Major colonisation of the intestinal microbiota starts at delivery when an infant comes into contact with microbes from the extrauterine environment. The microbial community co-evolves with its human host and after reaching the full complexity, outnumbers human cells by one or more orders of magnitude( Reference Zoetendal, Rajilić-Stojanović and de Vos 1 , Reference Scholtens, Oozeer and Martin 2 ). The collective genome of these microbes (also called microbiome) contributes significantly to our genetic coding capacity as they are found to contain over 3 million genes( Reference Qin 3 ). Unlike our own genome, the intestinal microbiome is not strictly vertically inherited. Moreover, this personalised ‘organ’ may be modified by a variety of foods, food components and pharmaceutical treatments targeting microbiota composition, stability and activity. While developing rapidly in diversity and complexity in the first years following birth, the intestinal microbiota becomes relatively stable in adulthood. A recent longitudinal study showed that the subject-specificity of the adult intestinal microbiota was maintained for over 10 years( Reference Rajilić-Stojanović, Heilig and Tims 4 ). This long-term stability provides further support for the cross-talk between the human host and microbiota( Reference Rajilić-Stojanović 5 ).
A series of changes in life style, such as improved hygienic conditions, reduced contact with companion or farm animals and a higher frequency of caesarean deliveries as well as expanded broad spectrum antibiotic use and dietary habits of both the infant and mother affect the intestinal colonisation process( Reference Rook, Raison and Lowry 6 ) (Fig. 1). Many of the factors described earlier are typical for an urban life style and are likely to lead to decreased exposure to microbes from the natural environment. Such exposure is thought to be essential both for the development of balanced microbiota diversity and composition and the optimal maturation of immune system( Reference Rook, Raison and Lowry 6 ), thus having lasting impact on later life health.
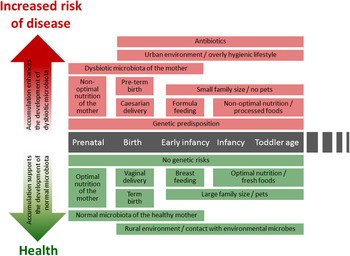
Fig. 1. (colour online) Modern life style factors associated with the development of intestinal microbiota and later life health.
Prenatal exposure to microbes and microbial compounds
Traditionally the fetus has been thought to be microbiologically sterile before birth. The presence of microbes in the amniotic fluid and placenta has mainly been associated with preterm deliveries due to maternal intrauterine infections and other pathological conditions( Reference Pararas, Skevaki and Kafetzis 7 – Reference DiGiulio, Romero and Amogan 9 ) and the presence of bacterial DNA in amniotic fluid has been associated with lower gestational age and with low mean birth weight( Reference DiGiulio, Romero and Amogan 9 , Reference DiGiulio, Romero and Kusanovic 10 ). However, recent studies utilising molecular methods have shown that DNA of non-pathogenic bacteria can be detected in placenta and amniotic fluid samples in normal conditions( Reference Satokari, Grönroos and Laitinen 11 , Reference Rautava, Collado and Salminen 12 ). Hence, ingestion of amniotic fluid during pregnancy continuously exposes the fetus to bacteria and/or microbe-associated molecular patterns (MAMP). The exact mechanism(s) of bacterial entry into the intrauterine environment remains elusive. However, ascension from the vagina, by retrograde spread from abdominal cavity, haematogeneously through placenta and contamination during medical procedures (such as amniocentesis) have been suggested as potential routes( Reference Goldenberg, Culhane and Iams 13 ). Also MAMP can induce the immunostimulatory effects, for example via the stimulation of Toll-like receptors (TLR), without the need for microbial cells to enter the amniotic cavity. This is supported by the study by Rautava et al. where the presence of bacterial DNA, indicative for the presence of other MAMP too, in placenta and amniotic fluid was associated with the induction of expression profiles of TLR, especially TLR2 and TLR5 in fetal intestine( Reference Rautava, Collado and Salminen 12 ).
Furthermore, the expression of different TLR, including TLR9, has been shown to change during the maturation of gut epithelial cells( Reference Nanthakumar, Meng and Goldstein 14 , Reference Gribar, Sodhi and Richardson 15 ). TLR9 recognises unmethylated CpG motifs in bacterial DNA and its signalling maintains the gut epithelial homeostasis by improving the barrier functions and by inducing tolerance towards other MAMP( Reference Kant, de Vos and Palva 16 ). In utero the intestinal expression of TLR9 of mouse embryos decreases from days 14 to 18 and then increases again during the postnatal period( Reference Gribar, Sodhi and Richardson 15 ). Thus, it appears that a full-term newborn is programmed to receive TLR9 stimulation, which will improve the tolerance towards commensal bacteria. Consistently with this, necrotising enterocolitis in preterm infants has been associated with decreased TLR9 and increased TLR4 expression of the intestinal epithelium( Reference Gribar, Sodhi and Richardson 15 ). Remarkably, TLR9 activation via CpG-DNA supplementation significantly reduced necrotising enterocolitis severity( Reference Gribar, Sodhi and Richardson 15 ) suggesting that microbiota rich in CpG motifs but poor in TLR4 ligands, such as lipopolysaccharide-carrying Gram-negative bacteria could be optimal for the prevention or alleviation of necrotising enterocolitis. In this regard human breast milk, which supports the growth of Bifidobacteria, organisms with high-guanine–cytosine content genomes that are especially rich in CpG-motifs, appears to be a ‘superfood’ for the newborns. A number of strains of commercially produced lactobacilli have also been found to be rich in CpG-DNA( Reference Kant, de Vos and Palva 16 ) and probiotic interventions have shown some promising results in the prevention and alleviation of necrotising enterocolitis ( Reference Downard, Renaud and St Peter 17 ). In a mouse model, TLR9 signalling was indeed observed to be an essential mediator of anti-inflammatory effects of probiotics( Reference Rachmilewitz, Katakura and Karmeli 18 ). Furthermore, DNA of Bifidobacterium and Lactobacillus spp., both rich in CpG motifs, have been found in human placenta( Reference Satokari, Grönroos and Laitinen 11 ). Thus, it seems that prenatal exposure to MAMP is an important step in programming the development of gut epithelium and immune system already in utero.
