Introduction
The detection of the zoonotic parasite Angiostrongylus cantonensis, the rat lungworm, in the city of Valencia (Spain) in 2021 marked a significant milestone (Galán-Puchades et al., Reference Galán-Puchades, Gómez-Samblás, Osuna, Sáez-Durán, Bueno-Marí and Fuentes2022). However, a review conducted in 2021 by Gonzálvez and Ruiz de Ybáñez (Reference Gonzálvez and Ruiz de Ybáñez2022) did not report this finding. Its significance cannot be overstated as it marks the initial documented occurrence of this zoonotic parasite in continental Europe. Previous observations of the parasite in European countries were limited to the Spanish islands of Tenerife in the Canary Islands (Foronda et al., Reference Foronda, López-González, Miquel, Torres, Segovia, Abreu-Acosta, Casanova, Valladares, Mas-Coma, Bargues and Feliu2010) and Mallorca in the Balearic Islands (Paredes-Esquivel et al., Reference Paredes-Esquivel, Sola, Delgado-Serra, Puig Riera, Negre, Miranda and Jurado-Rivera2019).
The expansion of A. cantonensis beyond its presumed native region in Southeast Asia is attributed to global phenomena, notably international trade and travel (Kliks and Palumbo, Reference Kliks and Palumbo1992; Gippet et al., Reference Gippet, Bates, Moulin and Bertelsmeier2023). Valencia, with a major port located less than 10 km from its city centre, plays a crucial role in this dynamic. This port is the 4th busiest in Europe and the 2nd largest in Spain and the entire Mediterranean region. It plays a pivotal role as a strategic maritime hub facilitating the flow of goods between Asia and Europe (Autoridad Portuaria de Valencia, 2024).
The life cycle of A. cantonensis revolves primarily around rats, in which adult parasites inhabit the pulmonary arteries and the right ventricle. However, in this part of its life cycle, the parasite usually produces only mild symptoms in rats (Morgan et al., Reference Morgan, Modry, Paredes-Esquivel, Foronda and Traversa2021). It completes its cycle in molluscs (snails and slugs) (Cowie et al., Reference Cowie, Ansdell, Panosian Dunavan and Rollins2022), which act as intermediate hosts, and uses crustaceans, planarians and frogs, among other taxa, as paratenic hosts (Turck et al., Reference Turck, Fox and Cowie2022). Domestic animals such as dogs (Cowie et al., Reference Cowie, Malik and Morgan2023) and wildlife can become accidentally infected through ingestion of 3rd-stage larvae (L3). In the specific case of humans, the L3 migrate to the brain, often leading to severe, sometimes fatal, neuroangiostrongyliasis that could produce eosinophilic meningitis or meningoencephalitis (Galán-Puchades et al., Reference Galán-Puchades, Gómez-Samblás, Osuna, Sáez-Durán, Bueno-Marí and Fuentes2023). The first association of A. cantonensis with eosinophilic meningitis (neuroangiostrongyliasis) in humans was documented by Beaver and Rosen (Reference Beaver and Rosen1964), based on a case in Taiwan in 1944. To date, the pathogenicity and pathophysiology of the disease remain poorly understood (Chang et al., Reference Chang, Wang, Wang, Lin, Hwu and Cheng2024). In the European continent, a case of neuroangiostrongyliasis was confirmed through a parasite antigen test in Paris in 2016 (Nguyen et al., Reference Nguyen, Rossi, Argy, Baker, Nickel, Marti, Zarrouk, Houzé, Fantin and Lefort2017), although the exact route of infection remains unknown. Various authors have warned about the global risk of neuroangiostrongyliasis emergence in countries where it had not been previously reported, designating it as an emerging parasitic disease (Cowie et al., Reference Cowie, Ansdell, Panosian Dunavan and Rollins2022; Gippet et al., Reference Gippet, Bates, Moulin and Bertelsmeier2023). Angiostrongylus cantonensis has now spread to many parts of the tropics and subtropics, and most recently to more temperate locations, not only to the Canary Islands (Spain) (Foronda et al., Reference Foronda, López-González, Miquel, Torres, Segovia, Abreu-Acosta, Casanova, Valladares, Mas-Coma, Bargues and Feliu2010), the Balearic Islands (Spain) (Paredes-Esquivel et al., Reference Paredes-Esquivel, Sola, Delgado-Serra, Puig Riera, Negre, Miranda and Jurado-Rivera2019) and mainland Spain (Galán-Puchades et al., Reference Galán-Puchades, Gómez-Samblás, Osuna, Sáez-Durán, Bueno-Marí and Fuentes2022), but also to Uganda (Mugisha et al., Reference Mugisha, Bwangamoi and Cranfield2012), the USA, namely Oklahoma and Georgia (York et al., Reference York, Creecy, Lord and Caire2015, Gottdenker et al., Reference Gottdenker, Nascimento Ramos, Hakimi, McHale, Rivera, Miller, Howerth, Burrel, Stilwell, McManamon and Verocai2023) and Argentina (Hancke et al., Reference Hancke, Guzman, Tripodi, Muschetto and Suárez2024). Cases of neuroangiostrongyliasis are likely to increase as the parasite spreads further. It is important to make medical practitioners more aware of this disease.
