Nutrients involved in one-carbon metabolism have been studied due to their active roles in DNA synthesis, repair and methylation. Folate has been long studied as a key nutrient in one-carbon metabolism. 5-Methyl-tetrahydrofolate (THF), a predominant cytoplasmic form of folate, provides one carbon to methylate homocysteine to methionine forming S-adenosylmethionine (SAM), a universal donor of methyl groups in DNA methylation. THF, a reduced form of folate, is converted to 5,10-methylene-THF by serine hydroxymethyl transferase (Reference Giovannucci1–Reference Kim3). Because of the important role of folate in DNA methylation and synthesis, folate has been suggested to be linked to carcinogenesis.
Alcohol intake increases the risk of colorectal cancer (CRC)(Reference Marmot, Atinmo and Byers4), and the metabolism of alcohol differs according to the expression of two major ethanol-metabolising genes, alcohol dehydrogenase 2 (ADH1B) (rs1229984) and aldehyde dehydrogenase 2 (ALDH2) (rs671)(Reference Seitz and Stickel5,Reference Crabb, Edenberg and Bosron6) . Ethanol is oxidised by the ADH1B (rs1229984) gene into acetaldehyde, which leads to the cleavage of folate(Reference Shaw, Jayatilleke and Herbert7). Acetaldehyde causes cytogenic abnormalities and produces reactive oxygen and nitrogen species (RONS) as well as DNA adducts. It also alters the structure and function of proteins, including enzymes involved in DNA repair and methylation and proteins involved in anti-oxidative defense, leading to DNA damage and inhibition of DNA methylation(Reference Seitz and Stickel5). Acetaldehyde is further converted to acetate by the ALDH2 (rs671) gene(Reference Seitz and Stickel8,Reference Seitz and Becker9) . The effect of alcohol intake on CRC risk may vary between individuals according to ADH1B (rs1229984) and ALDH2 (rs671) polymorphisms, and we also hypothesised that the association between methyl-related nutrients and CRC could be modified through alcohol metabolism.
Several epidemiological studies reported the potential link between methyl diet and CRC incidence. A US cohort study reported that a combination of high alcohol and a low intake of methionine and folate was associated with an increased CRC risk(Reference Giovannucci, Rimm and Ascherio10). Another case–control study found a positive association between low or medium folate combined with high alcohol intake and CRC, and the association was stronger for men than for women(Reference Freudenheim, Graham and Marshall11), whereas the Iowa Women’s Health Study found no association between high folate-low alcohol intake and the incidence of CRC risk(Reference Harnack, Jacobs and Nicodemus12).
One US cohort study has reported the association between a methyl-related diet and colorectal adenoma according to the aldehyde dehydrogenase 3 (ADH1C) and found an interaction by this genotype(Reference Giovannucci, Chen and Smith-Warner13). Because the proportion of A alleles in the ADH1B (rs1229984) and ALDH2 (rs671) genes is extremely low in Western European populations(Reference Goedde, Agarwal and Fritze14) (online Supplemental Fig. S1), research on polymorphisms in these genes is not feasible in these populations. However, an A allele in ADH1B (rs1229984) is the wild type in East Asian populations, and an A allele in ALDH2 (rs671) exists in East Asian populations. A few Asian studies have examined the association between ADH1B (rs1229984) and ALDH2 (rs671) polymorphisms and CRC risk, but they provided inconsistent results.
The World Health Organization Global Information System on Alcohol and Health reported that total alcohol consumption in 2013 was 9·33 litres per capita per year for the Korean population, 7·55 litres per capita per year for the Japanese population and 5·79 liters per capita per year for the Chinese population(15). Given the high alcohol consumption and high proportion of A alleles in ADH1B (rs1229984) and ALDH2 (rs671) genes in the Korean population, we aimed to investigate the interactions of alcohol or methyl diets (as defined by a combined low intake of folate and high intake of alcohol) by ADH1B (rs1229984) and ALDH2 (rs671) genotypes on the susceptibility of CRC development.
