Introduction
Bipolar disorder (BD) has a heritability of up to 70% and is associated with psychosocial impairment (Costanza et al., Reference Costanza, Vasileios, Ambrosetti, Shah, Amerio, Aguglia and Berardelli2022), with a relapse rate of up to 90% and a suicide rate that is 20 times greater than that of the general population. Although current pharmacological interventions, such as mood stabilizers (e.g. lithium) and antipsychotics, provide symptomatic relief, they have limited efficacy, especially in modulating the hypothalamic‒pituitary‒adrenal (HPA) axis (Berardelli et al., Reference Berardelli, Serafini, Cortese, Fiasche, O'Connor and Pompili2020), and more pronounced side effects, according to the most recent summary of FDA approved BD medications (Goes, Reference Goes2023) and the DRUGBANK database (https://go.drugbank.com/) (online Supplementary Table S1). Therefore, there is an urgent need for innovative treatments that can strike a balance between efficacy and tolerability. To address these issues, extensive research has explored the genetic and molecular basis of BD through genome-wide association studies (GWASs) and transcriptome-wide association studies. Several genetic loci associated with BD have been identified, such as NCAM1 (Patel et al., Reference Patel, Le-Niculescu, Koller, Green, Lahiri, McMahon and Niculescu2010), ITIH1 (Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011), and TIMP4 (Lu, Forgetta, Greenwood, Zhou, & Richards, Reference Lu, Forgetta, Greenwood, Zhou and Richards2023). However, the use of only genomic data and a single tissue source limits the interpretability of these findings. Furthermore, due to the complexity of the disease, a variety of these loci are located in noncoding genomic regions, which makes it difficult to pinpoint the exact biological mechanisms involved and the potential drug targets.
Mendelian randomization (MR), which incorporates genetic instrumental variables, has proven beneficial in determining the causal links between genetic loci and diseases. Compared with other quantitative trait loci (QTLs), protein QTLs (pQTLs) are potential therapeutic targets for BD because they are the end products of gene expression and important participants in biological processes. Recent advances have been made in identifying new drug targets for multiple diseases, including multiple sclerosis (Lin, Zhou, & Xu, Reference Lin, Zhou and Xu2023), ischemic stroke (Chong et al., Reference Chong, Sjaarda, Pigeyre, Mohammadi-Shemirani, Lali, Shoamanesh and Paré2019), and Alzheimer's disease (Wingo et al., Reference Wingo, Liu, Gerasimov, Gockley, Logsdon, Duong and Wingo2021), by combining GWAS and pQTL data. However, the application of this strategy in BD has not yet been fully explored.
Traditional pQTL studies have typically been limited to single tissue sources. In our research, we examined samples from the brain, cerebrospinal fluid (CSF), and plasma of patients with BD. The dorsolateral prefrontal cortex (DLPFC) is a crucial region associated with cognitive and executive impairments in patients with BD (Alonso-Lana et al., Reference Alonso-Lana, Moro, McKenna, Sarro, Romaguera, Monte and Pomarol-Clotet2019). Investigating DLPFC pQTLs is crucial for exploring abnormal brain changes in BD patients and identifying potential drug targets. CSF contains pivotal proteins for information transmission between brain regions, directly reflecting changes in circulating substances within the central nervous system (Tumani, Huss, & Bachhuber, Reference Tumani, Huss and Bachhuber2017). Recent proteomic analyses of CSF have highlighted proteins related to BD, including SPOCK1, CLEC1B, DRAXIN, and TNFSF21 (Göteson et al., Reference Göteson, Isgren, Jonsson, Sparding, Smedler, Pelanis and Landén2021). Additionally, peripheral fluid, such as plasma, is an important biological indicator with diagnostic and prognostic value in BD, as it can be collected easily with minimal risk (Kim et al., Reference Kim, Rhee, Lee, Han, Lee, Kim and Ha2021). Therefore, we aimed to integrate the strengths of different studies focusing on the DLPFC, CSF, and plasma to identify tissue-specific druggable proteins associated with BD.
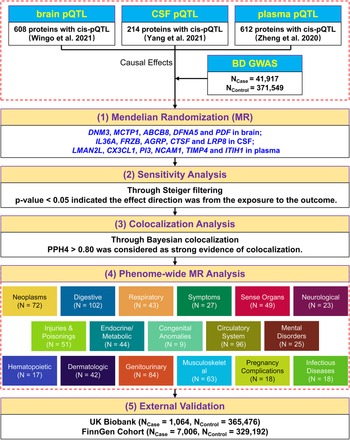
A five-step strategy was adopted to explore the relationship between protein profiles and BD. First, we performed two-sample MR by individually integrating pQTL datasets from the three tissues (brain (Wingo et al., Reference Wingo, Liu, Gerasimov, Gockley, Logsdon, Duong and Wingo2021), plasma (Zheng et al., Reference Zheng, Haberland, Baird, Walker, Haycock, Hurle and Gaunt2020), and CSF (Yang et al., Reference Yang, Farias, Ibanez, Suhy, Sadler, Fernandez and Cruchaga2021)) with the largest BD GWAS data from Mullins et al. (Mullins et al. Reference Mullins, Forstner, O'Connell, Coombes, Coleman, Qiao and Andreassen2021) to identify potential candidate proteins associated with BD. Second, sensitivity analysis was conducted to determine the causal direction between the candidate proteins and BD. Third, we applied Bayesian colocalization analysis to examine whether the two related signals (protein profile and BD phenotype) were influenced by a common causal single nucleotide polymorphism (SNP). Then, we conducted phenome-wide MR analysis to screen for possible side effects of the therapeutic targets for BD. To confirm our findings, we performed MR analysis with GWAS data from the UK Biobank and FinnGen cohorts for external validation. Figure 1 illustrates a graphical representation of the research design.