However, the precise timing and pathways of these invasions remain largely unknown. Several hypotheses have been proposed regarding the parasite's introduction to South America and the Canary Islands, its origins and the chronology of its arrival. It may have occurred through a single event, with a direct introduction from Asia to the Canary Islands, or through a series of intermediate steps involving Asia, other parts of the world, including other islands, such as Hawaii (Červená et al., Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019; Tian et al., Reference Tian, Chen, Duan, Qian, Li and Lv2023). Furthermore, little was known until 2015 (Martin-Alonso et al., Reference Martin-Alonso, Abreu-Yanes, Feliu, Mas-Coma, Bargues, Valladares and Foronda2015) about whether the parasite was completing its life cycle in Europe (political Europe) and which intermediate hosts were involved. In 2024, Fuentes et al. (Reference Fuentes, Gomez-Samblas, Richter, Sáez-Durán, Bueno-Marí, Osuna and Galán-Puchades2024) amplified the A. cantonensis genome in snails collected in several locations in Valencia. These snails serve as intermediate hosts in the parasite's life cycle, confirming the complete establishment of the nematode in this Spanish city. This conclusion was bolstered by the findings of a Mallorcan group (Jaume-Ramis et al., Reference Jaume-Ramis, Martínez-Ortí, Delgado-Serra, Bargues, Mas-Coma, Foronda and Paredes-Esquivel2023) and Martin-Carrillo et al. (Reference Martin-Carrillo, Baz-González, García-Livia, Amaro-Ramos, Abreu-Acosta, Miquel, Abreu-Yanes, Pino-Vera, Feliu and Foronda2023) from the Canary Islands, who also amplified the A. cantonensis genome in intermediate hosts. These investigations concluded that the snails Theba pisana and Cornu aspersum act as intermediate hosts for A. cantonensis in mainland Spain and Mallorca, while Cernuella virgata has been identified as a candidate host in mainland Spain only. Limacus flavus, Milax gagates, Insulivitrina emmersoni and Insulivitrina oromii were found infected by A. cantonensis in the Canary Islands of La Gomera and Gran Canaria.
Various genetic lineages of A. cantonensis have been identified across different endemic regions, although data on genetic and phenotypic diversity within invaded areas are limited (Červená et al., Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019). Lee et al. (Reference Lee, Chung, Wang, Lin, Wang, Tu, Wu and Yen2014) noted the potential variability in the pathogenicity of A. cantonensis across distinct genetic lineages. Additionally, it remains unknown whether a single host can be parasitized by 1 or multiple lineages, and, if so, whether these lineages would share a common origin.