Materials and methods
Study population
The current study included 1039 CRC cases and 997 cancer-free controls who were consecutively admitted to two university hospitals in Seoul, Korea, from 1998 to 2004. Newly diagnosed CRC cases and their cancer site locations were reconfirmed via pathology by medical record review. Controls were individuals who had been hospitalised during the same period of time with cholelithiasis, haemorrhoids, acute appendicitis, varicose, chronic otitis media, para nasal sinusitis, cataract or other non-neoplastic surgeries.
We excluded individuals who did not have information on ADH1B (rs1229984) and ALDH2 (rs671) genotypes (ten cases and twenty controls) or alcohol intake (twenty-nine cases and eighty-four controls). After exclusion, 1001 cases and 899 controls aged 30–79 years were analysed in the current study. Among 1900 patients, we partially excluded those with missing values on dietary information (five cases and 238 controls), and 996 cases and 661 controls were eligible for the analysis of methyl diets. The current study was conducted according to the guidelines laid down in the Declaration of Helsinki, and all procedures involving human subjects/patients were approved by the Institutional Review Board of the participating institution. Written informed consent was obtained from every participant.
Data collection
Sociodemographic status, alcohol intake, smoking status and family history of CRC were recorded by trained nurse interviewers using structured questionnaires. Height and weight taken 2 years before diagnosis or on interview date were reported. Body mass index (BMI, kg/m2) was calculated by dividing the weight taken 2 years before diagnosis in kilograms by the square of the height in metres.
Assessment of alcohol intake and methyl diet
Dietary intake was assessed by using a validated food frequency questionnaire(Reference Kim, Ahn and Paik16,Reference Kim, Kim and Ahn17) . Participants reported the frequency and amount of alcohol intake before symptoms began, and we calculated daily ethanol intake in grams per day based on the ethanol content of the beverage including beer, soju, Korean rice wine, wine and whiskey. Folate intake was categorised into tertiles based on the distribution of controls and adjusted for total energy intake by the residual regression method(Reference Willett and Stampfer18). Alcohol intake was categorised into <0·1 g/d of low intake, 0·1 to <30 g/d of moderate intake and ≥30 g/d of high intake. We categorised into a combination of the highest folate intake (≥333·78 μg/d) and low alcohol intake (<0·1 g/d) as high methyl diets, a combination of the lowest folate intake (<141·26 μg/d) and high alcohol intake (≥30 g/d) as low-methyl diets and moderate methyl diets otherwise.
Laboratory assays
The blood samples were collected at the time of interview, and DNA extraction from buffy coat cells was conducted using a commercial kit (Qiagen GmbH, Hilden, Germany). Genotyping for ADH1B (rs1229984) and ALDH2 (rs671) was performed by TaqMan assays (Applied Biosystems, Foster City, CA). The genotype distributions in controls were in agreement for Hardy-Weinberg equilibrium; P values were 0·26 for ADH1B (rs1229984) and 0·43 for ALDH2 (rs671).
Statistical analysis
t Tests for continuous variables and χ 2 tests for categorical variables were used to compare cases and controls. We estimated ORs and 95 % CIs to evaluate the associations of ADH1B (rs1229984) and ALDH2 (rs671) genotypes with CRC using multivariate logistic regression models. We adjusted for potential confounding factors by examining estimate change after including known risk factors for CRC; age (years, continuous); sex (men or women); pack-years of smoking (continuous); BMI, (kg/m2, continuous); and education levels (less than high school, high school and graduate or more) in the model. Total energy intake was included for analysis of methyl diets. We considered other potential confounders including a history of bowel disease, non-steroidal anti-inflammatory drugs (NSAID) use, fibre intake, red meat intake, physical activity or a family history of CRC, but did not include these variables because inclusion of these variables did not appreciably change the results in the final model.
