 continents produced an estimate of Cryptosporidium prevalence in pigs. As mentioned in a previous systematic review, Cryptosporidium prevalence in pigs was the highest in Asia, Africa and Europe (Hatam-Nahavandi et al., Reference Hatam-Nahavandi, Ahmadpour, Carmena, Spotin, Bangoura and Xiao2019). Compared with previous study, the prevalence of Cryptosporidium in pigs was the highest in Africa, Europe and Asia in our study. In Europe, the highest infection rate was in the UK (38.6%, 95% CI 33.2–44.1%) (Featherstone et al., Reference Featherstone, Marshall, Giles, Sayers and Pritchard2010), while the lowest rate was in Germany (0.4%, 95% CI 0.1–0.6%) (Wieler et al., Reference Wieler, Ilieff, Herbst, Bauer, Vieler, Bauerfeind and Zahner2001; Epe et al., Reference Epe, Coati and Schnieder2004). Cryptosporidium infection in pigs differs between countries and also in different regions of the same country. In China, 1 study reported an infection rate of only 0.9% (2/216) in pigs in Zhejiang (Liu et al., Reference Liu, Ni, Xu, Wang, Li, Shen and Yin2021), while another study found a much higher infection rate of 26.9% (101/375) in pigs in Shaanxi (Yao et al., Reference Yao, Wang, Wang, Li, Zhao, Song and Zhao2020).
Previous studies demonstrated that the rate of Cryptosporidium infection in pigs was related to age factors (Maddox-Hyttel et al., Reference Maddox-Hyttel, Langkjaer, Enemark and Vigre2006; Featherstone et al., Reference Featherstone, Marshall, Giles, Sayers and Pritchard2010). In our analysis, the Cryptosporidium infection rate in post-weaned pigs was significantly higher than that in pigs of other age groups. This is consistent with other studies (Wang et al., Reference Wang, Qiu, Jian, Zhang, Shen, Zhang and Xiao2010; Yui et al., Reference Yui, Shibahara, Kon, Yamamoto, Kameda and Taniyama2014a, Reference Yui, Nakajima, Yamamoto, Kon, Abe, Matsubayashi and Shibahara2014b; Petersen et al., Reference Petersen, Jianmin, Katakam, Mejer, Thamsborg, Dalsgaard and Enemark2015; Pettersson et al., Reference Pettersson, Ahola, Frössling, Wallgren and Troell2020; Qi et al., Reference Qi, Zhang, Xu, Zhang, Xing, Tao and Zhang2020). Post-weaned piglets may be more susceptible to Cryptosporidium infection due to reduced immunity resulting from the loss of maternal immunity, or it may be due to weaning stress (Maddox-Hyttel et al., Reference Maddox-Hyttel, Langkjaer, Enemark and Vigre2006; Li et al., Reference Li, Guo, Wen, Jiang, Ma and Han2018b). However, other studies revealed slightly divergent results. In Vietnam, the Cryptosporidium infection rate in pre-weaned pigs was higher (24.7%; 67/271) than that in post-weaned pigs (17.2%; 51/296), fattening pigs (7.1%; 7/98) or adult pigs (12.0%; 9/75) (Nguyen et al., Reference Nguyen, Honma, Geurden, Ikarash, Fukuda, Huynh, Nguyen and Nakai2012). In China, 2 studies showed higher rates of Cryptosporidium infection in finishing pigs than in pre-weaned, post-weaned and adult pigs (Chen and Huang, Reference Chen and Huang2007; Wang et al., Reference Wang, Li, Zou, Du, Song, Wang and Chen2022). In general, Cryptosporidium infection in post-weaned pigs has attracted greater attention. However, high rates of Cryptosporidium infection in pigs of other age groups suggest that different management measures among the geographical areas may be involved in infection.
The global prevalence of Cryptosporidium infection in pigs without diarrhoea was higher than that in pigs suffering from diarrhoea (P < 0.05). Most of the articles did not mention the presence or absence of diarrhoea in pigs. Insufficient data collection may also affect the stability of the results. Therefore, the relationship between Cryptosporidium infection and diarrhoea in pigs remains unclear. Experimental infection studies showed that pigs shed a high number of Cryptosporidium oocysts but had no or mild diarrhoea. When Cryptosporidium was co-infected with other enteric pathogens, pigs exhibited significant diarrhoea and had a high mortality rate (Enemark et al., Reference Enemark, Ahrens, Bille-Hansen, Heegaard, Vigre, Thamsborg and Lind2003). These results indicated that feces of apparently healthy pigs may also contain Cryptosporidium oocysts and that prevention of Cryptosporidium transmission in healthy pigs should be considered.
