Introduction
Leishmaniasis is a parasitological disease with worldwide distribution (World Health Organization, 2010). In the Mediterranean area, the disease is autochthonous, although imported cases are also found (Berriatua et al., Reference Berriatua, Maia, Conceição, Özbel, Töz, Baneth, Pérez-Cutillas, Ortuño, Muñoz, Jumakanova, Pereira, Rocha, Monge-Maillo, Gasimov, van der Stede, Torres and Gossner2021; Fernández-Arévalo et al., Reference Fernández-Arévalo, Ballart, Muñoz-Basagoiti, Basarte, Lobato, Arnau, Abras, Tebar, Llovet, Lami, Pratlong, Alsina, Roe, Puig, Muñoz and Gállego2022; Van der Auwera et al., Reference Van der Auwera, Davidsson, Buffet, Ruf, Gramiccia, Varani, Chicharro, Bart, Harms, Chiodini, Brekke, Robert-Gangneux, Cortes, Verweij, Scarabello, Karlsson Söbirk, Guéry, van Henten, Di Muccio, Carra, van Thiel, Vandeputte, Gaspari and Blum2022). In Spain, the incidence reported for 2018 was 0.65 cases/100 000 habitants (Centro Nacional de Epidemiología, 2019), and in 2017 the estimated incidence rate was 0.86 for visceral leishmaniasis (VL), 1.04 for cutaneous leishmaniasis (CL) and 0.12 for mucocutaneous leishmaniasis, with an estimated underreporting of 14.7–20.2% for VL and 50.4–55.1% for CL (Humanes-Navarro et al., Reference Humanes-Navarro, Herrador, Redondo, Cruz and Fernández-Martínez2021). VL and CL are the main clinical manifestations in the country, but occasionally atypical CL or mucosal presentations are also diagnosed (Aliaga et al., Reference Aliaga, Cobo, Mediavilla, Bravo, Osuna, Amador, Martín-Sánchez, Cordero and Navarro2003; Fernández Martínez et al., Reference Fernández Martínez, Gómez Barroso and Cano Portero2019). Although prevalence is higher in certain regions and seasons, the disease is present in most of Spain and cases are reported throughout the year (Fernández Martínez et al., Reference Fernández Martínez, Gómez Barroso and Cano Portero2019). The parasite responsible for autochthonous cases is Leishmania infantum, the principal domestic reservoir is the dog, and the main vectors are Phlebotomus perniciosus and Phlebotomus ariasi sand flies (Jiménez et al., Reference Jiménez, Ferrer-Dufol, Cañavate, Gutiérrez-Solar, Molina, Lagun, López-Vélez, Cercenado, Daudén, Blazquez, de Guevara, Gómez, de la Torre, Barros, Altes, Serra and Alvar1995; Gállego et al., Reference Gállego, Pratlong, Fisa, Riera, Rioux, Dedet and Portús2001; Martín-Sánchez et al., Reference Martín-Sánchez, Gramiccia, Di Muccio, Ludovisi and Morillas-Márquez2004; Berriatua et al., Reference Berriatua, Maia, Conceição, Özbel, Töz, Baneth, Pérez-Cutillas, Ortuño, Muñoz, Jumakanova, Pereira, Rocha, Monge-Maillo, Gasimov, van der Stede, Torres and Gossner2021). Phlebotomus langeroni has also been incriminated as a vector (Sáez et al., Reference Sáez, Morillas-Márquez, Merino-Espinosa, Corpas-López, Morales-Yuste, Pesson, Barón-López, Lucientes-Curdi and Martín-Sánchez2018) and different domestic and wild animals have been suspected or confirmed to be reservoirs (Azami-Conesa et al., Reference Azami-Conesa, Gómez-Muñoz and Martínez-Díaz2021; Cardoso et al., Reference Cardoso, Schallig, Persichetti and Pennisi2021; Martín-Sánchez et al., Reference Martín-Sánchez, Torres-Medina, Morillas-Márquez, Corpas-López and Díaz-Sáez2021).