We examined the joint association of ADH1B (rs1229984) and ALDH2 (rs671) genotypes with CRC according to alcohol intake. To investigate the joint association between methyl diets and CRC stratified by ADH1B (rs1229984) and ALDH2 (rs671) polymorphisms, we tested overall interaction using two-sample Wald test for top categories.
All analyses were performed using SAS 9.4 (SAS Institute Inc.). All statistical tests were two-sided, and P values < 0·05 were considered statistically significant.
Results
Baseline characteristics of the study population according to CRC status are shown in Table 1. The mean (±sd) values of age were 59·00 ± 9·60 years for cases and 55·20 ± 9·60 years for controls. Pack-years of smoking and the total amount of alcohol intake were significantly higher in cases than in controls. In addition, cases tended to have a family history of CRC than controls.
Table 1 Characteristics of study participants according to colorectal cancer status
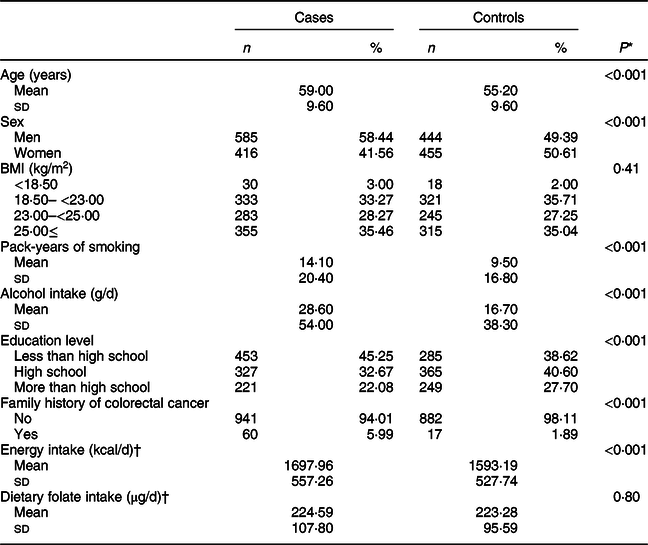
* t test for continuous variables and χ 2 test for categorical variables.
† A total of 996 cases and 661 controls were included.
The frequency of gene polymorphisms in controls reflected their East Asian origin; for ADH1B (rs1229984), 57·29 % were AA, 37·71 % were AG and 5·01 % were GG types. For ALDH2 (rs671) genotypes, 67·29 % were GG, 30·03 % were GA and 2·78 % were AA types (online Supplemental Fig. S1).
We found that total alcohol intake was significantly associated with an increased risk of CRC (Table 2); compared with the non-drinkers, ORs (95 % CI) were 1·60 (0·99, 2·58) for drinkers with 30–<45 g/d of alcohol intake, 2·08 (1·29, 3·36) for drinkers with 45–<60 g/d of alcohol intake and 2·02 (1·41, 2·87) for drinkers with ≥60 g/d of alcohol intake (P trend < 0·001). When we examined the association between alcohol intake and CRC risk among men and women separately, we still found an increased risk of CRC with increasing levels of alcohol intake. Compared with non-drinkers, ORs (95 % CIs) were 2·05 (1·39, 3·01) for drinkers with ≥60 g/d of alcohol intake (P trend < 0·001) among men and 3·95 (1·34, 11·66) for drinkers with 10–<20 g/d and 2·25 (0·87, 5·82) for drinkers with ≥20 g/d of alcohol intake among women (P trend = 0·02).