Early-life microbiota
Meconium is the very first faecal specimen produced by the infant after birth. It consists mainly of amniotic fluid but includes also mucus, intestinal epithelial cells and concentrate of metabolites such as bile acids and pancreatic secretions( Reference Kumagai, Kimura and Takei 19 ). Several reports have described meconium microbiota composition providing further evidence for the suggestion that microbiological colonisation may begin already in utero ( Reference Moles, Gomez and Heilig 20 – Reference Gosalbes, Llop and Valles 23 ).
Bacteria belonging to four major bacterial phyla in the intestine, Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria, are already detectable in the meconium. The predominant cultured bacteria seem to be bacilli within the Firmicutes phylum such as enterococci and staphylococci, or certain Proteobacteria such as Esherichia coli, Klebsiella and Enterobacter spp.( Reference Moles, Gomez and Heilig 20 – Reference Hu, Nomura and Bashir 24 ). This is in agreement with the reports that these facultative anaerobes are present in faeces of healthy newborn infants( Reference Adlerberth and Wold 25 , Reference Palmer, Bik and Digiulio 26 ). In addition, Enterococcus spp. are commonly present, ~40 and 50 % of infants colonised at day 3, respectively( Reference Adlerberth and Wold 25 ). Following the colonisation of facultative bacteria, anaerobic bacteria appear in the infant faeces within the first weeks of life, decreasing the abundance of facultative anaerobes and thus introducing a shift in microbiota community structure( Reference Avershina, Storro and Oien 27 ). It should be noted, however, that this shift may represent an outgrowth of specific groups of bacteria and does not preclude the fact that their colonisation might have already occurred at low level. Especially the abundance of Bifidobacteria increases rapidly from ~3·5–10 % in meconium( Reference Moles, Gomez and Heilig 20 , Reference Jimenez, Marin and Martin 22 , Reference de Weerth, Fuentes and Puylaert 28 ) to 50–70 % and even up to 90 % in the faeces of breast-fed infants at ages 1 month and 3 months, respectively( Reference Fallani, Young and Scott 29 – Reference Roger and McCartney 32 ). However, large inter-individual variations are characteristic for infant microbiota and the abundance of Bifidobacteria varies from 5 to 100 % in breast-fed infants( Reference Roger and McCartney 32 ). Considering formula-fed infants, Bifidobacteria (determined by fluorescent in situ hybridisation) may form a minor part of the microbiota, constituting ~25 % of the total microbiota( Reference Roger and McCartney 32 ). In addition to the individual variation, the Bifidobacterial abundance seems to vary greatly according to the geographic origin; infants from (northern) European countries harbour in general high numbers of Bifidobacteria( Reference Turroni, Peano and Pass 30 , Reference Roger and McCartney 32 , Reference Fallani, Amarri and Uusijärvi 33 ), whereas these bacteria are less predominant in Asian and American infants( Reference Palmer, Bik and Digiulio 26 , Reference Fan, Huo and Li 34 , Reference Koenig, Spor and Scalfone 35 ). This observation can be mainly explained by demographic differences and by differences in the rate and duration of breast-feeding between the countries and potentially also the differences in use of antibiotics.
After the introduction of solid foods and weaning the relative abundance of Bifidobacteria decreases gradually being ~60 % at 4 months, 25 % at 6 months and 10 % at 2 years( Reference Avershina, Storro and Oien 27 , Reference Nylund, Satokari and Nikkilä 36 , Reference Ringel-Kulka, Cheng and Ringel 37 ). Simultaneously, the relative abundance of lactobacilli decreases, whereas bacteria predominant in adult microbiota, such as Bacteroidetes and bacteria belonging to the Clostridium clusters XIVa and IV increase( Reference Roger and McCartney 32 , Reference Koenig, Spor and Scalfone 35 – Reference Ringel-Kulka, Cheng and Ringel 37 ). However, the early-life microbiota composition is characterised by high inter-individual variation( Reference Avershina, Storro and Oien 27 , Reference Nylund, Satokari and Nikkilä 36 , Reference Avershina, Storro and Oien 38 ). The bacterial abundances in healthy infant microbiota vary greatly in a subject-wise manner and fluctuate further in response to the changes in different life events such as antibiotic treatments and introduction of solid foods( Reference Palmer, Bik and Digiulio 26 , Reference Koenig, Spor and Scalfone 35 ) (Fig. 1). During and after weaning major changes occur in microbiota diversity and composition, this transitional phase being more pronounced in breast-fed than in formula-fed infants( Reference Roger and McCartney 32 ). The succession of Bacteroides spp. and bacteria belonging to the Clostridium clusters XIVa and IV proceeds rapidly while the relative proportion of Bifidobacteria decreases( Reference Roger and McCartney 32 , Reference Koenig, Spor and Scalfone 35 ).