In 2009, the largest outbreak of leishmaniasis due to L. infantum ever detected in Europe was declared in Fuenlabrada, in the southwest Madrid region (Spain) (Arce et al., Reference Arce, Estirado, Ordobas, Sevilla, García, Moratilla, de la Fuente, Martínez, Pérez, Aránguez, Iriso, Sevillano, Bernal and Vilas2013). At its peak (2010–2014), more than 600 human cases were reported (Dirección General de Salud Pública – Consejería de Sanidad de la Comunidad de Madrid, 2015; Fernández Martínez et al., Reference Fernández Martínez, Gómez Barroso and Cano Portero2019) and asymptomatic carriers were also detected (Molina et al., Reference Molina, Jiménez, García-Martínez, San Martín, Carrillo, Sánchez, Moreno, Alves and Alvar2020). The great majority of the patients were immunocompetent adults living in the area who presented cutaneous forms of the disease (Arce et al., Reference Arce, Estirado, Ordobas, Sevilla, García, Moratilla, de la Fuente, Martínez, Pérez, Aránguez, Iriso, Sevillano, Bernal and Vilas2013; Carrillo et al., Reference Carrillo, Moreno and Cruz2013). However, children and immunocompromised individuals were also affected, and VL cases and rare clinical manifestations were observed, as well as a high susceptibility of the immigrant population born in Sub-Saharan Africa (Arce et al., Reference Arce, Estirado, Ordobas, Sevilla, García, Moratilla, de la Fuente, Martínez, Pérez, Aránguez, Iriso, Sevillano, Bernal and Vilas2013; Gomez-Barroso et al., Reference Gomez-Barroso, Herrador, San Martín, Gherasim, Aguado, Romero-Maté, Molina, Aparicio and Benito2015; Horrillo et al., Reference Horrillo, San Martín, Molina, Madroñal, Matía, Castro, García-Martínez, Barrios, Cabello, Arata, Casas and Ruiz Giardin2015). Demographic and environmental changes, such as the construction of a vast green park connecting different urban zones, favoured the proliferation of P. perniciosus, hares (Lepus granatensis) and rabbits (Oryctolagus cuniculus), which were incriminated as reservoirs, and led to the outbreak (Molina et al., Reference Molina, Jiménez, Cruz, Iriso, Martín-Martín, Sevillano, Melero and Bernal2012; Carrillo et al., Reference Carrillo, Moreno and Cruz2013; Jiménez et al., Reference Jiménez, González, Martín-Martín, Hernández and Molina2014; González et al., Reference González, Molina, Iriso, Ruiz, Aldea, Tello, Fernández and Jiménez2021).
Over the years, the leishmaniasis cases, reservoirs, vectors and parasites associated with the outbreak have been extensively investigated. However, few studies have characterized the parasites beyond species level. Two studies used the internal transcriber spacer (ITS) to type 3 and 4 strains isolated from hares and rabbits, respectively (Molina et al., Reference Molina, Jiménez, Cruz, Iriso, Martín-Martín, Sevillano, Melero and Bernal2012; Jiménez et al., Reference Jiménez, González, Martín-Martín, Hernández and Molina2014). All strains shared the ITS sequence of the strain MHOM/ES/87/Lombardi (AJ000295), this strain corresponding to zymodeme MON-24 of L. infantum (Chicharro et al., Reference Chicharro, Llanes-Acevedo, García, Nieto, Moreno and Cruz2013). Similar results were obtained for 6 DNA samples and 67 isolates from sand flies when ITS2 was tested (Jiménez et al., Reference Jiménez, González, Iriso, Marco, Alegret, Fúster and Molina2013; González et al., Reference González, Jiménez, Hernández, Martín-Martín and Molina2017). In another study, 31 human isolates related to the outbreak were characterized by ITS and haspb (k26) markers (Chicharro et al., Reference Chicharro, Llanes-Acevedo, García, Nieto, Moreno and Cruz2013). All strains were identified as ITS-Lombardi, but when combined with the haspb (k26) results, 4 different genotypes were detected, L-920 being the most prevalent. Given this background, the aim of the present study was to dig deeper into the characterization of non-human strains from the leishmaniasis outbreak using multilocus enzyme electrophoresis (MLEE), the technique most extensively used in epidemiological studies of leishmaniasis foci and the gold standard recommended by the WHO for this purpose (World Health Organization, 2010). Additionally, strains were also analysed by PCR-sequencing of the heat shock protein 70 (hsp70), one of the main markers used for Leishmania characterization (Fernández-Arévalo et al., Reference Fernández-Arévalo, Ballart, Muñoz-Basagoiti, Basarte, Lobato, Arnau, Abras, Tebar, Llovet, Lami, Pratlong, Alsina, Roe, Puig, Muñoz and Gállego2022; Van der Auwera et al., Reference Van der Auwera, Davidsson, Buffet, Ruf, Gramiccia, Varani, Chicharro, Bart, Harms, Chiodini, Brekke, Robert-Gangneux, Cortes, Verweij, Scarabello, Karlsson Söbirk, Guéry, van Henten, Di Muccio, Carra, van Thiel, Vandeputte, Gaspari and Blum2022).
Materials and methods
Strains
Nineteen strains previously isolated from sand flies and leporids captured at different locations during the leishmaniasis outbreak area of Fuenlabrada (Spain) between 2011 and 2014 were selected for analysis. Eleven of them were isolated from 11 field-captured P. perniciosus in 2012 (n = 4), 2013 (n = 4) and 2014 (n = 3). The sand flies specimens had been captured in 4 collecting stations placed in institutional facilities: 3 located in the municipality of Fuenlabrada, in the border zone of the green park, (ATE-station n = 3, BOS-station n = 3, JIC-station n = 3) and 1 in the central area of the park in the municipality of Leganés (POL-station n = 2) (González et al., Reference González, Jiménez, Hernández, Martín-Martín and Molina2017). The other 8 strains were isolated from colony specimens of P. perniciosus used in direct xenodiagnosis of hares and rabbits. Five of them were isolated from 3 hares living in the park captured in 2011/12 and 3 were obtained from 3 rabbits captured in the border zone in 2013 (Molina et al., Reference Molina, Jiménez, Cruz, Iriso, Martín-Martín, Sevillano, Melero and Bernal2012; Jiménez et al., Reference Jiménez, González, Martín-Martín, Hernández and Molina2014). All the isolates had been previously characterized by ITS sequencing as the Lombardi type (Molina et al., Reference Molina, Jiménez, Cruz, Iriso, Martín-Martín, Sevillano, Melero and Bernal2012; Jiménez et al., Reference Jiménez, González, Iriso, Marco, Alegret, Fúster and Molina2013, Reference Jiménez, González, Martín-Martín, Hernández and Molina2014; González et al., Reference González, Jiménez, Hernández, Martín-Martín and Molina2017).