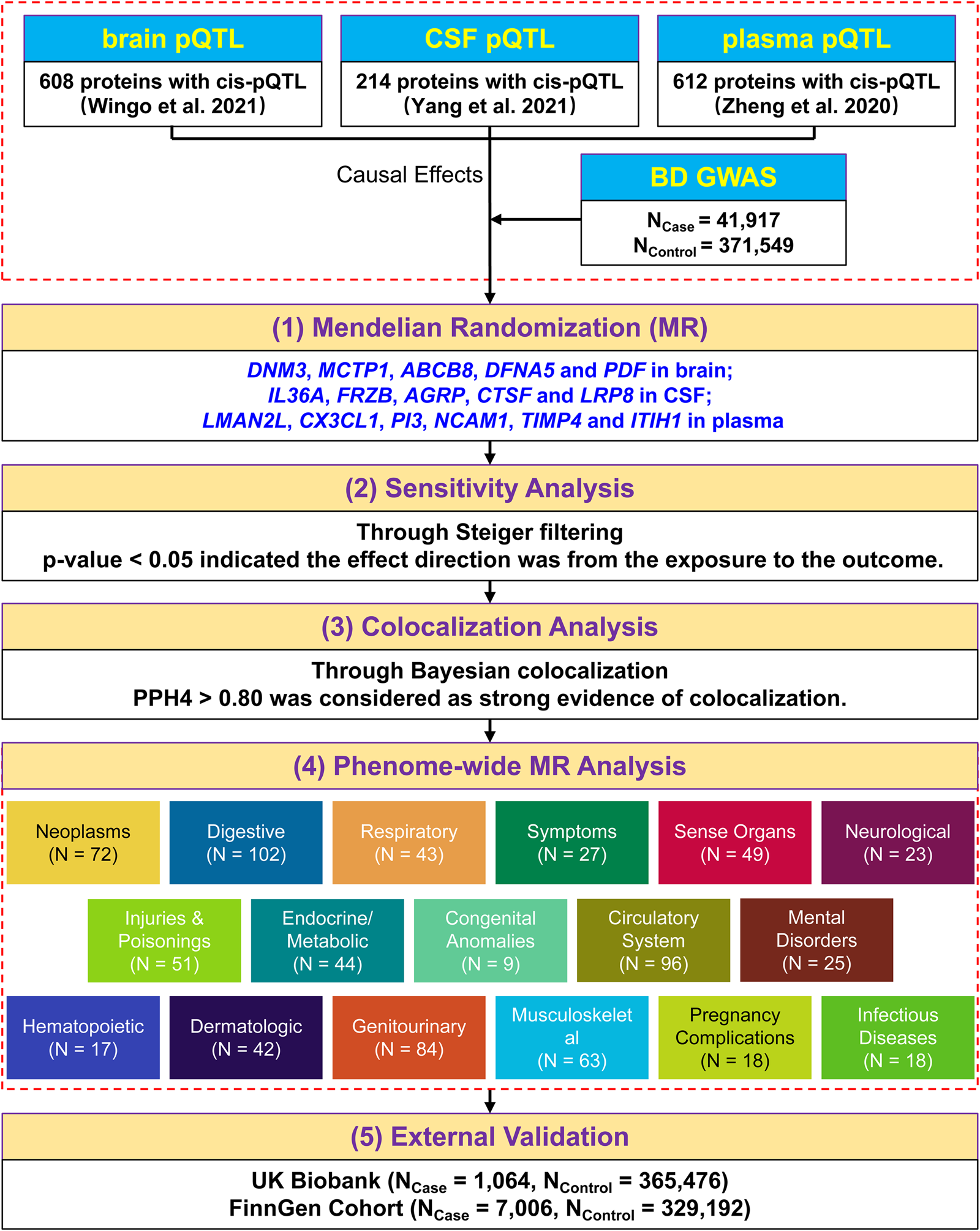
Figure 1. A schematic overview of the study design.
Methods
GWAS summary statistics
Summary statistics of BD were derived from the GWAS study conducted by Mullins et al. (Mullins et al. Reference Mullins, Forstner, O'Connell, Coombes, Coleman, Qiao and Andreassen2021) for our preliminary analysis. The GWAS meta-analysis included 57 BD cohorts comprising a total of 413 466 individuals (N Case = 41 917, N Control = 371 549). In addition, to confirm our findings, we used two other datasets obtained from the UK Biobank (N Case = 1064, N Control = 365 476) generated by Zhou et al. (Zhou et al. Reference Zhou, Nielsen, Fritsche, Dey, Gabrielsen, Wolford and Lee2018) and the FinnGen study (N Case = 7006, N Control = 329 192) generated by Kurki et al. (Kurki et al. Reference Kurki, Karjalainen, Palta, Sipila, Kristiansson, Donner and Palotie2023) for external validation.
Human brain, CSF and plasma pQTL data
The pQTL datasets for the brain, CSF and plasma were derived from studies conducted by Wingo et al. (Wingo et al. Reference Wingo, Liu, Gerasimov, Gockley, Logsdon, Duong and Wingo2021), Yang et al. (Yang et al. Reference Yang, Farias, Ibanez, Suhy, Sadler, Fernandez and Cruchaga2021), and Zheng et al. (Zheng et al. Reference Zheng, Haberland, Baird, Walker, Haycock, Hurle and Gaunt2020), respectively. Specifically, 616 cis-acting brain pQTLs linked to 608 proteins were chosen for our integrative analysis if they met the following criteria: (1) had a statistically significant genome-wide association (p < 5 × 10−8); (2) had linkage disequilibrium clumping (r 2 < 0.001); (3) were cis-acting pQTLs; and (4) contained robust SNPs with F-statistics >10. Similarly, the CSF pQTL data contained 233 cis-acting SNPs linked with 214 proteins, and the plasma pQTL data generated 616 cis-acting SNPs linked to 612 proteins. More detailed information on the pQTL and GWAS datasets is summarized in online Supplementary Table S2.
MR analysis
In this study, the SNPs linked to pQTLs in the brain, CSF, and plasma acted as exposure variables, and the BD GWAS data as the outcome variable. We performed MR analysis using the R package ‘TwoSampleMR’ (https://github.com/MRCIEU/TwoSampleMR) (Hemani et al., Reference Hemani, Zheng, Elsworth, Wade, Haberland, Baird and Haycock2018). The Wald ratio approach was employed when a protein had only one pQTL; otherwise, the inverse variance weighted (IVW) analysis method was used. Bonferroni correction was used to adjust the p value (0.05/608 for brain, 0.05/214 for CSF, and 0.05/612 for plasma).