Compared to other parasites, such as protozoans (e.g. Plasmodium falciparum has around 450 sequences in the GenBank), there are few A. cantonensis genome sequences available for phylogenetic and evolutionary studies. Partial sequences of the mitochondrial genes internal transcribed spacer (ITS)-1 and ITS-2 are the most numerous. However, it appears that researchers are increasingly recognizing the parasite's dispersal and zoonotic potential, which has led to an increase in research on the biology of A. cantonensis. For instance, Tian et al. (Reference Tian, Chen, Duan, Qian, Li and Lv2023) compiled and analysed all existing complete mitochondrial sequences, and several other partial mitochondrial and ITS markers of A. cantonensis published in various databases. Although Červená et al. (Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019), Jefferies et al. (Reference Jefferies, Shaw, Viney and Morgan2009), Liu et al. (Reference Liu, Zhang, Chen, Li, Ai, Wu, Zhu and Lin2011) and Rodpai et al. (Reference Rodpai, Intapan, Thanchomnang, Sanpool, Sadaow, Laymanivong, Aung, Phosuk, Laummaunwai and Maleewong2016) found that ITS1 and ITS2 are not suitable markers for resolving evolutionary relationships within A. cantonensis clades, as their low nucleotide diversity hinders phylogenetic reconstruction. Additionally, A. cantonensis has a notably low diversity mitochondrial genome, and hence longer sequences of the complete mitochondrial genome are needed for accurate phylogenetic resolution (Červená et al., Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019).
By using Illumina sequencing and assembly, the current study aims to provide insights into the temporal and geographical origins of A. cantonensis found for the first time in continental Europe, as well as to elucidate if the rat population studied in Valencia since 2021 is parasitized by 1 or more lineages of A. cantonensis.
Materials and methods
Collection of A. cantonensis
The A. cantonensis specimens used for sequencing in this study were collected from rats captured in Valencia between June 2021 and June 2022, specifically from rats trapped in 2 districts, one of them associated with orchards near the port (district 19-Pobles del Sud), and the other in the sewer system almost 15 km from the port (district 16-Benicalap), as described by Galán-Puchades et al. (Reference Galán-Puchades, Gómez-Samblás, Osuna, Sáez-Durán, Bueno-Marí and Fuentes2022). The infected rats selected for the study were randomly chosen, consisting of 2 Rattus norvegicus and 2 Rattus rattus individuals (Table S1). Angiostrongylus cantonensis adults and subadults were extracted from the rat pulmonary arteries and brain, respectively, and identified based on morphological and molecular data as described by Galán-Puchades et al. (Reference Galán-Puchades, Gómez-Samblás, Osuna, Sáez-Durán, Bueno-Marí and Fuentes2022). Upon examination of the nematode genitalia, 1 male and 1 female were selected from each of the 2 R. norvegicus and 1 R. rattus, and 1 male was selected from one of the R. rattus specimens (Table S1) for Illumina sequencing.
For this study, specimen codes were assigned as follows: Acan (A. cantonensis) RVNN (where RV represents Rattus Valencia and NN denotes rat number), followed by M (male) or F (female) to indicate the sex of the nematode (Table S1).
Isolation of genomic DNA from tissue segments of A. cantonensis adults
Genomic DNA (gDNA) from the 4 male and 3 female A. cantonensis was obtained using the Nucleospin Tissue Kit protocol (Macherey-Nagel, Düren, Germany, catalogue number 740952.250) with modifications. Considering the tough cuticle (Gasser et al., Reference Gasser, Chilton, Hoste and Beveridge1993) of nematodes, a mechanical disruption was performed by homogenizing the worms in tubes containing beads (MN Bead Tubes Type D, Macherey-Nagel, Düren, Germany, catalogue number 740814.5) and 200 μL of phosphate-buffered saline in a Precellys 24-Dual instrument (Bertin Technologies, Montigny-le-Bretonneux, France, catalogue number 03119-200-RD010) at 6500 rpm twice for 30 s. Subsequently, 200 μL of T1 buffer and 25 μL of proteinase K were added to each sample, mixed by vortexing, incubated overnight at 56°C and shaken every 3–4 h.
The following day, 200 μL of T3 buffer were added and incubated for 10 min at 70°C. DNA precipitation and washing steps were performed following the kit protocol, and elution was performed using 50 μL of MilliQ water. The quality and concentration of gDNA were assessed using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific Inc, Waltham, Massachusetts, United States). The samples were stored at −20°C until use.