The strains were thawed and cultured in Schneider's insect medium supplemented with 20% fetal bovine serum and 1% sterile human urine until the exponential growth phase (Fernández-Arévalo et al., Reference Fernández-Arévalo, Ballart, Muñoz-Basagoiti, Basarte, Lobato, Arnau, Abras, Tebar, Llovet, Lami, Pratlong, Alsina, Roe, Puig, Muñoz and Gállego2022).
Hsp70 gene sequencing
DNA from the cultured strains was obtained by the QIAmp DNA Mini Kit (Qiagen) following the manufacturer's instructions. Amplification was done using previously described primers and cycling conditions (Van der Auwera et al., Reference Van der Auwera, Maes, De Doncker, Ravel, Cnops, Van Esbroeck, Van Gompel, Clerinx and Dujardin2013; Fernández-Arévalo et al., Reference Fernández-Arévalo, Ballart, Muñoz-Basagoiti, Basarte, Lobato, Arnau, Abras, Tebar, Llovet, Lami, Pratlong, Alsina, Roe, Puig, Muñoz and Gállego2022). PCR products were enzymatically purified using EXOSAP-IT (Affymetrix USB) and double-strand sequenced by the Sanger method at the Scientific and Technologic Centres of the University of Barcelona. The obtained sequences were edited using MEGA X software (Kumar et al., Reference Kumar, Stecher, Li, Knyaz and Tamura2018).
MLEE
Protein extracts were obtained from mass cultures as previously described (Piarroux et al., Reference Piarroux, Trouvé, Pratlong, Martini, Lambert and Rioux1994). The strains were analysed by MLEE using a panel of 15 enzymes [malate dehydrogenase (MDH) E.C 1.1.1.37, malic enzyme (ME) EC 1.1.1.40, isocitrate dehydrogenase (ICD) EC 1.1.1.42, phosphogluconate dehydrogenase (PGD) EC 1.1.1.44, glucose-6-phosphate dehydrogenase (G6PD) EC 1.1.1.49, glutamate dehydrogenase (GLUD) EC 1.4.1.3, NADH diaphorase (DIA) EC 1.6.2.2, nucleoside phosphorylase purine (NP) − 1 and 2 − EC 2.4.2.1, glutamate oxaloacetate transaminase (GOT) − 1 and 2 − EC 2.6.1.1, phosphoglucomutase (PGM) EC 2.7.5.1, fumarate hydratase (FH) 4.2.1.2, mannose phosphate isomerase (MPI) EC 5.3.1.8 and glucose phosphate isomerase (GPI) EC 5.3.1.9] (Rioux et al., Reference Rioux, Lanotte, Serres, Pratlong, Bastien and Perieres1990). The samples were run on starch gel and revealed under suitable conditions for each enzyme. Migration distances were manually measured and compared with those of the marker strains (Supplementary Table S1) to assign the corresponding electromorphs and zymodemes. Once all enzyme mobilities were determined, the strains were tested again with a second batch of protein extract to confirm the results. The isoelectric focusing technique was also used to confirm the NP1 mobility (Piarroux et al., Reference Piarroux, Trouvé, Pratlong, Martini, Lambert and Rioux1994).
Results
Analysis of the hsp70 gene showed that all the analysed strains shared the same genotype, presenting the main sequence reported for L. infantum strains (GenBank accession numbers OQ747159–OQ747177). Neither SNPs nor heterozygous positions were detected.
The strains produced different MLEE patterns. All of them shared an electrophoretic mobility (EM) of 104 for the MDH enzyme (MDH104) and an EM of 100 for the other enzymes except NP1 (EM: 100, 130, 140, 150). Thus, NP1 was the only enzyme associated with enzymatic polymorphisms in Leishmania strains from the outbreak. NP1100 was observed in 11 out of the 19 strains, which were therefore identified as L. infantum zymodeme MON-34. Six strains shared NP1130 (MON-80), whereas NP1140 was observed in 1 strain (MON-24), and NP1150 in the remaining strain (MON-331; also known as GR-19 and MON-24 var NP1150) (Table 1).