Sensitivity analysis
Due to the limited number of instrumental variables (IVs), post-MR analysis methods such as Cochran's Q test, MR-Egger intercept test, MRPRESSO test, and I2GX test were unable to perform (Gleason, Yang, & Chen, Reference Gleason, Yang and Chen2021; Hemani et al., Reference Hemani, Zheng, Elsworth, Wade, Haberland, Baird and Haycock2018). Therefore, we utilized Steiger filtering to explore the causal direction between proteins and BD. A p value less than 0.05 indicated statistical significance.
Bayesian colocalization analysis
We performed Bayesian colocalization analysis using the R package ‘COLOC’ to determine whether the risk of BD and the changes in protein levels were attributed to the same single-nucleotide variations. Bayesian colocalization analysis calculated posterior probabilities for the following five crucial hypotheses: H0, neither associated with GWAS nor pQTL; H1, association with GWAS, not with pQTL; H2, association with pQTL, not with GWAS; H3, association with GWAS and pQTL, two independent SNPs; and H4, association with GWAS and pQTL, one shared SNP. A threshold of 0.80 for the probability of hypothesis H4 indicated strong evidence for colocalization (Zheng et al., Reference Zheng, Haberland, Baird, Walker, Haycock, Hurle and Gaunt2020).
Phenome-wide MR analysis
To elucidate the potential side effects (beneficial or adverse) of prior drug targets identified by aforementioned analyses, we performed an agnostic phenome-wide MR analysis of 783 disease traits. The SNPs linked to proteins were set as exposures (online Supplementary Table S3), and summary statistics of diseases from the UK Biobank cohort were set as outcomes. The Scalable and Accurate Implementation of Generalised Mixed Model (SAIGE V.0.29) method was applied to address the unbalanced case‒control ratio (Zhou et al., Reference Zhou, Nielsen, Fritsche, Dey, Gabrielsen, Wolford and Lee2018). To improve the interpretability of the results, we systematically selected 783 representative phenotypes for phenome-wide MR analysis, with more than 500 disease cases. Phenome-wide MR analysis was conducted using the same parameters as those used in the MR analysis. Bonferroni correction was applied (0.05/(4 × 783) = 1.60 × 10−5).
External validation
MR analysis was repeated only on the preliminarily identified proteins with the same-variant and significant-variant strategies and GWAS data from the UK Biobank and FinnGen datasets, respectively. The concordance of the odds ratios (ORs) between the primary and validation cohorts was considered successful, and therefore, a p value less than 0.05 was considered strong evidence of replication.
Results
Screening the three proteomes for BD causal proteins
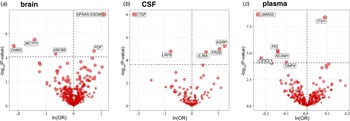
At different Bonferroni significance levels (p < 8.22 × 10−5, 0.05/608 for brain; p < 2.34 × 10−4, 0.05/214 for CSF; p < 8.17 × 10−5, 0.05/612 for plasma), MR analysis revealed that the altered protein abundances of 16 genes were associated with BD (Fig. 2a–2c), including DNM3, MCTP1, ABCB8, DFNA5, and PDF in the brain; IL36A, FRZB, AGRP, CTSF, and LRP8 in CSF; and LMAN2L, CX3CL1, PI3, NCAM1, TIMP4, and ITIH1 in plasma. In summary, decreased DNM3, MCTP1, ABCB8, CTSF, LRP8, LMAN2L, CX3CL1, PI3, NCAM1, and TIMP4 were risk factors for BD, whereas elevated DFNA5, PDF, IL36A, FRZB, AGRP, and ITIH1 increased the risk of BD (Fig. 2 and Table 1). We also observed that in plasma, CTSF (OR 0.84, 95% CI 0.77–0.92; p = 9.14 × 10−5) and AGRP (OR 1.11, 95% CI 1.05–1.17; p = 1.06 × 10−4) reached marginal significance for causal effects, and their effect directions were consistent with those in CSF (not shown in the figures and tables).
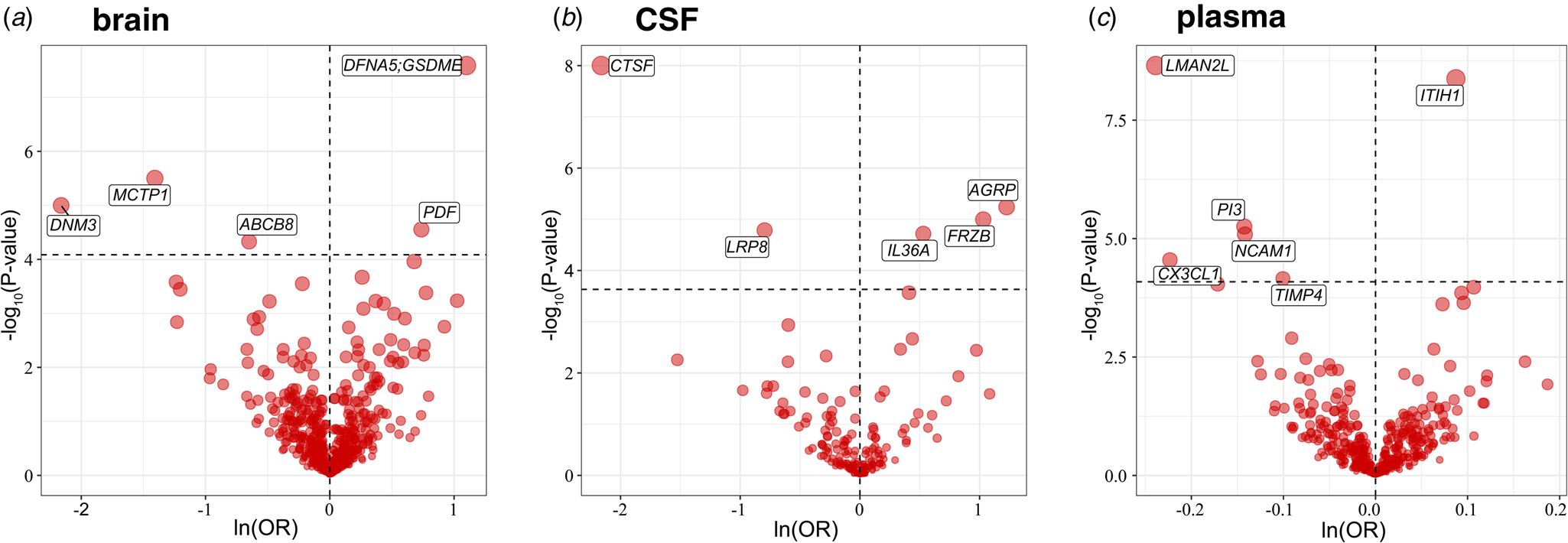
Figure 2. Volcano plot of the MR analysis results for proteins of three tissues. The horizontal dashed line indicates the diverse Bonferroni significance threshold (p < 8.22 × 10−5 in the brain, p < 2.34 × 10−4 in CSF, and p < 8.17 × 10−5 in plasma). The vertical dashed lines distinguish the effect direction. The proteins that passed the Bonferroni significance threshold are labeled with their names.