Whole genome sequencing of the rat lungworms
Library preparation and sequencing were performed by Biomarker Technologies GmbH (BMKGENE). Isolated gDNA (7.72–37.8 ng) was used for Illumina Reseq-M library preparation, followed by Illumina sequencing using Novaseq 6000 sequencing systems (150 bp, paired-end). The A. cantonensis genome size is estimated to be 290 Mb (Xu et al., Reference Xu, Xu, Sun, Xu, Zeng, Shan, Yuan, He, He, Yang, Luo, Wei, Wu, Liu, Xu, Dong, Song, Zhang, Yu, Wang, Zhang, Fang, Gao, Lv and Wu2019). We obtained around 3 Gb data per individual to achieve 10× coverage. Prior to further analyses, we performed a quality trimming with Trimmomatic (Bolger et al., Reference Bolger, Lohse and Usadel2014), using the options ‘ILLUMINACLIP:TruSeq3-PE.fa:2:30:10 LEADING:28 TRAILING:28 SLIDINGWINDOW:10:30 MINLEN:100’. After trimming, estimated coverage ranged between 7.5 and 9.6× (Table S1). Illumina reads are available in NCBI (Bioproject PRJNA1102741; SRA accession numbers in Table S1).
Assembly and phylogenetic analysis of the mitogenome
The full mitogenome of the 4 males and 3 females of A. cantonensis was assembled using 1 000 000 paired reads per species with the MITObim assembler (Hahn et al., Reference Hahn, Bachmann and Chevreux2013); see Table S1 for GenBank accession numbers. For the phylogenetic analysis of mitogenomes, 10 sequences of A. cantonensis were retrieved from GenBank (Table S2), and also 8 mitogenomes assembled and available as supplementary material from Tian et al. (Reference Tian, Chen, Duan, Qian, Li and Lv2023), listed in Table S2, were used. Sequences of Angiostrongylus costaricensis and Angiostrongylus malaysiensis were added to be used as outgroups for phylogenetic reconstruction (Table S2). COX-1 sequences were extracted from all mitogenomes, and 82 additional ones were obtained from GenBank (Table S3).
Sequences were aligned using mafft with the LINSI option (Katoh and Standley, Reference Katoh and Standley2013). Phylogenetic trees were constructed under Bayesian inference, performed in MrBayes version 3.2.7 (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001), with the GTR substitution model and Gamma rate variation (GTR-G), as in Tian et al. (Reference Tian, Chen, Duan, Qian, Li and Lv2023). The posterior probabilities were estimated using Markov chain Monte Carlo simulations. Haplotypes and minimum spanning trees were constructed using the R package pegas (Paradis, Reference Paradis2010). Number of nucleotide differences and P-distances were estimated with MEGA11 (Tamura et al., Reference Tamura, Stecher and Kumar2021).
Results
Two different haplotypes of A. cantonensis mtDNA from Valencia
Our mtDNA assemblies are collinear with those reported by Červená et al. (Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019). Length (13 505–13 509 bp) and gene content are also alike, with 12 protein-coding genes, 22 tRNA regions and 2 ribosomal subunits (12S and 16S).