Table 1. Results of MLEE Leishmania infantum characterization related to host, location and year of isolation

a ‘JIC’, ‘BOS’ and ‘ATE’ are the codes used to identify the collecting stations placed in the border zone of the park in the municipality of Fuenlabrada, and ‘POL’ the code the collecting station placed in the centre of the park in the municipality of Leganés.
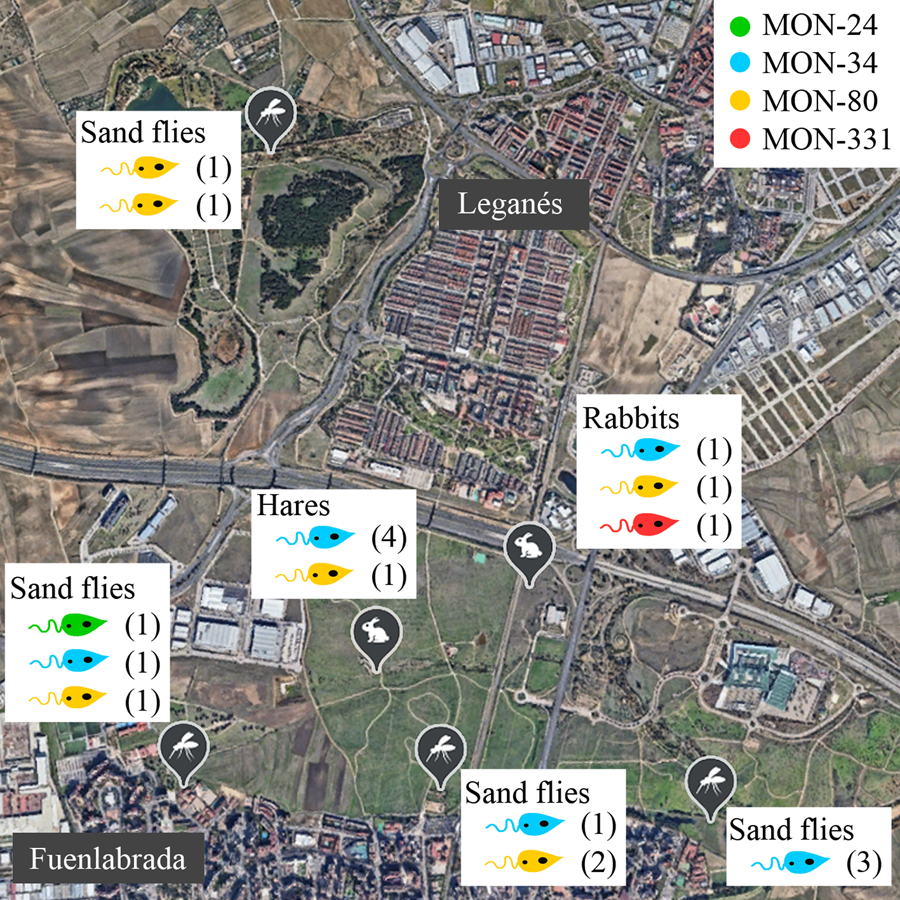
No correlations were apparent between the zymodeme and host, year or location (Table 1, Fig. 1). The only MON-24 strain was isolated from a P. perniciosus specimen in 2013. MON-34 and MON-80 zymodemes were found in rabbits, hares and sand flies from all collecting stations and in all years. Finally, the zymodeme MON-331 was detected in 1 rabbit. If the strains are grouped according to the collecting station and animal reservoir of origin, ‘JIC-station’, ‘POL-station’ and ‘rabbits’ present the most diversity, as each strain in these groups belonged to a different zymodeme. In contrast, all ‘ATE-station’ strains were MON-34 (Fig. 1).

Figure 1. Distribution of Leishmania infantum zymodemes and hosts analysed in the leishmaniasis outbreak from Madrid region (Spain). Each circle represents 1 strain coloured according to its zymodeme. POL (placed in the municipality of Leganés), JIC, BOS and ATE (in the municipality of Fuenlabrada) are sand flies collecting stations where L. infantum strains were isolated from P. perniciosus. Rabbits were captured all around the perimeter of the Bosquesur park (marked with a dotted line) while hares were captured inside the park. The map image was taken from Google Earth (https://earth.google.com).
Discussion
In an unexpected outbreak in an endemic area, identification of the aetiological agent gains importance. Human leishmaniasis can be caused by about 20 of the 56 Leishmania species described (Akhoundi et al., Reference Akhoundi, Kuhls, Cannet, Votýpka, Marty, Delaunay and Sereno2016) and in the current context of globalization and climate change, the presence of imported species cannot be ruled out. In the leishmaniasis outbreak in Fuenlabrada, although part of the immigrant population was highly affected (Arce et al., Reference Arce, Estirado, Ordobas, Sevilla, García, Moratilla, de la Fuente, Martínez, Pérez, Aránguez, Iriso, Sevillano, Bernal and Vilas2013; Gomez-Barroso et al., Reference Gomez-Barroso, Herrador, San Martín, Gherasim, Aguado, Romero-Maté, Molina, Aparicio and Benito2015), the causative species was promptly identified as the autochthonous L. infantum (Chicharro et al., Reference Chicharro, Llanes-Acevedo, García, Nieto, Moreno and Cruz2013).