Previously, it has been suggested that the microbiota diversity and composition stabilise and reach the level of adult microbiota within the first year or two( Reference Palmer, Bik and Digiulio 26 , Reference Mackie, Sghir and Gaskins 39 ). However, recent studies have shown that microbiota maturation will continue longer( Reference Nylund, Satokari and Nikkilä 36 , Reference Ringel-Kulka, Cheng and Ringel 37 , Reference Agans, Rigsbee and Kenche 40 ). Interestingly, the establishment of bacteria belonging to Clostridium cluster XIVa at a level similar to adults has been observed already in young children (age 1–4 years)( Reference Ringel-Kulka, Cheng and Ringel 37 ), while other bacterial groups still remain at low-level abundance. This indicates that the microbiota development is a gradual process, where some bacterial groups may reach the degree of stabilisation earlier than others. However, considering the major physiological changes taking place in the human body within childhood and adolescence it may be argued that the development of the intestinal microbiota continues throughout this time period and is not finished until the human host reaches adulthood. The first studies on adolescent microbiota also point to this direction as they reported significant differences between the microbiota composition of adolescent children (age 11–18 years) and adults, the most striking difference being the almost 2-fold higher abundance of Bifidobacteria in adolescent subjects (9 v. 5·5 % of total microbiota, respectively)( Reference Agans, Rigsbee and Kenche 40 ). However, in order to comprehensively understand the microbiota development and stabilisation, more longitudinal studies analysing time-series samples from the same individuals over a long-time period are needed.
Effect of breast milk on microbiota composition
Human milk oligosaccharides (HMO) have an essential role in the promotion of the development of normal physiology of intestine and immune system in infants. Human milk contains a complex mixture of oligosaccharides, their exact composition varying according to different extrinsic and intrinsic factors. These factors include the genetic background of the mother, maternal health status, diet, secretor status and Lewis blood group type( Reference Bode 41 – Reference Albrecht, Schols and van den Heuvel 43 ). Oligosaccharide molecules participate in the maintenance of a healthy gut microbiota in three ways. (1) They block the colonisation of pathogenic bacteria by acting as receptor analogues and binding to the bacterial surface, thus preventing the pathogens from binding to their target oligosaccharides on the epithelial cell surface( Reference Zivkovic, German and Lebrilla 44 ). (2) They act as prebiotic substrates promoting the growth of beneficial bacteria, notably Bifidobacteria, concurrently preventing the adherence of potentially harmful bacteria via colonisation resistance( Reference Bode 41 ). (3) They have also been suggested to modulate intestinal epithelial cells, lymphocyte cytokine production and leucocyte rolling and adhesion (comprehensively reviewed by Bode( Reference Bode 41 )).
Human infants lack the extensive set of enzymes needed for the digestion of glycan residues of HMO. Thus, these molecules pass undigested to the lower part of the intestinal tract, where they can be consumed by the specific members of infant gut microbiota( Reference Marcobal and Sonnenburg 45 ). Since a wide repertoire of enzymes are needed for the degradation and utilisation of the intricate structures of both HMO and plant polysaccharides, such processes most likely involve several different commensal bacteria acting synergistically. The two major bacterial genera described to have the capability for milk oligosaccharide utilisation are Bifidobacterium spp. and Bacteroides spp. Bifidobacteria, such as Bifidobacterium longum subsp. infantis and Bifidobacterium bifidum, typically abundant in infant microbiota, harbour a complex set of genes specifically related to HMO utilisation( Reference Sela, Garrido and Lerno 46 ). The B. longum subsp. infantis genome harbours entire gene clusters controlling the expression of glycosidases, membrane-spanning transporters and other proteins dedicated to human milk oligosaccharide utilisation( Reference Sela, Garrido and Lerno 46 , Reference Sela, Chapman and Adeuya 47 ). In contrast, B. longum subsp. longum, which is more abundant in adult microbiota, is unable to use diverse HMO, but has the capability to utilise short-chain oligosaccharides( Reference Sela, Garrido and Lerno 46 ). However, HMO have reported to up-regulate the expression of several pathways in B. longum subsp. Longum, such as genes involved in carbohydrate degradation and cell adherence( Reference Gonzalez, Klaassens and Malinen 48 ). Possibly, B. longum subsp. longum relies on cross-feeding with other bacteria, which first degrade complex polysaccharides to shorter units and thereby can also use HMO as a nutrient source.
Bacteroides spp. genomes harbour a specific gene cluster termed polysaccharide utilisation loci, enabling a wide range of saccharolytic ability( Reference Marcobal and Sonnenburg 45 , Reference Martens, Lowe and Chiang 49 ). For example, Bacteroides thetaiotaomicron can degrade more than a dozen different types of glycans( Reference Marcobal and Sonnenburg 45 ), most likely also HMO. In addition, in vitro utilisation of HMO by Bacteroides fragilis and Bacteroides vulgatus has been reported( Reference Marcobal and Sonnenburg 45 ). Consistently, B. fragilis and B. vulgatus are the predominant Bacteroides spp. found in breast-fed infants( Reference Tannock, Lawley and Munro 50 ). The abundance of bacterial groups, which have restricted capacity to utilise different polysaccharide compounds, are likely to fluctuate more in response to the type of incoming carbohydrates, whereas bacteria with a wide glycan-degrading capability may have a competitive advantage in the gut. Bacteroides spp. are among the first groups colonising the gut( Reference Palmer, Bik and Digiulio 26 , Reference Penders, Stobberingh and Thijs 51 ), increase further after the introduction of solid food and weaning( Reference Roger and McCartney 32 , Reference Koenig, Spor and Scalfone 35 ) and are part of the common core microbiota in adults( Reference Qin 3 , Reference Rajilić-Stojanović, Heilig and Molenaar 52 , Reference Huse, Ye and Zhou 53 ). Moreover, the ability of Bacteroides spp. to switch substrate specificity in response to the changing ingestion of nutrients indicates that they are adapted to the symbiotic life with human host and are permanent colonisers of the gut.