Table 1. MR, Steiger filtering analysis and Bayesian colocalization analysis results for proteins significantly associated with BD
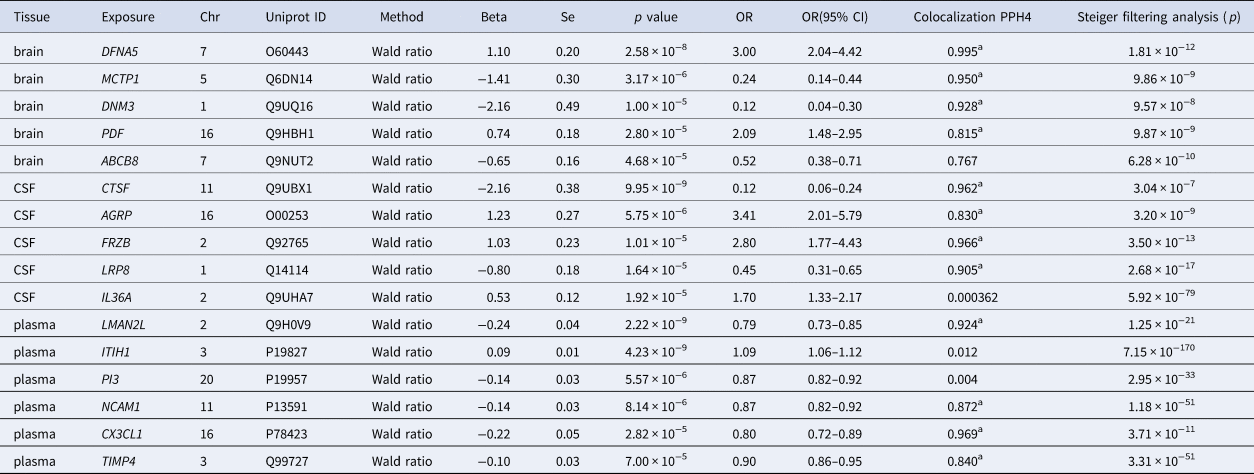
All SNPs used were cis-acting. Abbreviation: MR, Mendelian randomization; CSF, cerebrospinal fluid; BD, bipolar disorder; SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval.
a The results of PPH4 are over 0.80, which indicates that a genetic variant is shared by both traits of the protein level and BD.
Sensitivity analysis for causal proteins
Although the paucity of IVs constrained the possible sensitivity analyses, we verified the directionality of the causal effects via Steiger filtering. All the proteins detected by MR satisfied the Steiger filtering criterion (Table 1), implying that the genetic variants had stronger associations with the exposure than with the outcome and were suitable for instrumental variables to avoid reverse causality.
Bayesian colocalization analysis of BD causal proteins
To confirm whether the associations between BD and pQTLs discovered by MR analysis were driven by a shared causal SNP, we performed Bayesian colocalization analysis, and the results are shown in Table 1. In the brain, DFNA5, MCTP1, DNM3, and PDF had the same genetic variants associated with both BD and protein abundance. In CSF, CTSF, AGRP, FRZB, and LRP8 passed the Bayesian colocalization analysis, while LMAN2L, NCAM1, CX3CL1, and TIMP4 were successfully validated in plasma.
Phenome-wide MR analysis of BD prior druggable genes
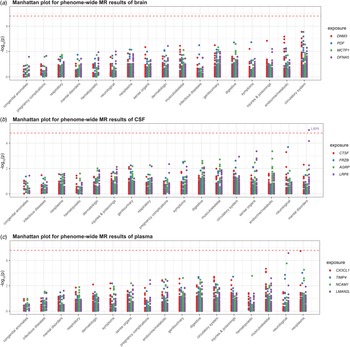
We conducted phenome-wide MR analysis of 783 non-BD diseases or traits in the UK Biobank to comprehensively characterize the side effect profiles of drug targets. Using the Wald ratio method, we detected no significant association for any potential drug target except for LRP8 (p < 1.60 × 10−5, 0.05/(4 × 783)) (Fig. 3). Decreased LRP8 expression was a risk factor for schizophrenia and other psychotic disorders (OR 0.013, 95% CI 0.002–0.091; p = 9.28 × 10−6).