When performing characterization studies within the L. donovani complex, and specifically for L. infantum strains, one of the major obstacles is the low intraspecific diversity (Maurício, Reference Maurício, Bruschi and Gradoni2018; Fernández-Arévalo et al., Reference Fernández-Arévalo, El Baidouri, Ravel, Ballart, Abras, Lachaud, Tebar, Lami, Pratlong, Gállego and Muñoz2020). Different techniques have been used for the intraspecific analysis of L. infantum, including MLEE, multilocus microsatellite typing, multilocus sequence typing (MLST), randomly amplified polymorphic DNA, PCR-RFLP, and PCR-sequencing (Toledo et al., Reference Toledo, Martín-Sánchez, Pesson, Sanchiz-Marín and Morillas-Márquez2002; Ferroglio et al., Reference Ferroglio, Romano, Trisciuoglio, Poggi, Ghiggi, Sacchi and Biglino2006; Montoya et al., Reference Montoya, Gállego, Gavignet, Piarroux, Rioux, Portús and Fisa2007; Aït-Oudhia et al., Reference Aït-Oudhia, Harrat, Benikhlef, Dedet and Pratlong2011; Van der Auwera and Dujardin, Reference Van der Auwera and Dujardin2015). Depending on the technique and genetic markers used, a different resolution of genetic diversity is achieved. For instance, microsatellite typing provides extremely variable results, whereas single gene PCR-sequencing or PCR-RFLP techniques often show limited diversity (Van der Auwera and Dujardin, Reference Van der Auwera and Dujardin2015). Accordingly, the 19 studied strains from the outbreak were genetically identical by both hsp70 and ITS sequencing. The hsp70 marker is very reliable for Leishmania species identification, but its resolution is too low to perform intraspecies characterization within the Leishmania donovani complex (Fernández-Arévalo et al., Reference Fernández-Arévalo, El Baidouri, Ravel, Ballart, Abras, Lachaud, Tebar, Lami, Pratlong, Gállego and Muñoz2020). The ITS region can differentiate strains below the species level, and it has been correlated with the geographical origin. Evidence for this is the distinction of the Lombardi genotype from the ITS-A genotype, which is also common in the area. However, in L. infantum or L. donovani complex strains, ITS shows a lower degree of polymorphism than in other complexes, not being sufficiently resolving for outbreak studies (Kuhls et al., Reference Kuhls, Mauricio, Pratlong, Presber and Schönian2005; Fernández-Arévalo et al., Reference Fernández-Arévalo, Ballart, Muñoz-Basagoiti, Basarte, Lobato, Arnau, Abras, Tebar, Llovet, Lami, Pratlong, Alsina, Roe, Puig, Muñoz and Gállego2022). Sequencing results can be improved in terms of resolution and robustness by a MLST approach (Kuhls and Mauricio, Reference Kuhls, Mauricio and Clos2019). However, the lack of such studies in the area, together with the absence of a consensus scheme providing sufficient resolution for L. infantum sensu stricto strains from the same geographical origin, hinders, in this case, an integrative understanding of the results. As a side note, it may be interesting to mention that a recent study based on complete maxicircle coding regions, which could be considered kind of MLST, detected 2 distinct L. infantum populations among humans in the Madrid region, in almost perfect agreement with ITS typing (Solana et al., Reference Solana, Chicharro, García, Aguado, Moreno and Requena2022).
MLEE offers an intermediate and manageable level of discrimination for these species (Fernández-Arévalo et al., Reference Fernández-Arévalo, Ballart, Muñoz-Basagoiti, Basarte, Lobato, Arnau, Abras, Tebar, Llovet, Lami, Pratlong, Alsina, Roe, Puig, Muñoz and Gállego2022) and it has been extensively used in epidemiological studies to assess the geographical distribution of isolates and clinical presentations, and to elucidate vectors, and reservoirs (Pratlong et al., Reference Pratlong, Rioux, Marry, Faraut-Gambarelli, Dereure, Lanotte and Dedet2004; Montoya et al., Reference Montoya, Gállego, Gavignet, Piarroux, Rioux, Portús and Fisa2007; Aït-Oudhia et al., Reference Aït-Oudhia, Harrat, Benikhlef, Dedet and Pratlong2011; Haouas et al., Reference Haouas, Chaker, Chargui, Gorcii, Belhadj, Kallel, Aoun, Akrout, Ben Said, Pratlong, Dedet, Mezhoud, Lami, Zribi, Azaiez and Babba2012). However, nowadays, this methodology is falling out of use. The requirement of mass culture of the strains and its manual and laborious protocol make it a costly and time-consuming technique, limiting its use to specific situations. MLEE is not the most suitable technique for inferring taxonomy, as it relies on phenotypic traits that may not be consistent with genetics. But, when applied for L. infantum intraspecific typing, MLEE can provide valuable information on reservoirs and vectors, allowing comparison between foci, and providing data to fill epidemiological gaps. Here lies the interest in using MLEE to characterize the strains isolated from the vectors and reservoirs implicated in the leishmaniasis outbreak in Madrid.