Human milk is also a source of bacteria to the infant. The predominant bacteria observed in human milk samples are Bacilli, such as Streptococcus spp. and Staphylococcus spp.( Reference Hunt, Foster and Forney 54 , Reference Jost, Lacroix and Braegger 55 ). In addition, Bifidobacterium spp. are present and Bacteroidetes and specific Clostridia such as butyrate-producing bacteria Faecalibacterium and Roseburia spp. have been detected( Reference Hunt, Foster and Forney 54 , Reference Jost, Lacroix and Braegger 56 , Reference Martin, Jimenez and Heilig 57 ). The bacterial composition of breast milk varies depending on the genetic background, maternal dietary habits and demographic differences between the mothers. For example, European mothers commonly harbour Lactobacillus and Bifidobacterium spp. in their breast milk, whereas these bacteria were rarely detected in mothers from the USA, possibly as a result of technical differences and drawbacks in DNA extraction( Reference Jost, Lacroix and Braegger 55 , Reference Jost, Lacroix and Braegger 56 , Reference Ward, Hosid and Ioshikhes 58 ). Furthermore, the mode of delivery has been shown to affect the milk microbiota composition( Reference Cabrera-Rubio, Collado and Laitinen 59 ). Milk samples from mothers who delivered their infants vaginally contained more Leuconostocaceae and less Carnobacteriaceae than milk samples from mothers who had gone through an elective caesarean section.
Maternal health status seems to have a major effect on the milk microbiota composition. For example, milk microbiota of overweight mothers differs from that of normal weight mothers( Reference Cabrera-Rubio, Collado and Laitinen 59 , Reference Collado, Laitinen and Salminen 60 ). The bacterial composition of breast milk seems to be stable at intra-individual level over time, while representing a great inter-individual variation. This suggests that human milk microbiota is highly personalised, in a manner similar to intestinal microbiota( Reference Costello, Lauber and Hamady 61 – Reference Ursell, Clemente and Rideout 63 ).
Recently, the existence of a ‘core’ milk microbiota has been suggested( Reference Hunt, Foster and Forney 54 ). The milk core microbiota consisted of nine operational taxonomic units, corresponding to Staphylococcus, Streptococcus (Firmicutes), Corynebacterium, Propionibacterium (Actinobacteria) and Serratia, Pseudomonas, Ralstonia, Sphingomonas and Bradyrhizobiaceae (Proteobacteria), constituting approximately half the total bacterial community. It is noteworthy that many of the core microbiota genera are typically found from the skin and it seems likely that some part of the breast milk microbiota originates from the skin. Another origin of bacteria in human milk may be the intestinal tract of the mother. It has been suggested that intestinal bacteria could transfer within the phagocytosing cells from the gut to human milk via entero-mammary circulation of immune cells( Reference Grönlund, Gueimonde and Laitinen 64 ). Interestingly, Bifidobacterium breve is one of the most commonly detected Bifidobacterial species in human milk samples( Reference Martin, Jimenez and Heilig 57 , Reference Boesten, Schuren and Ben Amor 65 – Reference Solis, de Los Reyes-Gavilan and Fernandez 69 ) and it produces exopolysaccharide, which masks other surface antigens and presents an ability to remain immunologically ‘silent’( Reference Fanning, Hall and Cronin 70 ). The production of exopolysaccharide seems to be important for the persistence of B. breve in the gut( Reference Fanning, Hall and Cronin 70 ). Speculatively, exopolysaccharide may also play a role in the survival of this bacterium within immune cells, enabling its transfer via the enteromammary circulation route. Moreover, B. breve and other Bifidobacteria are known to produce specific pili that are assumed to play a role in colonisation( Reference O'Connell Motherway, Zomer and Leahy 71 ). While the origin of bacteria in human milk remains an open question, human milk bacteria should be considered as an important source of bacteria in the establishment of intestinal microbiota during early life (Fig. 1).
Perturbations in the development of microbiota diversity and composition
Antibiotic treatments
The administration of antibiotics has been considered the most remarkable extrinsic factor affecting the microbiota composition and development during early infancy and childhood (Fig. 1). The majority of antibiotics used to treat early-life infections have a rather broad-spectrum antimicrobial activity, inhibiting the growth of both pathogenic bacteria and the beneficial members of commensal microbiota. Especially in children, the major effect is the reduction of bacteria considered to have health-promoting properties such as Bifidobacterium spp. and Lactobacillus spp., which may have long-term effects on the infants’ health later in life.
After the antibiotic treatment, the overall microbiota diversity is generally decreased and the microbiota composition is often characterised by a dominance of a few bacterial groups( Reference Fouhy, Guinane and Hussey 72 , Reference Dethlefsen and Relman 73 ). In a recent study, the effect of early-life antibiotic treatment (ampicillin and gentamicin) on microbiota composition of newborn infants was analysed using a high-throughput sequencing( Reference Fouhy, Guinane and Hussey 72 ). The authors reported higher proportions of Proteobacteria and reduced abundances of genera Bifidobacterium and Lactobacillus in antibiotic-treated infants when compared with untreated controls 1 month after the cessation of treatment( Reference Fouhy, Guinane and Hussey 72 ). These findings are in line with previous observations considering the microbiota deviations after the administration of different types of antibiotics( Reference Fallani, Young and Scott 29 , Reference Rea, Dobson and O'Sullivan 74 ). Moreover, a declined prevalence of Bacteroidetes and higher abundances of enterobacteria and enterococci have been reported in antibiotic-treated infants when compared with healthy controls( Reference Fallani, Young and Scott 29 , Reference Fouhy, Guinane and Hussey 72 , Reference Tanaka, Kobayashi and Songjinda 75 ). In addition, a decreased abundance of Bifidobacteria have been reported in patients who have received antibiotics( Reference Hussey, Wall and Gruffman 76 , Reference Mangin, Leveque and Magne 77 ). However, the sensitivity for antibiotics seems to be a strain-specific feature and thus different Bifidobacterium species may be distinctly affected. In a study by Mangin et al. ( Reference Mangin, Suau and Gotteland 78 ), the total number of Bifidobacteria was not decreased after amoxicillin treatment for 7 d. Instead, the diversity of Bifidobacterium spp. population and a shift in species composition was observed. Specifically, a complete loss of Bifidobacterium adolescentis group and a decreased amount of B. bifidum were detected, whereas B. longum and B. catenulatum group bacteria were not affected( Reference Mangin, Suau and Gotteland 78 ). Thus, high diversity of Bifidobacterial species may protect the infant from more extensive effects of specific antibiotics.