Table 2 ORs and 95 % CIs for colorectal cancer status according to total alcohol intake
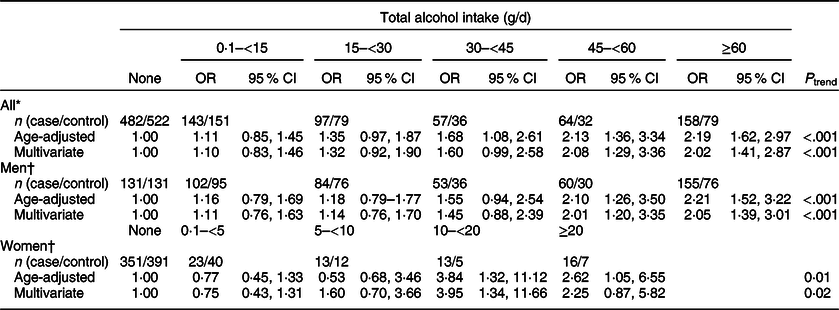
* Multivariate models were adjusted for age (years, continuous), sex (men or women), pack-years of smoking (continuous), BMI (kg/m2, continuous) and education level (less than high school, high school and more than high school).
† Multivariate models were adjusted for age (years, continuous), pack-years of smoking (continuous), BMI (kg/m2, continuous) and education level (less than high school, high school and more than high school).
We found that alcohol intake varied in individuals with ADH1B (rs1229984) or ALDH2 (rs671) polymorphisms in our study population (Table 3). The mean values of alcohol intake were 17·37 g/d for the AA genotype, 17·02 g/d for the AG genotype and 6·80 g/d for the GG genotype of ADH1B (rs1229984). Koreans who had the AA genotype of ALDH2 (rs671) had a mean alcohol intake of 0 g/d, whereas those who had the GG genotype of ALDH2 (rs671) had a mean alcohol intake of 21·67 g/d. We found that the ADH1B (rs1229984) polymorphism was not associated with CRC risk, but there was a suggestive inverse association for the A allele of ALDH2 (rs671); comparing ALDH2 (rs671) GA/AA with the GG genotype, the OR (95 % CI) was 0·83 (0·68, 1·02).
Table 3 ORs and 95 % CIs for colorectal cancer according to ADH1B (rs1229984) and ALDH2 (rs671) polymorphisms*
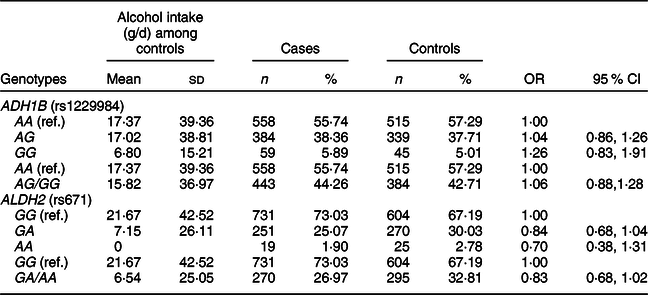
* Models were adjusted for age (years, continuous), sex (men or women), and alcohol intake (g/d, continuous).
For the association between alcohol intake and CRC risk according to the combined genotypes for ADH1B and ALDH2, we found relatively increased risk in CRC among individuals with ADH1B AG/GG and ALDH2 GA/AA (online Supplemental Fig. S2). In a sensitivity analysis where we excluded individuals who did not have information on diet, we observed the similar results (data not shown).
We examined whether the associations of alcohol intake with CRC varied by ADH1B (rs1229984) and ALDH2 (rs671) polymorphisms or folate intake (Table 4). Increasing alcohol intake was associated with an increased risk of CRC regardless of types of ADH1B (rs 1229984) and ALDH2 (rs 671). Participants who drank ≥30 g/d of alcohol had a higher risk of CRC compared with non-drinkers in both strata of low and high folate (P interaction = 0·12).