The use of MLEE to type L. infantum has shown that strains zymodemes in humans may belong to viscerotropic, dermotropic or both (Pratlong et al., Reference Pratlong, Rioux, Marry, Faraut-Gambarelli, Dereure, Lanotte and Dedet2004). In the canine reservoir, zymodeme MON-1 is predominant and few others have been isolated, giving rise to the suspicion of anthroponotic transmission cycles in some foci (Aït-Oudhia et al., Reference Aït-Oudhia, Harrat, Benikhlef, Dedet and Pratlong2011). MON-1 is also the main zymodeme identified in isolates from other reservoir hosts, such as cats, foxes, rats, horses and even rabbits (Gállego et al., Reference Gállego, Pratlong, Fisa, Riera, Rioux, Dedet and Portús2001; Martín-Sánchez et al., Reference Martín-Sánchez, Gramiccia, Di Muccio, Ludovisi and Morillas-Márquez2004; Campino et al., Reference Campino, Pratlong, Abranches, Rioux, Santos-Gomes, Alves-Pires, Cortes, Ramada, Cristovão, Afonso and Dedet2006; Díaz-Sáez et al., Reference Díaz-Sáez, Merino-Espinosa, Morales-Yuste, Corpas-López, Pratlong, Morillas-Márquez and Martín-Sánchez2014). In contrast, greater variation has been observed in strains isolated from sand flies (Gállego et al., Reference Gállego, Pratlong, Fisa, Riera, Rioux, Dedet and Portús2001; Martín-Sánchez et al., Reference Martín-Sánchez, Gramiccia, Di Muccio, Ludovisi and Morillas-Márquez2004). In the strains included in the present study, MLEE analysis revealed 4 different zymodemes (MON-24, MON-34, MON-80 and MON-331), which vary from each other by a single enzyme (NP1). Except for MON-331, these zymodemes are commonly found in the Madrid region, as well as in the rest of Spain and the Mediterranean basin (Jiménez et al., Reference Jiménez, Ferrer-Dufol, Cañavate, Gutiérrez-Solar, Molina, Lagun, López-Vélez, Cercenado, Daudén, Blazquez, de Guevara, Gómez, de la Torre, Barros, Altes, Serra and Alvar1995; Chicharro et al., Reference Chicharro, Jiménez and Alvar2003, Reference Chicharro, Llanes-Acevedo, García, Nieto, Moreno and Cruz2013; Pratlong et al., Reference Pratlong, Lami, Ravel, Balard, Dereure, Serres, El Baidouri and Dedet2013). Nevertheless, the frequency of the zymodemes detected here differs from what is usually found in Spain. None of the strains analysed belonged to MON-1, the predominant L. infantum zymodeme, which may be found in ITS-Lombardi strains. Moreover, only a single strain was identified as MON-24, the second most frequent human L. infantum zymodeme including in the area of the outbreak (Jiménez et al., Reference Jiménez, Ferrer-Dufol, Cañavate, Gutiérrez-Solar, Molina, Lagun, López-Vélez, Cercenado, Daudén, Blazquez, de Guevara, Gómez, de la Torre, Barros, Altes, Serra and Alvar1995; Chicharro et al., Reference Chicharro, Jiménez and Alvar2003, Reference Chicharro, Llanes-Acevedo, García, Nieto, Moreno and Cruz2013), where it was isolated from a P. perniciosus. Other studies carried out in the Mediterranean basin have identified MON-24 in strains from cases of human VL and CL and canine leishmaniasis, as well as from sand flies (Gállego et al., Reference Gállego, Pratlong, Fisa, Riera, Rioux, Dedet and Portús2001; Martín-Sánchez et al., Reference Martín-Sánchez, Gramiccia, Di Muccio, Ludovisi and Morillas-Márquez2004; Campino et al., Reference Campino, Pratlong, Abranches, Rioux, Santos-Gomes, Alves-Pires, Cortes, Ramada, Cristovão, Afonso and Dedet2006; Haouas et al., Reference Haouas, Chaker, Chargui, Gorcii, Belhadj, Kallel, Aoun, Akrout, Ben Said, Pratlong, Dedet, Mezhoud, Lami, Zribi, Azaiez and Babba2012). Surprisingly, the most prevalent zymodeme in our study was MON-34 (11/19 strains), followed by MON-80 (6/19 strains), both detected in hares, rabbits and sand flies captured at all the collection points. According to the literature, zymodeme MON-34 has been isolated in human CL and VL cases and in animals (dogs and racoons), but it has not been reported in sand flies until now (Jiménez et al., Reference Jiménez, Ferrer-Dufol, Cañavate, Gutiérrez-Solar, Molina, Lagun, López-Vélez, Cercenado, Daudén, Blazquez, de