The microbiota recovery begins shortly after the cessation of antibiotic administration but it seems to be rather slow and gradual process and the recovery remains often incomplete( Reference Dethlefsen and Relman 73 , Reference Jernberg, Lofmark and Edlund 79 , Reference Jakobsson, Jernberg and Andersson 80 ). In a recent study, the elevated amounts of Proteobacteria could still be detected 2 months after the termination of antibiotic medication, whereas Bifidobacteria and Lactobacilli abundances were more or less recovered( Reference Fouhy, Guinane and Hussey 72 ). The overgrowth of Proteobacteria, especially Enterobacteriaceae after the antibiotic treatment(s) has been widely reported. This effect can be explained by the competitive advantage obtained by the production of β-lactamases. These enzymes degrade the β-lactam antibiotic structure, thus providing resistance against several antibiotics such as amoxicillin, ampicillin and gentamicin.
Interestingly, early-life antibiotic treatment(s) have been associated with the increased risk for health problems later in life such as the risk for coeliac disease (CD) development( Reference Mårild, Ye and Lebwohl 81 ), allergic diseases( Reference Foliaki, Pearce and Björksten 82 ) and the increased risk of obesity at school age( Reference Ajslev, Andersen and Gamborg 83 ). Furthermore, prenatal exposure to antibiotics may have long-term effects on the health later in life, since maternal antibiotic use during pregnancy has been associated with an increased risk of cow's milk allergy, asthma, eczema and hay fever in their infants( Reference McKeever, Lewis and Smith 84 , Reference Metsälä, Lundqvist and Virta 85 ). However, only a limited number of studies utilising high-throughput analysis methods on the evaluation of microbiota diversity and composition after antibiotic therapies have been published. Further studies are required to assess the impact of antibiotics on host-microbe cross-talk and interactions. Furthermore, the long-term effects of antibiotics on both microbiota and on later health status of the paediatric patients need to be urgently assessed.
Colic
Colic crying is one of the most common problems in early life confronting ~10–25 % of otherwise healthy infants within the first months of life( Reference Pärtty, Luoto and Kalliomäki 86 , Reference Milidou, Sondergaard and Jensen 87 ). Colic cry is characterised by inexplicable, excessive crying >3 h/d for 3 d or more in 1 week, whereas it does not respond to any interventions such as feeding, diaper change or other solicitude procedures( Reference Wessel, Cobb and Jackson 88 ). Usually colic crying starts from 2-week-old to 3-month-old infants and declines after a few months. Although its aetiology and pathogenesis remain obscure, an association between colic cry and immaturity of intestinal function and/or neurodevelopmental maturity as well as excessive colonic gas production has been suggested( Reference Savino, Cordisco and Tarasco 89 , Reference Rhoads, Fatheree and Norori 90 ). In addition, an aberrant microbiota composition has been suggested to promote colicky symptoms. Microbiota diversity and stability have been observed to be lower in infants suffering from colic than in healthy control subjects( Reference de Weerth, Fuentes and Puylaert 28 , Reference Rhoads, Fatheree and Norori 90 ).
The most consistent finding is the decreased amounts of Bifidobacterium spp. and Lactobacillus spp. in infants with colic or extensive crying( Reference de Weerth, Fuentes and Puylaert 28 , Reference Pärtty, Kalliomäki and Endo 91 ). Conversely, elevated numbers of these bacteria have been linked to decreased colicky symptoms( Reference Roos, Dicksved and Tarasco 92 ). A recent study reported an association between delayed colonisation by Bifidobacterium infantis and increased risk of irritability in preterm infants( Reference Pärtty, Luoto and Kalliomäki 86 ). Previously, the colonisation of B. infantis has been associated with normal development of immune tolerance and the species has been shown to be capable of normalising the permeability of intestinal mucosa( Reference Chichlowski, De Lartigue and German 93 ). This effect is most likely mediated by bioactive factors secreted by B. infantis, which have been shown to induce the expression of tight junction proteins, thus tightening the connections between enterocytes( Reference Ewaschuk, Diaz and Meddings 94 ). Moreover, Bifidobacteria have been associated with reduced abdominal pain and discomfort in adults( Reference Jalanka-Tuovinen, Salonen and Nikkilä 95 ).
Bacteria that are increased in infants with excessive crying and colic symptoms include anaerobic Gram-negative and coliform bacteria( Reference de Weerth, Fuentes and Puylaert 28 , Reference Savino, Cordisco and Tarasco 89 , Reference Rhoads, Fatheree and Norori 90 , Reference Savino, Cordisco and Tarasco 96 ). It has been speculated that coliform bacteria such as E. coli may overtake the beneficial bacteria in colicky infants resulting in reduced induction of regulatory T cells by beneficial commensals and increased production of cytokines by antigen-presenting cells, thus leading to immune dysregulation and increased permeability of intestinal epithelium( Reference Savino, Cordisco and Tarasco 89 ). A recent study utilising high-throughput microarray analysis reported a negative association between crying and butyrate-producing bacteria such as Butyrivibrio crossotus, Eubacterium rectale and Eubacterium hallii, which were found to be 1·5-fold more abundant in healthy infants without colic symptoms( Reference de Weerth, Fuentes and Puylaert 28 ). Butyrate-producing bacteria have been shown to reduce the pain sensation( Reference Vanhoutvin, Troost and Kilkens 97 ) and proposed to reinforce gut defense barrier by increasing the production of mucins( Reference Burger-van Paassen, Vincent and Puiman 98 ). Butyrate also up-regulates the expression of tight junction proteins, thus leading to decreased intestinal permeability( Reference Ma, Fan and Li 99 , Reference Wang, Wang and Wang 100 ).