Table 4 Association between alcohol intake and colorectal cancer risk according to ADH1B (rs1229984) and ALHD2 (rs671) polymorphisms* and folate intake†
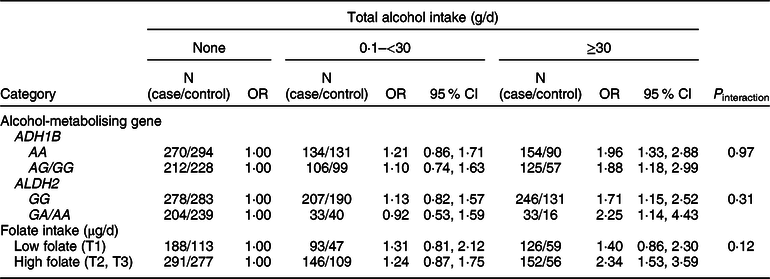
* Models were adjusted for age (years, continuous), sex (men or women), pack-years of smoking (continuous), BMI (kg/m2, continuous) and education level (less than high school, high school and more than high school).
† A total of 996 cases and 661 controls were included; models were adjusted for age (years, continuous), sex (men or women), pack-years of smoking (continuous), BMI (kg/m2, continuous), education level (less than high school, high school and more than high school) and total energy intake (kcal/d, continuous); folate intake was adjusted for energy by residual methods and categorised into tertiles (<141·26, 141·26 to <333·78 and ≥333·78 μg/d).
We found an apparent interaction of the ALDH2 (rs671) polymorphism but not the ADH1B (rs1229984) polymorphism regarding the association between methyl diets and CRC risk (Fig. 1). For ADH1B (rs1229984) polymorphisms, ORs (95 % CIs) comparing low-methyl with high-methyl diets were 1·93 (1·10, 3·39) among individuals with the ADH1B (rs1229984) AA genotypes and 1·51 (0·79, 2·88) among those with ADH1B (rs1229984) AG/GG genotypes (P interaction = 0·58) (Fig. 1A). For ALDH2 (rs671) polymorphisms, ORs (95 % CIs) comparing low-methyl with high-methyl diets were 1·36 (0·82, 2·23) among those with the ALDH2 (rs671) GG genotypes and 9·08 (1·93, 42·60) among those with the ALDH2 (rs671) GA/AA genotypes (P interaction = 0·02) (Fig. 1B). In the additional analysis, we categorised methyl diets by tertiles of folate intake and alcohol intake into <0·1 and ≥10 g/d and found that individuals with low-methyl diet (the lowest tertile and ≥10 g/d of alcohol) had a 4·86 times higher risk of CRC among ALDH2 A allele group compared with high-methyl diets, but no association in GG group (online Supplemental Table S2).