Guevara, Gómez, de la Torre, Barros, Altes, Serra and Alvar1995; Harrat et al., Reference Harrat, Pratlong, Belazzoug, Dereure, Deniau, Rioux, Belkaid and Dedet1996; Gállego et al., Reference Gállego, Pratlong, Fisa, Riera, Rioux, Dedet and Portús2001; Martín-Sánchez et al., Reference Martín-Sánchez, Gramiccia, Di Muccio, Ludovisi and Morillas-Márquez2004; Montoya et al., Reference Montoya, Gállego, Gavignet, Piarroux, Rioux, Portús and Fisa2007; Pratlong et al., Reference Pratlong, Lami, Ravel, Balard, Dereure, Serres, El Baidouri and Dedet2013; Fernández-Arévalo et al., Reference Fernández-Arévalo, Ballart, Muñoz-Basagoiti, Basarte, Lobato, Arnau, Abras, Tebar, Llovet, Lami, Pratlong, Alsina, Roe, Puig, Muñoz and Gállego2022). MON-80 is less frequent in Spain, and it has been isolated mainly in humans with VL and CL throughout its distribution range (Gállego et al., Reference Gállego, Pratlong, Fisa, Riera, Rioux, Dedet and Portús2001; Chicharro et al., Reference Chicharro, Jiménez and Alvar2003; Martín-Sánchez et al., Reference Martín-Sánchez, Gramiccia, Di Muccio, Ludovisi and Morillas-Márquez2004; Campino et al., Reference Campino, Pratlong, Abranches, Rioux, Santos-Gomes, Alves-Pires, Cortes, Ramada, Cristovão, Afonso and Dedet2006; Pratlong et al., Reference Pratlong, Lami, Ravel, Balard, Dereure, Serres, El Baidouri and Dedet2013). To the best of our knowledge, only 1 study in North Africa has reported MON-80 in dogs (Benikhlef et al., Reference Benikhlef, Aoun, Bedoui, Harrat and Bouratbine2009), and no previous reports describe it in sand flies, although it has been demonstrated experimentally that P. perniciosus could be a potential vector in the outbreak area (Remadi et al., Reference Remadi, Jiménez, Chargui, Haouas, Babba and Molina2018). Therefore, this is the first time that MON-34 and MON-80 strains have been isolated from P. perniciosus and leporids.
Regarding the zymodeme MON-331, found here in a rabbit, only 1 strain has been described before (GR-19, MON-24 var NP1150), in an HIV-positive human with VL (Martín-Sánchez et al., Reference Martín-Sánchez, Gramiccia, Di Muccio, Ludovisi and Morillas-Márquez2004). Although rare, NP1150 has been detected in other zymodemes of the L. donovani complex in isolates from Kenia, Sudan and China (Pratlong et al., Reference Pratlong, Lami, Ravel, Balard, Dereure, Serres, El Baidouri and Dedet2013).
Unfortunately, no human isolates from this outbreak have been typed by MLEE. Only 1 study has characterized human strains from the Fuenlabrada outbreak (Chicharro et al., Reference Chicharro, Llanes-Acevedo, García, Nieto, Moreno and Cruz2013), using ITS and haspb (k26) as molecular markers, also identifying 4 strain types (by haspb (k26)) that shared the ITS-Lombardi sequence. There is no apparent correlation between the isoenzymes and ITS sequences. Besides MON-34, MON-80 and MON-331, reported here, ITS-Lombardi has been described in MON-1, MON-24 and MON-27 strains (Chicharro et al., Reference Chicharro, Llanes-Acevedo, García, Nieto, Moreno and Cruz2013; Fernández-Arévalo et al., Reference Fernández-Arévalo, Ballart, Muñoz-Basagoiti, Basarte, Lobato, Arnau, Abras, Tebar, Llovet, Lami, Pratlong, Alsina, Roe, Puig, Muñoz and Gállego2022), whereas ITS sequence types other than Lombardi have been observed in MON-1, MON-24 and MON-34 strains (Kuhls et al., Reference Kuhls, Mauricio, Pratlong, Presber and Schönian2005; Chicharro et al., Reference Chicharro, Llanes-Acevedo, García, Nieto, Moreno and Cruz2013; Fernández-Arévalo et al., Reference Fernández-Arévalo, Ballart, Muñoz-Basagoiti, Basarte, Lobato, Arnau, Abras, Tebar, Llovet, Lami, Pratlong, Alsina, Roe, Puig, Muñoz and Gállego2022). As far as we know, there is no correlation between haspb (k26) and MLEE either. Based on the available data, it seems that human, reservoir and vector strains from the Fuenlabrada outbreak are similar, but it is not possible to ascertain any relationship between them.