The role of aberrant microbiota composition in colic is supported by the observation that colicky symptoms could be alleviated by probiotic supplementation of Lactobacillus reuteri DSM 17938( Reference Savino, Cordisco and Tarasco 101 ). The authors suggested the improvement of gut motility and function and the reduction of visceral pain as possible mechanisms of probiotic action. Interestingly, the probiotic strain L. reuteri DSM 17938 has also been shown to reduce gastric distension and accelerate gastric emptying rate, which could potentially alleviate colic symptoms( Reference Indrio, Riezzo and Raimondi 102 ). Furthermore, two L. reuteri strains have been shown to inhibit the growth of colic-associated coliforms in vitro ( Reference Savino, Cordisco and Tarasco 96 ). This inhibition was mediated by bacteriocins and other inhibitory molecules produced by L. reuteri. Moreover, the potential of Lactobacillus rhamnosus GG in alleviation of colic symptoms have been reported( Reference Pärtty, Luoto and Kalliomäki 86 , Reference Pärtty and Isolauri 103 ). In contrast, a recent study has shown that L. reuteri strain DSM 17938 was not effective in protecting newborns from colic( Reference Sung, Hiscock and Tang 104 ). Thus, these studies warrant further assessment and well-planned intervention studies to characterise the potential effects of other probiotic strains in addition to L. reuteri and L. rhamnosus GG.
Atopic diseases
Atopic diseases are chronic and relapsing disorders usually starting in early childhood. Atopy has been characterised as a genetic disposition to develop an allergic reaction and produce elevated levels of IgE upon exposure to an environmental antigen( Reference Bieber 105 ). Atopic diseases include eczema (atopic dermatitis), allergic rhinitis (hay fever), allergic conjunctivitis and allergic asthma. In early life, the most common form of atopic disease is eczema, its prevalence being ~15–30 % depending on the country studied( Reference Deckers, McLean and Linssen 106 ). The incidences of eczema and other allergic diseases are more common in industrialised countries, the highest prevalence typically found in Northern Europe( Reference Bieber 105 ). During the past decades, associations between the composition of intestinal microbiota and atopic diseases have been studied intensively.
Some of the studies evaluating the associations between microbiota composition and atopy have also addressed the microbiota composition preceding the development of disease. Reduced diversity at early life (i.e. at ages 1 week, 1 month or 4 months) has been associated with an increased risk of developing atopy or allergic disease( Reference Abrahamsson, Jakobsson and Andersson 107 – Reference Kalliomäki, Kirjavainen and Eerola 112 ). However, after age 1 year the total microbiota diversity in children either developing or having eczema is comparable or even higher than that of healthy children( Reference Nylund, Satokari and Nikkilä 36 , Reference Abrahamsson, Jakobsson and Andersson 107 ). In addition, the pathogenesis of atopic diseases is associated with an impaired gut barrier function and increased intestinal permeability and gastrointestinal symptoms are common among the patients( Reference Rosenfeldt, Benfeldt and Valerius 113 ). Thus, it seems that sufficient diversity of microbiota in early infancy is essential for modulation of the expression of genes involved in the normal pattern of intestinal development such as postnatal intestinal maturation and maintenance of mucosal barrier( Reference Sjögren, Tomicic and Lundberg 114 , Reference Hooper and Macpherson 115 ). However, microbiota development and diversification should not happen too expeditiously, since prematurely occurring changes towards an adult-type microbiota may predispose infants, e.g. with eczema( Reference Nylund, Satokari and Nikkilä 36 ). It is possible that an infant-type microbiota supports an adequate gut barrier function and tolerance against allergens in an immature gut and affects the maturation of the gut epithelium and immune functions in a way that results in reinforcement of the normal mucosal barrier function( Reference Rosenfeldt, Benfeldt and Valerius 113 , Reference Maynard, Elson and Hatton 116 ).
The results on specific bacteria either increasing or decreasing the risk of developing atopic diseases or associated with their onset are still conflicting( Reference Nylund, Satokari and Nikkilä 36 , Reference Penders, Stobberingh and Thijs 51 , Reference Sepp, Julge and Mikelsaar 117 – Reference Mah, Chin and Wong 121 ). Aberrations in Bifidobacterial community have been associated with children with atopic diseases, most often characterised by either reduced total abundance or shifts in species community( Reference Maynard, Elson and Hatton 116 , Reference Sepp, Julge and Mikelsaar 117 , Reference Johansson, Sjögren and Persson 119 , Reference Gore, Munro and Lay 120 ). Furthermore, decreased amounts of Bacteroides spp. and increased amounts of specific Firmicutes such as Staphylococcus aureus and different clostridial groups, have been associated with the development and onset of allergic diseases( Reference Nylund, Satokari and Nikkilä 36 , Reference Abrahamsson, Jakobsson and Andersson 107 , Reference Björksten, Sepp and Julge 122 – Reference Thompson-Chagoyan, Fallani and Maldonado 125 ). Interestingly, both Bifidobacterium spp. and Bacteroides spp. have been reported to have anti-inflammatory properties via their ability to direct the cellular and physical maturation of the developing immune system( Reference Hooper, Wong and Thelin 126 , Reference Pagnini, Saeed and Bamias 127 ). For example, polysaccharide A from B. fragilis is able to direct the development of CD4+ T cells, thus inducing the differentiation of T helper (Th) 1 lineage and correction of the Th1/Th2 imbalance( Reference Mazmanian, Liu and Tzianabos 128 ). Furthermore, this polysaccharide has been shown to promote immunologic tolerance through induction of regulatory T cells, resulting in suppression of IL-17 responses( Reference Round, Lee and Li 129 ). Moreover, both Bifidobacterium and Bacteroides spp. have high frequency of immunostimulatory CpG motifs in their genomes, thus being rich in TLR9 ligands( Reference Kant, de Vos and Palva 16 ). TLR9 stimulation is known to both enhance epithelial integrity and direct immune responses towards Th1 type (reviewed in Kant et al.( Reference Kant, de Vos and Palva 16 )). These effects may be diminished in allergic subjects, who have reduced numbers of Bifidobacterium and Bacteroides spp.