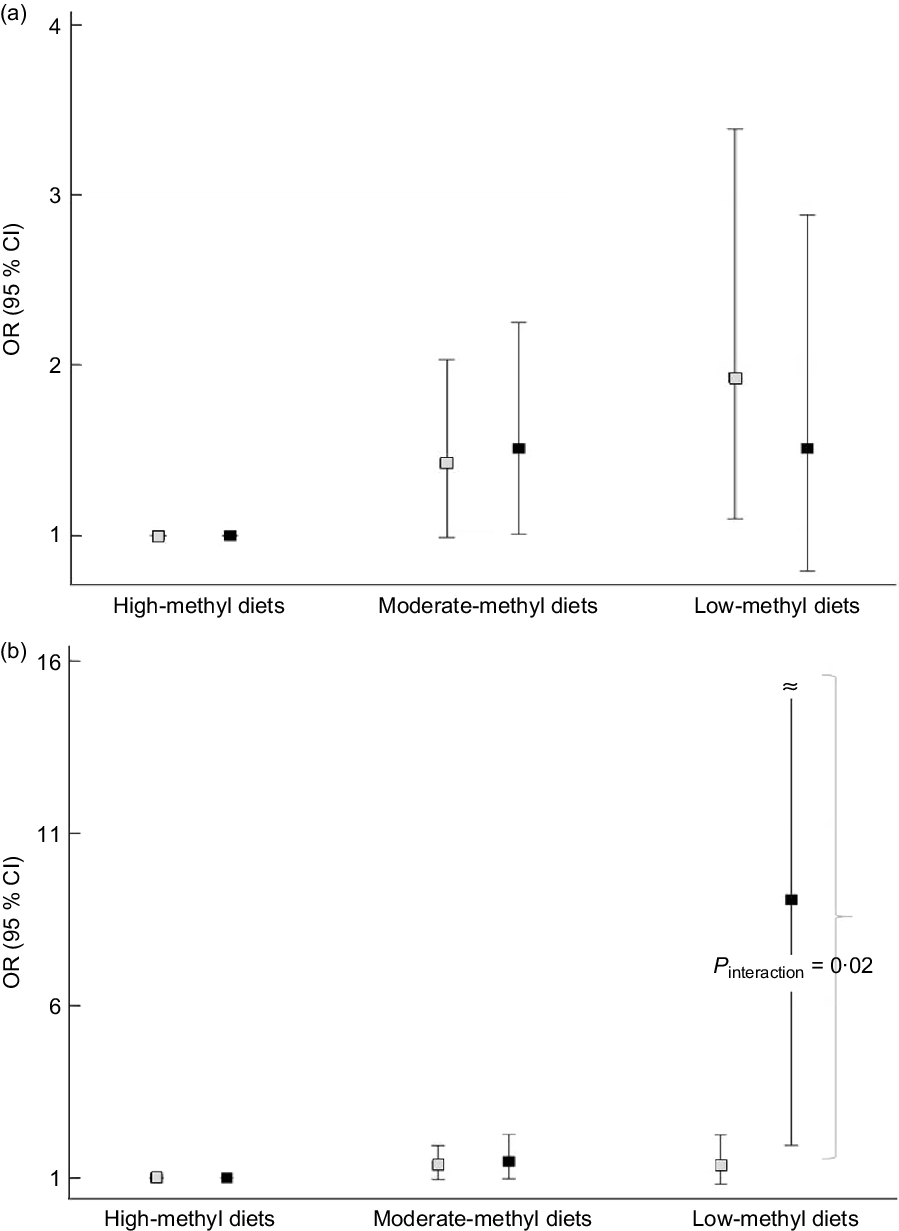
Fig. 1. Association between methyl diets and colorectal cancer risk according to AHD1B (rs1229984) and ALHD2 (rs671) polymorphisms. The squares indicate the study-specific ORs, and the horizontal lines indicate the 95 % CIs. Models were adjusted for age (years, continuous), sex (men or women), pack-years of smoking (continuous), BMI (kg/m2, continuous), education level (less than high school, high school, and more than high school) and total energy intake (kcal/d, continuous). (a) Comparing low-methyl diets with high-methyl diets, ORs (95 % CIs) were 1·93 (1·10, 3·39) among those with the ADH1B (rs1229984) AA types, 1·51 (0·79, 2·88) among those with the ADH1B (rs1229984) AG/GG genotypes (P interaction = 0·58). (b) Comparing low-methyl diets with high-methyl diets, ORs (95 % CIs) were 1·36 (0·82, 2·23) among those with the ALDH2 (rs671) GG types, 9·08 (1·93, 42·60) among those with the ALDH2 (rs671) GA/AA genotypes (P interaction = 0·02). All the values are presented in online Supplemental Table S1. (a) ![]() , ADH1B AA;
, ADH1B AA; ![]() , ADH1B AG/GG; (b)
, ADH1B AG/GG; (b) ![]() , ALDH2 GG;
, ALDH2 GG; ![]() , ALDH2 GA/AA
, ALDH2 GA/AA
Discussion
In our case–control study, we found that alcohol intake increased the risk of CRC. When we examined the interaction of ADH1B (rs1229984) or ALDH2 (rs671) polymorphisms between methyl diets and CRC, we found that the association was remarkably strong in individuals with ALDH2 (rs671) GA/AA polymorphisms with low-methyl diets compared with individuals with high-methyl diets.