According to MLEE background records, it is likely that the human strains from the outbreak could correspond to MON-34, MON-80 and MON-24. Regarding clinical manifestations, all these zymodemes have been detected in immunocompetent and immunocompromised individuals with VL and CL (Jiménez et al., Reference Jiménez, Ferrer-Dufol, Cañavate, Gutiérrez-Solar, Molina, Lagun, López-Vélez, Cercenado, Daudén, Blazquez, de Guevara, Gómez, de la Torre, Barros, Altes, Serra and Alvar1995; Gállego et al., Reference Gállego, Pratlong, Fisa, Riera, Rioux, Dedet and Portús2001; Chicharro et al., Reference Chicharro, Jiménez and Alvar2003; Martín-Sánchez et al., Reference Martín-Sánchez, Gramiccia, Di Muccio, Ludovisi and Morillas-Márquez2004; Pratlong et al., Reference Pratlong, Lami, Ravel, Balard, Dereure, Serres, El Baidouri and Dedet2013), cases of which were also reported in the outbreak (Arce et al., Reference Arce, Estirado, Ordobas, Sevilla, García, Moratilla, de la Fuente, Martínez, Pérez, Aránguez, Iriso, Sevillano, Bernal and Vilas2013; Carrillo et al., Reference Carrillo, Moreno and Cruz2013; Horrillo et al., Reference Horrillo, San Martín, Molina, Madroñal, Matía, Castro, García-Martínez, Barrios, Cabello, Arata, Casas and Ruiz Giardin2015). However, despite providing data of unquestionable interest, it is unlikely that MLEE would be performed in human samples due to the performance drawbacks mentioned above.
When gathering all the available information about L. infantum in the Fuenlabrada outbreak, various questions arise. On one hand, the typing results reflect a certain homogeneity among the strains involved, although they belong to different zymodemes, and the strain types found are not new or exclusive to the area, which casts doubt on the emergence and transmission of a specific strain type as the detonator of the outbreak. On the other hand, the strains isolated from P. perniciosus in the area have shown higher virulence in the transmission of L. infantum to hamsters in ex vivo tests with experimentally infected P. perniciosus and in in vivo studies with the hamster and mouse model (Domínguez-Bernal et al., Reference Domínguez-Bernal, Jiménez, Molina, Ordóñez-Gutiérrez, Martínez-Rodrigo, Mas, Cutuli and Carrión2014; Martín-Martín et al., Reference Martín-Martín, Jiménez, González, Eguiluz and Molina2015; Mas et al., Reference Mas, Martínez-Rodrigo, Orden, Molina, Jiménez, Jiménez, Carrión and Domínguez-Bernal2021). However, the strains do not seem to cause disease in leporids (Molina et al., Reference Molina, Jiménez, Cruz, Iriso, Martín-Martín, Sevillano, Melero and Bernal2012; Jiménez et al., Reference Jiménez, González, Martín-Martín, Hernández and Molina2014), nor were they found in local dogs, and some exhibit less virulent behaviour in canine cells (Mas et al., Reference Mas, Martínez-Rodrigo, Orden, Viñals, Domínguez-Bernal and Carrión2020). It remains uncertain if there was a selection of these strain types in the leporid reservoir or if they were simply the strain types present in the area at the onset of the outbreak, which arose from the interaction between large populations of vectors, reservoirs and humans. Furthermore, it is unknown if these strains are circulating among leporids in other endemic regions.
The lack of comparable characterization studies hampers a global understanding of the situation. To fully determine the epidemiological characteristics of the Fuenlabrada outbreak and the circulating strains in the region, more comprehensive studies with a higher number of isolates from humans, vectors and reservoirs from a range of L. infantum endemic regions are needed, as well as more robust and discriminative techniques.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182023001336
Data availability statement
Sequences are available in GenBank under accession numbers OQ747159-OQ747177.
Acknowledgements
The authors wish to thank Dr Francine Pratlong and Patrick Lami for their help with queries and for assigning a MON-code to the zymodeme with NP1150. The participating UB and ISGlobal investigators are part of the GREPIMER group (Grup de Recerca en Patología Importada i Malaties Emergents i Re-emergents) recognized by the Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR, 2017 SGR 924 and 2021 SGR 01562). ISGlobal research was supported by the Tropical Disease Cooperative Research Network (RICET) (RD12/0018/0010) and by CIBER -Consorcio Centro de Investigación Biomédica en Red- (CB 2021), Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación and Unión Europea – NextGenerationEU. We acknowledge support from the grant CEX2018-000806-S funded by MCIN/AEI/ 10.13039/501100011033, and support from the Generalitat de Catalunya through the CERCA Program.
Author's contributions
Conceptualization: Anna Fernández-Arévalo, Estela González, Montserrat Gállego, Maribel Jiménez and Ricardo Molina. Investigation: Anna Fernández-Arévalo, Estela González, Cristina Ballart, Inés Martín-Martín, Silvia Tebar and Montserrat Gállego. Formal analysis and interpretation: Anna Fernández-Arévalo, Estela González, Montserrat Gállego, Carme Muñoz, Maribel Jiménez and Ricardo Molina. Writing – original draft preparation: Anna Fernández-Arévalo, Estela González, Montserrat Gállego, Carme Muñoz, Maribel Jiménez and Ricardo Molina; Writing – review and editing: all authors. Final approval: all authors.
Financial support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Competing interests
None.
Ethical standards
Not applicable.