In recent studies, increased levels of IL-17 have been associated with asthma( Reference Alyasin, Karimi and Amin 130 , Reference Ramirez-Velazquez, Castillo and Guido-Bayardo 131 ). Furthermore, one of the most important defence mechanisms in the epithelial barrier is IgA, which is present at high concentrations in the intestinal mucus layer( Reference Brandtzaeg 132 ). Low levels of IgA predisposes infants to increased binding of antigens to mucosal membrane, to increased mucosal leakiness and an increased uptake of dietary antigens( Reference Johansen, Pekna and Norderhaug 133 ). Low levels of IgA have also been associated with increased risk for the development of IgE-mediated allergic diseases in children( Reference Kukkonen, Kuitunen and Haahtela 134 ). Furthermore, it has been suggested that high numbers of Clostridium spp. may be associated with degradation of antigen-specific IgA, which could debilitate the immature gut barrier( Reference Pärtty, Luoto and Kalliomäki 86 ). The protective role of specific bacteria and their compounds against atopy and allergic diseases is further supported by several clinical studies reporting the effects of probiotic strains on the alleviation of allergic symptoms even when the probiotics failed to modify the microbiota composition or diversity( Reference Nermes, Kantele and Atosuo 135 , Reference Elazab, Mendy and Gasana 136 ). These effects can be related to the probiotic effects on the hosts’ immunological functions such as improvement of the barrier function and increasing allergen-specific IgA levels, which are essential for the development of tolerance and can be considered as a marker for immune maturation( Reference Rosenfeldt, Benfeldt and Valerius 113 , Reference Kukkonen, Kuitunen and Haahtela 134 , Reference Nermes, Kantele and Atosuo 135 , Reference Mantis, Rol and Corthesy 137 – Reference Di Mauro, Neu and Riezzo 139 ). Furthermore, probiotics have been suggested to have immunomodulatory impacts that affect the Th1/Th2 balance such as stimulation of Th1-type immune responses, induction of apoptosis of Th2 cells and induction of regulatory T and dendritic cells( Reference Kant, de Vos and Palva 16 , Reference Rautava, Arvilommi and Isolauri 138 , Reference Kwon, Lee and So 140 – Reference West, Hammarstrom and Hernell 144 ).
Coeliac disease
CD is an autoimmune disorder of the small intestine that occurs in genetically predisposed individuals. It is caused by a reaction to dietary gluten and related prolamines, which are proteins found in maize such as wheat, barley and rye. Upon exposure to gluten, inflammatory cascade is induced in the small intestinal epithelium leading to a villous atrophy and crypt hyperplasia( Reference Green and Jabri 145 ). Typical symptoms include different gastrointestinal symptoms such as diarrhoea, abdominal pain and distension( Reference Murch, Jenkins and Auth 146 ). Untreated CD may lead to weight loss, malabsorption and growth disturbances in paediatric patients( Reference Murch, Jenkins and Auth 146 ). The CD is a multifactorial disorder and its pathogenesis involves both genetic and environmental factors. For example, a high frequency of infectious episodes early in life( Reference Stene, Honeyman and Hoffenberg 147 , Reference Myleus, Hernell and Gothefors 148 ), antibiotic treatments( Reference Mårild, Ye and Lebwohl 81 ) as well as the timing of gluten introduction into the diet( Reference Sellitto, Bai and Serena 149 , Reference Ivarsson, Persson and Nystrom 150 ) have been associated with the onset of CD in genetically susceptible infants (Fig. 1).
A specific role for the intestinal microbiota in CD development has been suggested( Reference Sellitto, Bai and Serena 149 , Reference Sanz, De Pama and Laparra 151 , Reference De Palma, Nadal and Medina 152 ). Indeed, deviations in faecal and duodenal microbiota associated with CD have been reported( Reference Sellitto, Bai and Serena 149 , Reference Sanz, De Pama and Laparra 151 – Reference Schippa, Iebba and Barbato 153 ), although recent studies utilising high-throughput methods have reported comparable microbiota compositions in patients and healthy controls( Reference de Meij, Budding and Grasman 154 – Reference Cheng, Kalliomäki and Heilig 157 ). A recent study utilising a high-throughput microarray method in analysing duodenal biopsies of paediatric CD patients in Finland found that while the overall microbiota composition was comparable between CD and healthy subjects, a profile of eight bacterial groups was observed to distinguish patients from healthy controls( Reference Cheng, Kalliomäki and Heilig 157 ). This profile was characterised by higher abundances of bacteria related to Prevotella melaninogenica, Haemophilus and Serratia spp., whereas those related to P. oralis, P. cinnamivorans, Ruminococcus bromiii, Proteus and Clostridium stercorarium were decreased in CD patients( Reference Cheng, Kalliomäki and Heilig 157 ). Also the total abundance of Prevotella spp. was found to be slightly increased (did not reach a statistical significance)( Reference Cheng, Kalliomäki and Heilig 157 ), which supports the previous findings by a Swedish research group, who found an association of elevated total abundance of Prevotella spp. and CD( Reference Forsberg, Fahlgren and Horstedt 158 , Reference Ou, Hedberg and Horstedt 159 ). Microbiota dysbiosis of CD patients may be characterised by an increased microbiota diversity( Reference De Palma, Nadal and Medina 152 , Reference Schippa, Iebba and Barbato 153 , Reference Nadal, Donat and Ribes-Koninckx 160 , Reference Sanchez, Donat and Ribes-Koninckx 161 ), but these findings have been contradicted recently( Reference Cheng, Kalliomäki and Heilig 157 ). Furthermore, patients with active CD seem to have an increased inter-individual similarity when compared with patients in remission state or healthy controls( Reference Schippa, Iebba and Barbato 153 , Reference Forsberg, Fahlgren and Horstedt 158 ). It has been suggested that altered glycosylation patterns observed in mucosa of CD patients( Reference Forsberg, Fahlgren and Horstedt 158 ) may create a more selective pressure leading to a more homogenous microbial colonisation( Reference Schippa, Iebba and Barbato 153 ).