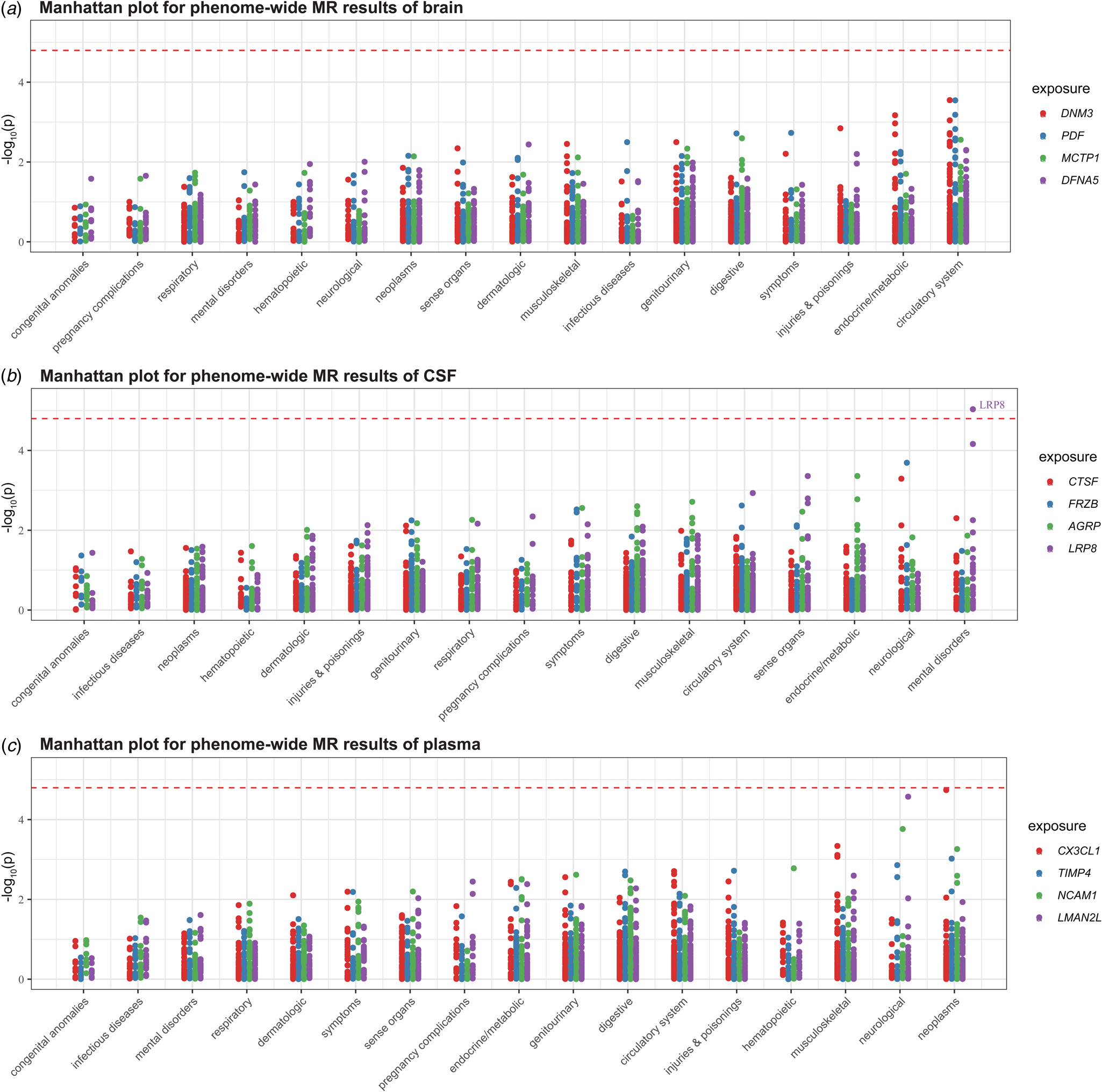
Figure 3. Manhattan plot for phenome-wide MR results of three tissues. To enhance visualization, the y-axis shows −log10(p). A dot represents a disease trait, and different colors represent the MR results of different proteins. Only Bonferroni-significant disease associations are illustrated (p < 0.05/(4 × 783) = 1.60 × 10−5).
External validation of BD causal proteins
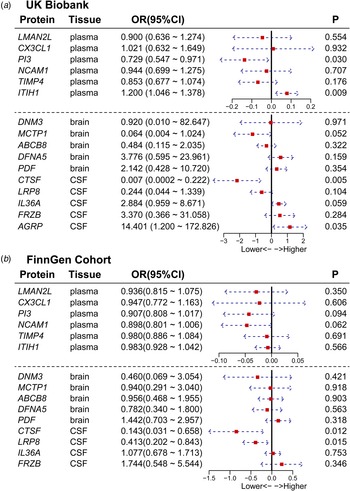
We conducted a replication two-sample MR analysis utilizing the same-variant and significant-variant strategies for the UK Biobank and FinnGen cohorts, which included sufficient data to perform two-sample MR analysis for the 16 proteins we identified. For all 16 candidate proteins, the directional effects of 13 (81.25%) proteins were concordant between the primary and validation cohorts. Furthermore, the results for 5 proteins were replicated at p < 0.05 (Fig. 4a and 4b), among which CTSF was confirmed in both datasets. The results for other proteins were replicated in one dataset, such as AGRP, PI3, and ITIH1 in UK Biobank and LRP8 in FinnGen. From the perspective of tissues, LRP8, AGRP, CTSF in CSF; PI3, and ITIH1 in plasma showed strong evidence of replication (Table 2 and Fig. 4).