We previously reported that low-methyl diets were associated with an increased risk of CRC among CC/CT carriers of methylenetetrahydrofolate reductase (MTHFR)(Reference Kim, Cho and Kim19). In the current study, we observed that this risk was nine times higher for low-methyl diets v. high-methyl diets among participants with the A allele of ALDH2 (rs671). Likewise, a US prospective cohort study found that a combination of high alcohol and low folate intake had seventeen times higher risk of colorectal adenoma among those with ADH1C slow metabolized than those with fast metabolised(Reference Giovannucci, Chen and Smith-Warner13), and a Japanese case–control study reported an 11·9 times higher risk of oral and pharyngeal cancer among high drinkers (≥4 units/d) with low to median folate intake compared with never drinkers with high folate intake in ALDH2 (rs671) GA/AA (Reference Matsuo, Rossi and Negri20). A Taiwanese, multicenter, case–control study reported that individuals with the combination of an AA genotype of ALDH2 (rs671) and ingesting 1200 or greater g of alcohol per year had a 39·76-fold higher risk of oesophageal cancer than did individuals with the combination of the GG genotype of ALDH2 (rs671) and those who abstained from drinking(Reference Chen, Chen and Wu21). In the same Taiwanese study, for the ADH1B (rs1229984) polymorphism, a 147·43 times higher risk of oesophageal cancer risk was observed for participants with the GG genotype of ADH1B (rs1229984) who drank 1200 or greater g of alcohol per year compared to non-drinkers with the AA genotype of ADH1B (rs1229984). A US Health Professional’s Follow-up Study found that an increased risk of colorectal adenoma among individuals with ADH1C slow metaboliser and a combination of high alcohol and low folate intake(Reference Giovannucci, Chen and Smith-Warner13).
Individuals with inactive AA alleles in ALDH2 (rs671) have a low conversion of acetaldehyde to acetate and experience side effects such as facial flushing, palpitations and dizziness(Reference Borras, Coutelle and Rosell22). This reaction could lead to low alcohol intake among those with the A allele of ALDH2 (rs671)(Reference Matsuo, Wakai and Hirose23), which is in agreement with the findings from our study. We found that Koreans who had the GG alleles of ADH1B (rs1229984) drank lower amounts of alcohol than did those with the AA or AG alleles, but previous European and East Asian studies reported higher alcohol consumption among participants who had the GG alleles of ADH1B (rs1229984)(Reference Matsuo, Wakai and Hirose23,Reference Zhang, Hou and Terry24) . We cannot rule out a chance finding to explain this inconsistency.
A potential mechanism explaining the high risk of CRC with high alcohol intake is attributed to the prolongation of ethanol’s contact with colon mucosa and the slow conversion of the carcinogenic acetaldehyde to acetate. Slow digestion of ethanol could lead to an unfavourable response to ethanol such as intestinal inflammation. For example, ethanol in the intestinal tract destroys proteins that make a tight junction linking cells in the intestinal wall, making it easier for intestinal bacteria to pass through the mucosal plasma membrane. This process makes the intestinal wall vulnerable to inflammation, which may be related to cancer development(Reference Elamin, Masclee and Dekker25,Reference Bishehsari, Magno and Swanson26) .
In contrast, folate has a key nutrient as an essential coenzyme for the pathway in DNA methylation, repair and synthesis. THF, the active form of folate, is required to convert homocysteine into methionine, forming SAM for DNA methylation. THF is used to convert to 5,10-methylene-THF, which is required for DNA synthesis involved in the synthesis of thymidylate and purine (Reference Kim3,Reference Choi and Mason27) . Therefore, folate deficiency can contribute to impaired DNA methylation and generate abnormal DNA synthesis and repair(Reference Counts and Goodman28,Reference Blount, Mack and Wehr29) .