Such microbiota deviations are only partly restored after long-term treatment with gluten-free diet. A higher diversity and a complete rearrangement in Eubacterium species community as well as changed metabolomic profiles were observed in CD patients who had followed gluten-free diet for 2 years when compared to healthy controls( Reference Di Cagno, De Angelis and De Pasquale 162 ). In contrast, the proportions of E. coli and Staphylococcus were observed to normalise after treatment with gluten-free diet( Reference Collado, Donat and Ribes-Koninckx 163 ).
In the active phase of CD, the reduction of Gram-positive bacteria population, especially the numbers or proportion of Bifidobacterium spp. has been reported( Reference De Palma, Nadal and Medina 152 , Reference Collado, Donat and Ribes-Koninckx 163 ). Such findings may be of interest, since Bifidobacteria have been suggested to alleviate gastrointestinal symptoms of adult coeliac patients( Reference Smecuol, Hwang and Sugai 164 ) and have been associated with reduced abdominal pain and discomfort in healthy adults( Reference Jalanka-Tuovinen, Salonen and Nikkilä 95 ). In contrast to declined proportions of Gram-positives, Gram-negative bacteria such as Clostridium groups( Reference Nadal, Donat and Ribes-Koninckx 160 , Reference Collado, Donat and Ribes-Koninckx 163 ), Prevotella spp.( Reference Cheng, Kalliomäki and Heilig 157 , Reference Ou, Hedberg and Horstedt 159 ) and E. coli ( Reference Schippa, Iebba and Barbato 153 , Reference Collado, Donat and Ribes-Koninckx 163 ) seem to be increased in paediatric CD patients. The most constant finding is the higher abundance of Bacteroides spp. in faeces and duodenal biopsies of CD patients( Reference De Palma, Nadal and Medina 152 , Reference Schippa, Iebba and Barbato 153 , Reference Collado, Donat and Ribes-Koninckx 163 ), although a complete lack of the members of phylum Bacteroidetes was observed in CD predisposed infants in a prospective study( Reference Sellitto, Bai and Serena 149 ).
Furthermore, another study reported a reduction in IgA-coated bacteria, especially IgA-coated Bacteroides in faeces of untreated and treated CD patients when compared to healthy controls( Reference Sanz, De Pama and Laparra 151 ). The authors stated that host defences against this bacterial group might be reduced in coeliac disease, thus allowing its increased colonisation. Moreover, shifts in Bacteroides spp. composition in early-life microbiota have been reported in infants with high genetic risk for CD development compared to infants with low genetic risk( Reference Collado, Donat and Ribes-Koninckx 163 ). In detail, Bacteroides uniformis, B. ovatus and B. plebeius were associated with a low genetic risk, whereas B. vulgatus seems to be more prevalent both in high-risk infants( Reference Sanchez, De Palma and Capilla 165 ) and in infants with active CD( Reference Schippa, Iebba and Barbato 153 ). Furthermore, B. fragilis has been associated with an increased risk for CD development in genetically predisposed infants who were formula-fed( Reference Palma, Capilla and Nova 166 ). Interestingly, polysaccharide A produced by B. fragilis has been shown to induce the differentiation of Th1-type immune cells( Reference Mazmanian, Liu and Tzianabos 128 ). In addition, a decreased duodenal expression of TLR2 and increased expression of TLR9 and IL-8 have been observed in infants with CD( Reference Kalliomäki, Satokari and Lähteenoja 155 ). It has been suggested that increased TLR9 signalling in the duodenum may contribute to the Th1 response found in the small intestinal mucosa of CD subjects( Reference Kalliomäki, Satokari and Lähteenoja 155 , Reference Cheng, Kalliomäki and Heilig 157 ). Furthermore, the expression of tight junction protein coding ZO-1 is significantly down-regulated in untreated CD patients when compared to patients with treated CD( Reference Cheng, Kalliomäki and Heilig 157 ). Thus, a synergistic effect of Bacteroides spp. and increased TLR9 signalling may lead to an excessive induction of Th1-type immune response, which may contribute to the onset and/or remission of the coeliac disease. Collectively, it seems that both altered microbiota composition and dysregulated host–microbe interaction may have a role in CD.
Conclusions
The microbiota development is a gradual process, which begins during early phases of pregnancy. During the succession of microbes some bacterial groups reach the degree of maturation earlier than others. The development of the intestinal microbiota is likely to continue throughout childhood and adolescence and may not be completed until the human host reaches adulthood. The course of development is affected by both life-style factors and medical practices that direct the intestinal colonisation and have an impact on health later in life. Our understanding of both the compositional development and the diversity and function of the intestinal microbiota and its effects on health and disease is constantly improving but further studies are still needed to address the long-term influence of early-life gut microbiota on intestinal, systemic immunity and other organ systems.
Acknowledgements
None.
Financial Support
This work was supported by The Finnish Graduate School on Applied Bioscience: Bioengineering, Food and Nutrition, Environment (for L. N.), The Academy of Finland (Grant no. 258438 for R. S. and Grant numbers 137389 and 141140 for W. de V.) and the ERC Advanced Grant no. 250172 (Microbes Inside) of the European Research Council.
Conflicts of Interest
None.
Authorship
L. N. wrote the paper; R. S. designed Fig. 1; all authors corrected and approved the manuscript.