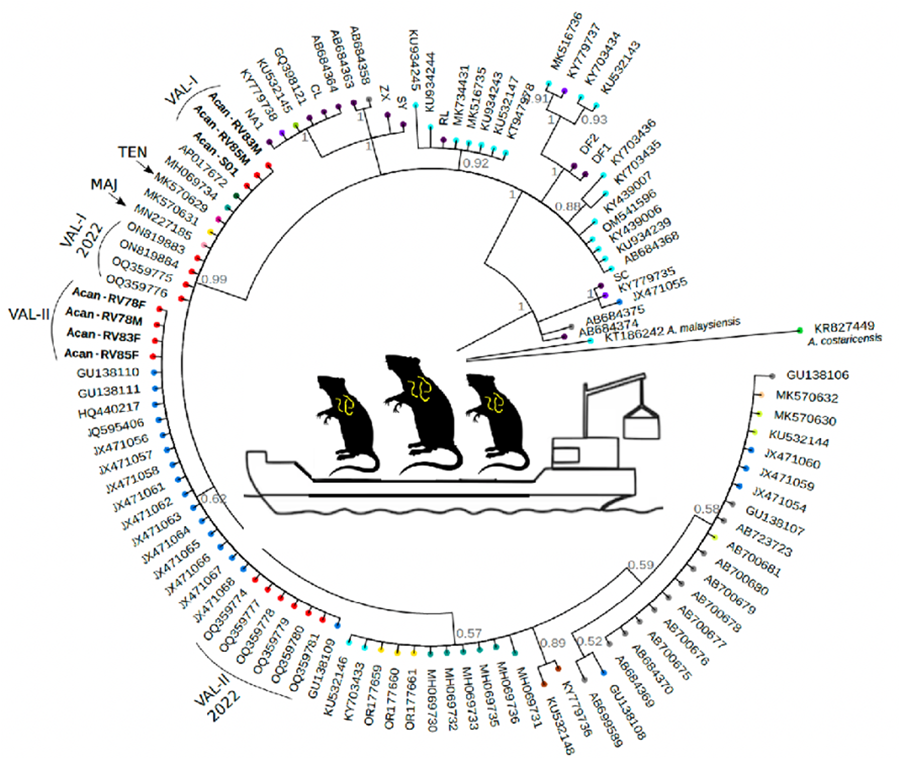
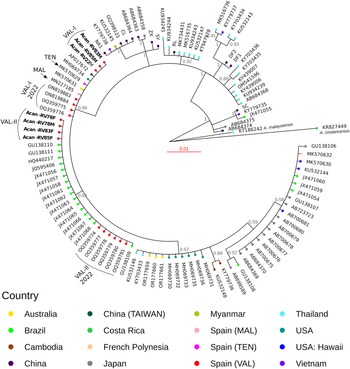
We found 2 different haplotypes of mtDNA in the samples of A. cantonensis from Valencia (Fig. 1A, B). Both haplotypes belong to clade II, defined by Tian et al. (Reference Tian, Chen, Duan, Qian, Li and Lv2023), which includes sequences from French Polynesia, Hawaii, Tenerife (Spain) and Sydney (Australia), sequenced by Červená et al. (Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019), and from Taiwan (Kikuchi et al., Reference Kikuchi, Afrin and Yoshida2016) (Table S2).

Figure 1. (A) Phylogenetic tree of complete mtDNA sequences, built using MrBayes. Numbers in the nodes indicate posterior probabilities. (B) Valencia samples branch shown at a smaller scale for better visualization.
Haplotypes from different locations in mainland China show a larger genetic distance among them than among haplotypes from elsewhere, even though the latter come from widely separated places (e.g. Spain and Australia; see Fig. 2).

Figure 2. Minimal spanning network of complete mtDNA sequences. Numbers indicate the number of nucleotide differences between haplotypes.
The sequences of the individuals found in Valencia were split into 2 groups: 3 of them (Val-I) are very similar (0–2 nucleotide changes) to the Sydney (MK570631), Tenerife (MK570629) and Taiwan (AP017672) isolates; and the other 4 sequences (Val-II) group together in a different branch, with none of the sequences from other countries (Fig. 1B).
Val-I is identical to the haplotype found in Tenerife (Fig. 2). There were more differences between Val-I and Val-II (14 nucleotide changes) than between Val-I and one of the haplotypes found in Sydney (AUS-I, 2 nucleotide changes) and Taiwan (1 nucleotide change). There is almost no variation within the Val-I and Val-II haplotypes, and neither were any intermediate haplotypes found.
In each of the 2 rats, RV83 (R. norvegicus) and RV85 (R. rattus), we found A. cantonensis individuals of 2 different mitochondrial haplotypes (Table S1, Fig. 1).
Cytochrome c oxidase I (COI) indicates the origin of the Val-II haplotype in Brazil
A phylogenetic analysis using 107 sequences of A. cantonensis from various locations, plus 1 A. costaricensis and 1 A. malaysiensis as outgroup taxa (Fig. 3) indicates that the Val-I sequences (Valencia, Tenerife, Hawaii, Taiwan and Sydney) are the same as those in Mallorca (Paredes-Esquivel et al., Reference Paredes-Esquivel, Sola, Delgado-Serra, Puig Riera, Negre, Miranda and Jurado-Rivera2019) and New Orleans (Rael et al., Reference Rael, Peterson, Ghersi-Chavez, Riegel, Lesen and Blum2018).

Figure 3. COI sequence phylogenetic analyses performed using MrBayes. Numbers on the nodes indicate posterior probabilities.