Pre-weaned pigs shed significantly more Cryptosporidium oocysts than older pigs, and this was associated with C. suis infection (Kvác et al., Reference Kvác, Hanzlíková, Sak and Kvetonová2009b). Piglets were more susceptible to C. suis infection, while older pigs were more susceptible to C. scrofarum (Yin et al., Reference Yin, Yuan, Cai, Shen, Jiang, Zhang and Cao2013). Compared with previous studies, C. suis and C. scrofarum are still the dominant species in pigs. Other Cryptosporidium species (C. parvum, C. muris, C. tyzzeri, C. andersoni, C. struthioni) have occasionally been reported in pigs. House mice were the main hosts of C. muris and C. tyzzeri (Feng et al., Reference Feng, Ryan and Xiao2018), and mice on pig farms may be involved in transmitting Cryptosporidium. Cryptosporidium parvum infection in pigs mainly occurred in Europe (Wieler et al., Reference Wieler, Ilieff, Herbst, Bauer, Vieler, Bauerfeind and Zahner2001; Zintl et al., Reference Zintl, Neville, Maguire, Fanning, Mulcahy, Smith and De Waal2007; Kvác et al., Reference Kvác, Sak, Hanzlíková, Kotilová and Kvetonová2009a; García-Presedo et al., Reference García-Presedo, Pedraza-Díaz, González-Warleta, Mezo, Gómez-Bautista, Ortega-Mora and Castro-Hermida2013; Němejc et al., Reference Němejc, Sak, Květoňová, Kernerová, Rost, Cama and Kváč2013a; Rzeżutka et al., Reference Rzeżutka, Kaupke, Kozyra and Pejsak2014; Pettersson et al., Reference Pettersson, Ahola, Frössling, Wallgren and Troell2020), Asia (Katsuda et al., Reference Katsuda, Kohmoto, Kawashima and Tsunemitsu2006; Qi et al., Reference Qi, Zhang, Xu, Zhang, Xing, Tao and Zhang2020; Yao et al., Reference Yao, Wang, Wang, Li, Zhao, Song and Zhao2020; Liu et al., Reference Liu, Ni, Xu, Wang, Li, Shen and Yin2021; Resnhaleksmana et al., Reference Resnhaleksmana, Wijayanti and Artama2021) and North America (Atwill et al., Reference Atwill, Sweitzer, Pereira, Gardner, Van Vuren and Boyce1997; Farzan et al., Reference Farzan, Parrington, Coklin, Cook, Pintar, Pollari and Dixon2011; Budu-Amoako et al., Reference Budu-Amoako, Greenwood, Dixon, Barkema, Hurnik, Estey and McClure2012). Cryptosporidium parvum may play a role in zoonotic transmission on pig farms. Therefore, necessary measures should be taken to reduce contact between breeders and pigs to reduce the transmission of Cryptosporidium from pigs to humans.
Oocysts can survive for a long time under many environmental conditions (Rose et al., Reference Rose, Huffman and Gennaccaro2002; Gorospe, Reference Gorospe2005; Alum et al., Reference Alum, Absar, Asaad, Rubino and Ijaz2014), and a single oocyst is sufficient to infect and cause disease in a susceptible host (Ramirez et al., Reference Ramirez, Ward and Sreevatsan2004). The prevalence of Cryptosporidium in pigs in regions with −30° to 0° latitude range (22.9%, 193/872) and 0°–60° longitude range (29.3%, 774/5729) was higher than that in pigs in other regions. Jagai et al. predicted that climate change would increase the spread of cryptosporidiosis infection, and that this spread would vary by season and location (Jagai et al., Reference Jagai, Castronovo, Monchak and Naumova2009). The prevalence of Cryptosporidium in pigs was higher in areas with a mean yearly precipitation of 800–1200 mm (20.7%, 2006/10 586), mean yearly temperature of 5–10 °C (25.4%, 603/4991) and mean yearly relative humidity of < 60% (21.5%, 627/3921). These results indicated that cryptosporidiosis was more likely to occur in warm and rainy areas. Factors such as rainfall, temperature and humidity influence the life cycle of Cryptosporidium and may influence the timing and intensity of disease outbreaks (Patz et al., Reference Patz, Graczyk, Geller and Vittor2000).
Limitations
The current study has the following limitations:
1. Some countries had only 1 publication of Cryptosporidium infecting pigs in the past 30 years.
2. Unpublished data were not included in the analysis.
3. Data of some conference abstracts were not included in the analysis.
4. Some publications lacked full text, and these articles were excluded.
5. Analysis of the factors involved was limited. Factors such as season, feeding model and pig breed may also be sources of heterogeneity.
Even so, we believe that the results of this study are close to the true global prevalence of Cryptosporidium in pigs.
Conclusions
This analysis shows that Cryptosporidium infection in pigs is widespread worldwide. Cryptosporidium can cause high levels of disease, particularly in Africa where infection rates are as high as 40.8%. Cryptosporidium suis is the dominant species in pre-weaned pigs while C. scrofarum is the dominant species in fattening and adult pigs. Pig age is an important risk factor associated with cryptosporidiosis. Age should be considered so that farmers can implement effective management plans based on geographical area and environmental factors and prevent zoonotic transmission. These findings highlight the role of pigs as possible potential hosts of zoonotic cryptosporidiosis and the need for additional studies on the prevalence, transmission and control of Cryptosporidium in pigs.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182023000276
Data availability
All data generated or used during the study appear in the submitted article.
Acknowledgements
We thank Accdon-LetPub Editor for editing the English text of a draft of this manuscript.
Author's contribution
L. Z. conceived and designed the study; Y. C., H. Q. and J. L. conducted the study; J. H., Y. C., H. X. and Y. W. collected and analysed the data; Y. C. and L. Z. wrote the manuscript. All the authors have read and approved the final version of the manuscript.
Financial support
This research was funded by the NSFC-Henan Joint Fund Key Project (U1904203) and the Leading Talents of the Central Plains Thousand Talents Program (19CZ0122).
Conflict of interest
None.
Ethical standards
Not applicable.