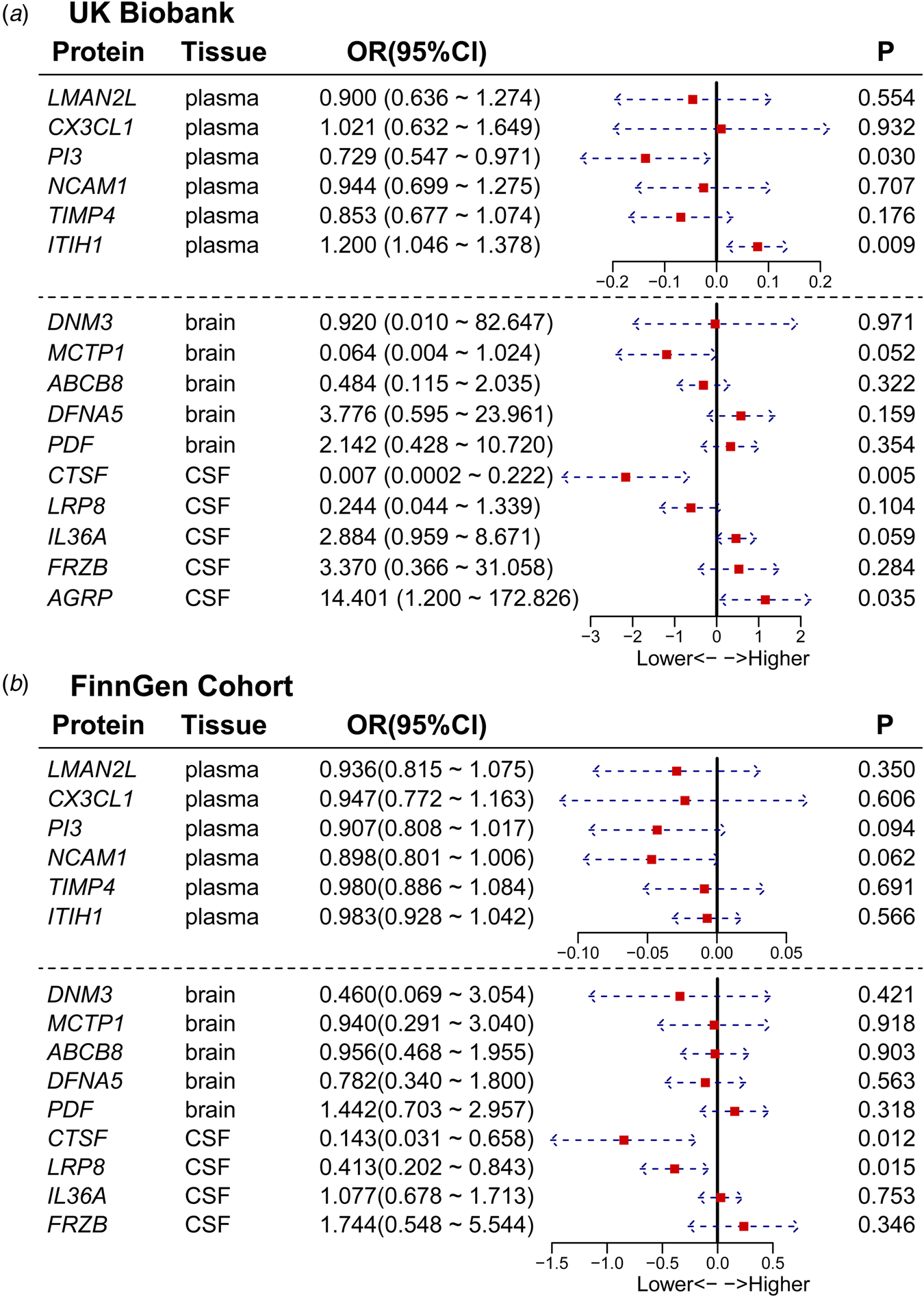
Figure 4. Forest plot of the results of external validation. The figure shows the effect size and 95% confidence interval of the MR analysis for proteins on BD risk. To facilitate the visualization of the OR forest plot results, we log10-transformed the OR values.
Table 2. Summary results of five-step stragety of BD associated genes
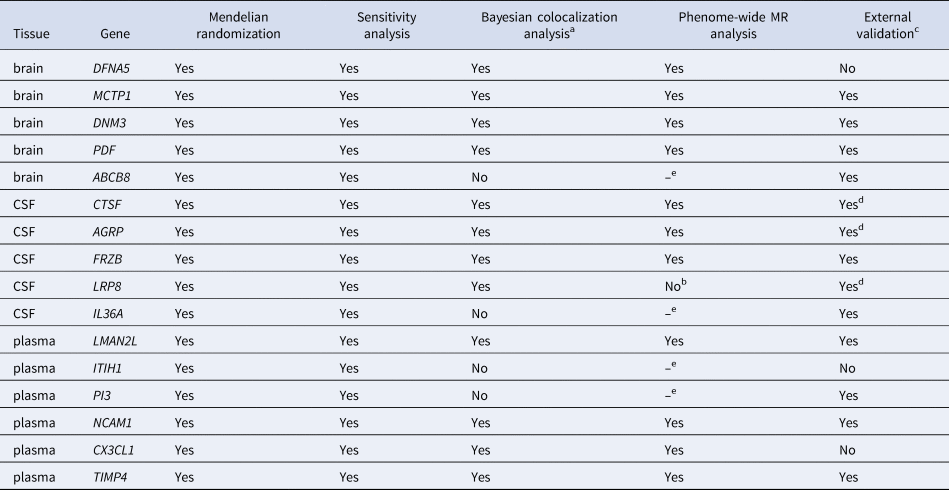
Abbreviation: MR, Mendelian randomization; BD, bipolar disorder.
a Values above 0.80 were considered as strong evidence of colocalization.
b Potential drugs targeting LRP8 showed a potential curable effect on schizophrenia and other psychotic disorders.
c The consistent directionality of OR values between primary and validation cohorts were considered as successful replication.
d These proteins passed Bayesian colocalization analysis and were not only consistent in the directionality of OR values between primary and both validation cohorts, but also had p values less than 0.05 in at least one dataset, which were considered as strong evidence of replication.
e The phenome-wide MR analysis was limited to proteins exhibiting strong colocalization.
Discussion
In the present study, we employed a five-step strategy that leveraged the strengths of various analyses to integrate proteomic data with BD GWAS and identified a total of 9 proteins that confer BD risk, including upregulated PDF, downregulated MCTP1 and DNM3 in the brain; upregulated AGRP and FRZB, downregulated CTSF in CSF; and downregulated LMAN2L, NCAM1, TIMP4 in plasma. Risk proteins in plasma and CSF may be able to aid in the diagnosis of BD and the development of drugs targeting peripheral body fluid due to the availability of samples during the clinical process, while brain-specific risk proteins may be used to determine the molecular mechanisms of BD because of their presence in in situ lesions. Phenome-wide MR analysis was used to examine the safety and tolerability of the identified protein targets. Moreover, to explore their potential as clinical drug targets, we collected data on current medications targeting these causal proteins from the DRUGBANK database (https://go.drugbank.com/) (online Supplementary Table S4).