The Val-II sequences form a cluster with 16 sequences from different locations in Brazil (Fig. 3), belonging to the one defined as the ac8 haplotype by Monte et al. (Reference Monte, Simões, Oliveira, Novaes, Thiengo, Silva, Estrela and Júnior2012), which groups sequences from the states of São Paulo, Espirito Santo and Rio de Janeiro (southeastern Brazil), Pará (northern Brazil) and Pernambuco (northeastern Brazil) (Table S2) (Simões et al., Reference Simões, Monteiro, Sánchez, Thiengo, Garcia, Costa-Neto, Luque and Maldonado2011; Monte et al., Reference Monte, Simões, Oliveira, Novaes, Thiengo, Silva, Estrela and Júnior2012; Moreira et al., Reference Moreira, Giese, Melo, Simões, Thiengo, Maldonado and Santos2013).
Discussion
This study is the first comprehensive sequencing of the complete mitochondrial genome of A. cantonensis from 7 individuals derived from continental Europe. Červená et al. (Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019) aimed to elucidate the global geographic movements of A. cantonensis and recognized that, until recently, sequencing the mitochondrial genome was both complex and expensive. Even now, the number of published A. cantonensis sequences remains low. In their investigation, Červená et al. (Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019) examined 4 individuals, each originating from geographically distant countries. According to our results, the genome assembly obtained mirrors the characteristics and features outlined by Červená et al. (Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019) in their study.
Previous studies on A. cantonensis mtDNA revealed a greater nucleotide diversity among individuals in Asia compared with the mere 3 nucleotides differing across the entire mtDNA in individuals isolated from geographically distant regions such as Sydney and Tenerife. Our study corroborates this phenomenon when analysing all sequences isolated from China, compiled by Tian et al. (Reference Tian, Chen, Duan, Qian, Li and Lv2023). As suggested by Červená et al. (Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019), it appears that only a few individuals have managed to migrate from mainland China, and the genetic variability observed across different and distant regions of the world is likely to stem from a single invasion event from mainland China.
Among the 7 individuals sequenced, we identified 2 distinct haplotypes (Figs 1 and 2). All of our individuals belong to clade II, as defined by Tian et al. (Reference Tian, Chen, Duan, Qian, Li and Lv2023). This clade includes most of the sequences that occur outside mainland China, encompassing sequences from French Polynesia (MK570632), Hawaii (MK570630), Tenerife (MK570629), Sydney (MK570631) and Taiwan (AP017672), among others.
Additionally, 3 of our individuals from 3 different rats (Acan-RV22M, Acan-RV83M and Acan-RV85M) correspond to haplotype Val-I, clustering with specimens isolated from Australia, Tenerife and Taiwan. Meanwhile, 4 individuals isolated from 3 different rats (Acan-RV78F, Acan-RV78M, Acan-RV83F and Acan-RV85F), among which 1 harboured both male and female nematodes, were grouped into haplotype Val-II, closely related to specimens isolated from Brazil.
This study is the first to demonstrate the coexistence of 2 distinct strains of A. cantonensis within a single host. Specifically, we found 2 rats with different haplotypes: RV83 parasitized by Acan-RV83M (Val-I) and Acan-RV83F (Val-II), and RV85 parasitized by Acan-RV85M (Val-I) and Acan-RV85F (Val-II). Such an observation was made possible through comprehensive mitochondrial genome sequencing, an approach seldom employed in previous investigations. Our findings underscore that the haplotype of A. cantonensis is not contingent upon the host species, as evidenced by the presence of the same haplotype parasitizing different rat species (i.e. Acan-RV22 isolated from R. rattus and Acan-RV83 from R. norvegicus). Notably, this phenomenon has been observed in districts near the maritime port.
The notion that these 2 haplotypes diverged from a common origin upon arrival in Valencia is refuted when considering the genetic distances between them. Val-I aligns with the haplotype found in Tenerife, whereas there are more differences between Val-I and Val-II (14 nucleotide changes) than between Val-I and the haplotypes found in Sydney and Taiwan. As there was almost no variation within the Val-I and Val-II haplotypes, and intermediate haplotypes were not observed, it is unlikely that these haplotypes diverged in situ in Valencia.