Alcohol may disrupt the intestinal absorption of dietary folate and folate supply to tissues, blocking the normal storage and release by the liver(Reference Hillman and Steinberg30). Genetic polymorphisms in ADH1B (rs1229984) and ALDH2 (rs671) determined the efficiency of these enzymes in oxidising alcohol after drinking. Among individuals with the A allele of ADH1B (rs1229984), alcohol may be absorbed into the stomach in a short time and distributed into the bloodstream, resulting in rapid production of acetaldehyde in the liver. The low activity of ALDH2 (rs671) polymorphisms delays the processing of acetaldehyde in the body, thus leading to longer acetaldehyde exposure in the colon(Reference Seitz and Becker9,Reference Druesne-Pecollo, Tehard and Mallet31) . Prolonged exposure of acetaldehyde among individuals with low activity of ALDH2 (rs671) polymorphisms may lead to elevate antagonistic activity of alcohol against folate, resulting in inhibition of folate absorption and disturbance in DNA synthesis, repair and methylation. The disturbance could be worsened when folate intake is low.
There are several strengths and limitations in our study. Our study included a large sample size and is the first, to our knowledge, to explore the joint association between methyl diets and CRC risk according to the ADH1B (rs1229984) or ALDH2 (rs671) polymorphisms in East Asian populations. The relatively higher variation in alcohol intake and the high proportion of ADH1B (rs1229984) and ALDH2 (rs671) variants in the Korean population allowed us to explore the interaction by these genotypes even among individuals who experience side effects such as facial flushing. However, we had limitations in our study. First, relatively low prevalence of ALDH2 A allele among those with low-methyl diets led to the wide range of CIs. However, given that individuals with ALDH2 A allele rarely drank high amount of alcohol, increased risk of CRC among those with low-methyl diets and ALDH2 A allele may warrant the presentation of the data in this group. Further studies need to be replicated. Second, we did not consider nutrients involved in one-carbon metabolism other than folate due to a lack of information. Third, measurement error in alcohol intake and other covariates may exist. We cannot rule out the possibility that unmeasured or residual confounding factors were present, although we adjusted for potential confounding factors. Forth, we cannot rule out the possibility of selection bias due to the exclusion of imbalanced missing values between cases and controls in the dietary analysis. However, we found the similar results for alcohol and genetic polymorphisms in the sensitivity analysis where we excluded individuals who did not have information on diet. Lastly, given that 74 % of women were non-drinkers in our study, we could not conduct sex-specific analyses for the joint effects of alcohol and genetic polymorphisms on CRC risk.
In summary, our data suggest that low-methyl diets increased the risk of CRC among individuals with a rare variant (inactive form) of ALDH2 (rs671) genotypes. Our study results need to be replicated in further epidemiologic studies.
Acknowledgements
Acknowledgements: The authors would like to thank all the participants for their contribution to the study. Financial support: This research was supported by the MSIP (Ministry of Science, ICT and Future Planning), Korea, under the ITRC (Information Technology Research Center) support program (IITP-2019-2014-1-00720) supervised by the IITP (Institute for Information & communications Technology Promotion). Conflict of interest: There are no conflicts of interest. Authorship: Conception and design: D.-H.K. Development of methodology: J.K., B.-H.L., D.-Y.H., J.J., H.-J.L., Y.-O.A. and D.-H.K. Acquisition of data: J.K., B.-H.L., D.-Y.H., J.J., H.-J.L., Y.-O.A. and D.-H.K. Analysis and interpretation of data: J.E.S., J.E.L., J.J. and D.-H.K. Review: J.K., B.-H.L., D.-Y.H., J.J., H.-J.L., Y.-O.A. and D.-H.K. Writing and/or revision of the manuscript: J.E.S., J.E.L. and D.-H.K. Administrative, technical or material support: J.E.S., J.E.L. and D.-H.K. Study supervision: D.-H.K. Ethics of human subject participation: The current study was conducted according to the guidelines laid down in the Declaration of Helsinki, and all procedures involving research study participants were approved by the Institutional Review Board of Seoul National University Hospital and Hallym University Sacred Heart Hospital. Written informed consent was obtained from every participant.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S136898001900452X