In brain tissue, we identified upregulated PDF and downregulated MCTP1 and DNM3 as risk factors for BD, among which PDF and DNM3 were reported for the first time in this disease. PDF is a peptide release factor that regulates mitochondrial protein synthesis by removing the N-terminal formyl group (Bögeholz, Mercier, Wintermeyer, & Rodnina, Reference Bögeholz, Mercier, Wintermeyer and Rodnina2021). Impaired mitochondrial pathways are an important hallmark of BD patients (Cuperfain, Zhang, Kennedy, & Goncalves, Reference Cuperfain, Zhang, Kennedy and Goncalves2018). However, the association between mental disorders and PDF has not been investigated, and further studies are warranted to elucidate its mechanism. DNM3 encodes a dynamin protein involved in synaptic vesicle endocytosis (Raimondi et al., Reference Raimondi, Ferguson, Lou, Armbruster, Paradise, Giovedi and De Camilli2011). A recent exon-focused GWAS reported the association of DNM3 with obsessive-compulsive disorder and schizophrenia (Costas et al., Reference Costas, Carrera, Alonso, Gurriarán, Segalàs, Real and Carracedo2016), suggesting that this gene may cause cross-disorder risk. The BD risk gene MCTP1 (Scott et al., Reference Scott, Muglia, Kong, Guan, Flickinger, Upmanyu and Boehnke2009) is a transmembrane protein that regulates calcium ion binding activity. Basic research has shown that dysregulation of MCTP1 might cause altered synaptic vesicle recycling and oxidative stress resulting from glutamate toxicity (Qiu, Yu, & Liang, Reference Qiu, Yu and Liang2015). This evidence supports its connection with BD risk. Interestingly, both MCTP1 and DNM3 are involved in the regulation of synaptic vesicles, and the dysregulation of synaptic proteins in the DLPFC has been extensively discussed in BD patients (Aryal et al., Reference Aryal, Bonanno, Song, Mani, Keshishian, Carr and Dejanovic2023). Therefore, mitochondrial pathways and synaptic vesicle-related pathways are important directions for future research on the molecular mechanisms of BD pathogenesis.
For cerebrospinal fluid, we found that upregulated AGRP and FRZB, downregulated CTSF increased BD risk. AGRP, a biased agonist of melanocortin receptors coexpressed with neuropeptide Y and gamma-aminobutyric acid (GABA) in the hypothalamus, mediates neuroendocrine responses to immune (Boutagouga Boudjadja et al., Reference Boutagouga Boudjadja, Culotta, De Paula, Harno, Hunter, Cavalcanti-de-Albuquerque and D'Agostino2022) and inflammatory regulation (Klima et al., Reference Klima, Kruger, Goldstein, Pulido, Low, Assenmacher and Betley2023; Xiao, Xia-Zhang, Vulliemoz, Ferin, & Wardlaw, Reference Xiao, Xia-Zhang, Vulliemoz, Ferin and Wardlaw2003) by AGRP neurons. A previous case‒control study reported that AGRP levels were higher in euthymic bipolar disorder patients than in healthy controls (Özkorumak Karagüzel et al., Reference Özkorumak Karagüzel, Kural, Tiryaki, Keleş Altun, Özer and Civil Arslan2018), which is consistent with our findings. Currently, there are no drugs targeting AGRP (online Supplementary Table S4). Hence, drug development targeting AGRP may be promising, and future studies are needed to clarify the specific pathogenic mechanism and the potential of using AGRP as a clinical drug target. FRZB is a known secreted Wnt antagonist (Leyns, Bouwmeester, Kim, Piccolo, & De Robertis, Reference Leyns, Bouwmeester, Kim, Piccolo and De Robertis1997), and the Wnt signaling pathway is closely involved in the microenvironment of the central nervous system by regulating blood‒brain barrier homeostasis (Liebner, Dijkhuizen, Reiss, Plate, & Agalliu, Reference Liebner, Dijkhuizen, Reiss, Plate and Agalliu2018). CTSF is a cysteine protease that is involved in the lysosomal protein degradation system (Turk, Turk, & Turk, Reference Turk, Turk and Turk2000). A lack of CTSF causes lysosomal substrate degradation disorders, leading to neurological lysosomal storage diseases (Berkovic et al., Reference Berkovic, Oliver, Canafoglia, Krieger, Damiano, Hildebrand and Carpenter2019; Tang et al., Reference Tang, Lee, Galvez, Robillard, Mole and Chapman2006). A previous GWAS reported that CTSF was located in the linkage disequilibrium region of the BD risk locus rs10896135, with an OR of 0.89 (Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011), which is consistent with our findings that downregulated CTSF may confer BD risk. Notably, strong evidence of CTSF replication was obtained in two different independent validation cohorts, suggesting that CTSF is a high-confidence risk gene for BD. We found that proteins dysregulated in CSF mainly affect the homeostasis of the nervous system and neuroendocrine system, which not only suggests the importance of body fluid to internal homeostasis but also reveals the value of these proteins as biomarkers of disease.
In plasma, downregulation of BD risk genes LMAN2L (Chen et al., Reference Chen, Jiang, Akula, Shugart, Wendland, Steele and Strauss2013; Lim et al., Reference Lim, Zain, Reynolds, Zain, Roffeei, Zainal and Mohamed2014; Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011), NCAM1 (Arai et al., Reference Arai, Itokawa, Yamada, Toyota, Arai, Haga and Yoshikawa2004; Atz, Rollins, & Vawter, Reference Atz, Rollins and Vawter2007; Ayalew et al., Reference Ayalew, Le-Niculescu, Levey, Jain, Changala, Patel and Niculescu2012), and TIMP4 (Lu et al., Reference Lu, Forgetta, Greenwood, Zhou and Richards2023) results in increased disease risk. NCAM1, also known as CD56, is a neural cell adhesion molecule involved in synaptic plasticity, neurodevelopment, and neurogenesis (Kiss & Muller, Reference Kiss and Muller2001). It regulates inflammatory cascades mediated by mitogen-activated protein kinase (MAPK) (Krushel, Tai, Cunningham, Edelman, & Crossin, Reference Krushel, Tai, Cunningham, Edelman and Crossin1998) and nuclear factor-κB (NF − κB) (Krushel, Cunningham, Edelman, & Crossin, Reference Krushel, Cunningham, Edelman and Crossin1999) that are activated by neuroinflammatory environments. A clinical study revealed that the level of NCAM1 in whole blood is related to the severity of BD (Jesudas, Nandeesha, Menon, & Allimuthu, Reference Jesudas, Nandeesha, Menon and Allimuthu2020). NCAM1 is also considered a common causative gene of schizophrenia and BD (Ayalew et al., Reference Ayalew, Le-Niculescu, Levey, Jain, Changala, Patel and Niculescu2012), and a recent genome-wide pleiotropic analysis identified NCAM1 as a shared potential pleiotropic locus in gastrointestinal diseases and psychiatric diseases, including BD (Gong et al., Reference Gong, Guo, Li, Liu, Yan, Liu and Yuan2023). These findings highlight the outstanding potential of NCAM1 as a disease marker. TIMP4 is an important inhibitor of matrix metalloproteinases, and the latter is closely related to blood‒brain barrier damage caused by neuroinflammation (Han & Jiang, Reference Han and Jiang2021). A previous MR study proposed that an increase in the genetically predicted circulating TIMP4 level was associated with a reduced risk of BD (minimum OR 0.88, 95% CI 0.82–0.94) (Lu et al., Reference Lu, Forgetta, Greenwood, Zhou and Richards2023) LMAN2L encodes a lectin mannose-binding protein that mediates the trafficking and secretion of glycoproteins in the endoplasmic reticulum. LMAN2L may promote the trafficking of neuroreceptors under endoplasmic reticulum stress conditions (Fu, Zhang, & Mu, Reference Fu, Zhang and Mu2019; Qin et al., Reference Qin, Kawasaki, Hu, Tozawa, Matsumoto and Yamamoto2012). Similar to those in CSF, dysregulated proteins in plasma are mostly related to homeostasis imbalances caused by inflammatory stimuli and endoplasmic reticulum stress. These studies corroborated our conclusion that circulating protein levels have the potential to be an auxiliary diagnostic instrument for disease.