Our hypothesis regarding the invasion and distribution of A. cantonensis,, associated with its first detection in continental Europe, suggests that haplotype Val-I (clade II) originated from Asia via Taiwan (clade II), aligning with the hypothesis of Červená et al. (Reference Červená, Modrý, Fecková, Hrazdilová, Foronda, Alonso, Lee, Walker, Niebuhr, Malik and Šlapeta2019). In our case, this may be attributed to the influx of cargo ships arriving on a daily basis at the port of Valencia, carrying containers that may also introduce rats from Asia. Although A. cantonensis originated from the Chinese mainland (clade I), our minimum spanning network analysis (Fig. 2) suggests that its outward spread was initially via Thailand (clade I), which are the sequences belonging to clade I that are molecularly closest to clade II. Still, haplotypes from Thailand (clade I) have 190 nucleotide differences compared to the clade II group of samples from Spain (Tenerife and Valencia), French Polynesia, Hawaii, Sydney and Taiwan. However, it is difficult to infer a clear timeline from Thailand because of both incomplete sampling and low differentiation among haplotypes in clade II.
Regarding the haplotype Val-II (clade II), our results from the analysis of COI sequences suggest that it entered continental Spain (Valencia) from Brazil. The absence of polymorphisms among individuals of the Val-II haplotype in Valencia, coupled with a divergence of 14 mutations from the closest Spanish haplotype (Tenerife and Val-I from Valencia), supports this inference. The exact time of arrival in Valencia could not be determined due to the lack of complete sequences from specimens isolated in Brazil, the potential source of introduction.
However, Val-I is more widely spread among European places frequently connected by boat (Valencia, Mallorca, Tenerife) and its sequences are also more variable, which supports the inference that Val-I was the first haplotype to enter Spain, and the arrival of Val-II from Brazil must have been more recent.
This study concludes that the A. cantonensis specimens isolated from rats in the European continent so far, specifically in the city of Valencia, belong to clade II as defined by Tian et al. (Reference Tian, Chen, Duan, Qian, Li and Lv2023). This clade encompasses the other A. cantonensis specimens spread worldwide outside mainland China. Additionally, in Valencia, 2 haplotypes are found, differing by 14 nucleotides. Our results also indicate that the same rat, regardless of the species (R. norvegicus or R. rattus), can be parasitized by both haplotypes. Val-I is mostly identical to the specimens described from Tenerife and has been found both in rats captured near the port and in a city location 15 km away from the port. Val-II, a haplotype seemingly originating from Brazil, has only been found in rats captured near the port. Therefore, sequencing individuals from all sampled districts in Valencia with infected rats, as well as from other locations further from the coast, would be necessary to determine the true extent of this Val-II haplotype. This approach would enable the correlation of haplotype distribution with specific sampling locations relative to the port, which probably serves as the most important point of entry of the parasite to mainland Europe. Additionally, it would be worthwhile to obtain mitogenomes from A. cantonensis individuals found in Mallorca and to extend the study to other relevant ports in Spain, such as Algeciras, Spain's number 1 port, as the rat lungworm was not found in Barcelona (Galán-Puchades et al., Reference Galán-Puchades, Sanxis-Furió, Pascual, Bueno-Marí, Franco, Peracho, Montalvo and Fuentes2018), the 3rd busiest port in Spain after Valencia.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182024001318.
Data availability statement
All sequencing data generated in this study have been deposited in GenBank. Accession numbers for all the generated data are provided in the supplementary tables. All the information required to replicate the analyses presented in this study are included within the main text and supplementary materials.
Acknowledgements
We would like to acknowledge the Health Service of Valencia City Council for overseeing and promoting this research in the city.
Author contributions
M. G.-S., B. N.-D. and A. O. conceived and designed the study and conducted data gathering. M. T. G.-P., M. V. F. and S. S.-D. conducted experimental work, collected the samples and identified parasites. R. B.-M. reviewed the draft. M. G.-S., B. N.-D., A. O., M. T. G.-P. and M. V. F. wrote the article. M. Gómez-Samblás and B. Navarro-Dominguez contributed equally to this work.
Financial support
The authors acknowledge the Granada University Research and Knowledge Transfer Fund (PPIT) for supporting Beatriz Navarro-Dominguez, as well as funding for open access charge: Universidad de Granada/CBUA.
Competing interests
None.