We acknowledge some limitations of our study. First, the proteins we identified in different tissues were distinct, which may be attributed to the following reasons: (1) pQTLs are specific to different tissues. (2) Due to the restriction of sample sources in the original studies, the MR analysis included limited number of pQTLs. However, we still found that CTSF and AGRP, which were identified in CSF, showed a marginal causal effect in plasma, and their effect directions in both tissues were concordant. Second, our analysis was mainly based on data from European populations, due to the lack of ethnic diversity in the proteomic samples and may not be applicable to other ethnic groups. In addition, post-MR analysis methods such as Cochran's Q test and MR-Egger intercept test were not performed due to the limitation of IVs. Additional relevant analyses are still needed to validate the associations between risk genes and BD, and other post-GWAS analyses, such as proteome-wide association studies (Brandes, Linial, & Linial, Reference Brandes, Linial and Linial2020), are also good methodologies. Finally, the causal mechanisms of most of these candidate proteins in the pathogenesis of BD are still unclear, and biological validation is required to confirm our findings.
This study has several strengths. Firstly, we focused on pQTLs from 3 different tissue sources, the advantages of which could complement each other and have varied clinical significance (Luykx et al., Reference Luykx, Bakker, Visser, Verhoeven-Duif, Buizer-Voskamp, den Heijer and Ophoff2015; Yang et al., Reference Yang, Farias, Ibanez, Suhy, Sadler, Fernandez and Cruchaga2021). Risk genes found in the brain may be related to disease pathogenesis, while risk genes in CSF and plasma may serve as diagnostic and prognostic markers. Secondly, the phenome-wide MR analysis results showed that most of the risk genes we identified were promising for drug development and did not affect other vital systems or organs, increasing the application value of our findings. Interestingly, several studies have identified LRP8 as a common risk gene for schizophrenia and BD (Li et al., Reference Li, Huang, Grigoroiu-Serbanescu, Bergen, Landen and Hultman2016; Xiao et al., Reference Xiao, Yu, Li, Wang, Li, Chang and Li2020). The effect direction of the risk gene LRP8 on schizophrenia found in our phenome-wide MR analyses was consistent with that for BD, suggesting that drugs developed against this target may have potential curative effects in both diseases. Finally, the introduction of external validation cohorts confirmed the consistency of the effect directions of most risk genes, which increased the reliability of our conclusions. Intervention targeting these genes to increase (or decrease) their protein abundance is expected to become an important method for the treatment of BD in the future.
Conclusions
In conclusion, we identified nine candidate druggable proteins for BD, including MCTP1, DNM3, and PDF in the brain; CTSF, AGRP, and FRZB in CSF; and LMAN2L, NCAM1, and TIMP4 in plasma. Our study provides novel insights into the molecular mechanisms of BD and highlights promising candidate proteins for therapeutic interventions.
Abbreviations
BD: bipolar disorder; CSF: cerebrospinal fluid; GWAS: genome-wide association studies; pQTLs: protein quantitative trait loci; CI: confidence interval; IVW: inverse variance weighted; LD: linkage disequilibrium; MR: Mendelian randomization; OR: odds ratio; SNP: single nucleotide polymorphism; DNM3: dynamin 3; MCTP1: multiple C2 and transmembrane domain containing 1; ABCB8: ATP binding cassette subfamily B member 8; DFNA5: deafness, autosomal dominant 5; PDF: peptide deformylase; CTSF: cathepsin F; LRP8: LDL receptor related protein 8; IL36A: interleukin 36 alpha; FRZB: frizzled related protein; AGRP: agouti related neuropeptide; LMAN2L: lectin, mannose binding 2 like; CX3CL1: C-X3-C motif chemokine ligand 1; PI3: peptidase inhibitor 3; NCAM1: neural cell adhesion molecule 1; TIMP4: TIMP metallopeptidase inhibitor 4; ITIH1: inter-alpha-trypsin inhibitor heavy chain H1
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0033291724001077.
Acknowledgements
We would like to thank the authors of the original GWAS and pQTL studies included in this article. All the authors are grateful for their participation in our research.
Funding statement
This work was partly funded by National Nature Science Foundation of China Project (82001413); Key R & D projects of Science and Technology Department of Sichuan Province (2021YFS0248); Postdoctoral Foundation of West China Hospital (2020HXBH163).
Competing interests
The authors declare no competing interests or financial relationships with commercial interests.
Ethical standards
The present study is a secondary analysis of publicly available data. Ethical approval was granted for each of the original GWASs. In addition, no individual-level data were used in this study. Therefore, no new ethical review board approval was needed.