Introduction
Sorghum [Sorghum bicolor (L.) Moench] is a drought-tolerant crop used for human and animal consumption (Srinivasa Rao et al., Reference Srinivasa Rao, Reddy, Nagaraj, Upadhyaya, Wang, Upadhyaya and Kole2014). It is an important dietary staple cereal cultivated in the dry regions of multiple countries. Today, the crop is the fifth most important cereal worldwide behind maize (Zay mays L.), wheat (Triticum aestivum L.), rice (Oryza sativa L.) and barley (Hordeum vulgare L.) (FAOSTAT, 2022). Sorghum was domesticated in Northeast Africa, an area that extends from the arid regions of Ethiopia to Sudan (Mann et al., Reference Mann, Kimber and Miller1983), although its expansion to the West and South regions of Africa lead to multiple re-domestication events. This genetically diverse crop is classified into five major races (Kafir, Durra, Bicolor, Caudatum and Guinea) based on their inflorescence architecture and kernel morphology (Snowden, Reference Snowden1936). These races have been associated with specific environmental adaptations and enclose substantial phenotypic and genetic variation (Cuevas et al., Reference Cuevas, Rosa-Valentin, Hayes, Rooney and Hoffmann2017; Olatoye et al., Reference Olatoye, Hu, Maina and Morris2018; Faye et al., Reference Faye, Maina, Hu, Fonceka, Cisse and Morris2019; Cuevas and Prom, Reference Cuevas and Prom2020). The compact panicle shape of the Durra race was domesticated in the Ethiopia surrounding regions and it is associated with drought tolerance (Mutava et al., Reference Mutava, Prasad, Tuinstra, Kofoid and Yu2011; Fracasso et al., Reference Fracasso, Trindade and Amaducci2016). In contrast, the Guinea race was domesticated in the humid regions of West Africa, where an open panicle shape that provided a better air flow was selected to reduce the incidence of grain mould (Cuevas et al., Reference Cuevas, Prom and Rosa-Valentin2018). However, phenotypic and genotypic diversity studies in Ethiopian germplasm found high frequency of admixture accessions and the presence of a large number of rare alleles that shows its large diversity (Cuevas et al., Reference Cuevas, Rosa-Valentin, Hayes, Rooney and Hoffmann2017; Girma et al., Reference Girma, Nida, Seyoum, Mekonen, Nega, Lule, Dessalegn, Bekele, Gebreyohannes, Adeyanju and Tirfessa2019, Reference Girma, Nida, Tirfessa, Lule, Bejiga, Seyoum, Mekonen, Nega, Dessalegn, Birhanu, Bekele, Gebreyohannes, Ayana, Tesso, Ejeta and Mengiste2020; Nida et al., Reference Nida, Girma, Mekonen, Lee, Seyoum, Dessalegn, Tadesse, Ayana, Senbetay, Tesso, Ejeta and Mengiste2019).
Crop improvement requires effective use of genetic diversity and the identification of the most valuable germplasm in an ex-situ collection (Byrne et al., Reference Byrne, Volk, Gardner, Gore, Simon and Smith2018). The National Plant Germplasm System (NPGS), belonging to USDA – Agriculture Research Service, maintains a sorghum germplasm collection that includes >41,000 accessions from 114 countries. This germplasm collection is a publicly global genetic resource for sorghum breeding programmes. The large size of this germplasm collection and the fact that tropical accessions flower during short-day length (<12 h; photoperiod sensitive) limits its effective screening for many economically important traits. Today, phenotypic analysis of NPGS sorghum germplasm collection mainly relies on descriptors traits taken during the regeneration process in tropical regions. To encourage the use of this germplasm collection, a core collection of 2438 accessions was established using passport information (Dahlberg et al., Reference Dahlberg, Burke and Rosenow2004), later by a genomic characterization for core sets from Ethiopia (Cuevas et al., Reference Cuevas, Rosa-Valentin, Hayes, Rooney and Hoffmann2017), Sudan (Cuevas and Prom, Reference Cuevas and Prom2020), Senegal (Faye et al., Reference Faye, Maina, Hu, Fonceka, Cisse and Morris2019), Nigeria and Niger (Olatoye et al., Reference Olatoye, Hu, Maina and Morris2018). Phenotypic and genetic analysis found the existence of structure populations within each core sets that could be related to adaptation to different agroclimatic regions and human selection. Moreover, these genomic characterizations were suitable for genome-wide association analysis (GWAS) providing a new resource for the study of agronomically important traits. Certainly, the screening of NPGS tropical sorghum germplasm is necessary to identify most valuable accessions that can be used to broaden the genetic diversity of breeding programmes.
The NPGS Ethiopian core set includes 376 accessions and a genomic characterization revealed 148,476 single nucleotide polymorphism (SNPs) (Cuevas et al., Reference Cuevas, Rosa-Valentin, Hayes, Rooney and Hoffmann2017). Population structure analysis found that its genetic diversity comprises 11 populations associated with morphologic traits. Likewise, a genomic and population structure analysis of 1425 Ethiopian sorghum landraces maintained at the Ethiopian Biodiversity Institute (EBI) in Ethiopian found this germplasm can also be stratified into 11 populations (Girma et al., Reference Girma, Nida, Seyoum, Mekonen, Nega, Lule, Dessalegn, Bekele, Gebreyohannes, Adeyanju and Tirfessa2019). A germplasm collection project across Ethiopia farmers field collected a total of 304 accessions that represent the different production regions (Wondimu et al., Reference Wondimu, Dong, Paterson, Worku and Bantte2021). The population structure analysis found six populations with strong correlations with the geographic distribution of the accessions. Nevertheless, these populations showed continuous gene flow that preserves genetic diversity and increases the adaptability of sorghum populations to different agriculture niches. In fact, Ethiopian germplasm with cold (Singh, Reference Singh1985) and drought tolerance (Adugna and Tirfessa, Reference Adugna and Tirfessa2014), resistance to anthracnose [Colletotrichum sublineola (formerly C. graminicola P. Henn in Kabàt and Bubk)] (Cuevas et al., Reference Cuevas, Prom and Cruet-Burgos2019b), grain mould caused by a complex of fungal pathogens (Nida et al., Reference Nida, Girma, Mekonen, Lee, Seyoum, Dessalegn, Tadesse, Ayana, Senbetay, Tesso, Ejeta and Mengiste2019) and nutritional quality (Rhodes et al., Reference Rhodes, Hoffmann, Rooney, Herald, Bean, Boyles, Brenton and Kresovich2017) have been reported. The high genetic and phenotypic diversity of Ethiopian germplasm is a valuable resource for sorghum improvement. The association of population structure or genomic regions with important traits is a strategy that could be used to facilitate the screening of these Ethiopian germplasm collection.
Grain mould is one of the most important diseases affecting sorghum production worldwide (Thakur et al., Reference Thakur, Reddy, Indira, Rao, Navi, Yang and Ramesh2006). The disease is manifested by discolouring of the inside and outside of the grain, softening the endosperm, and reducing the acceptability for food and feed processing (Rooney and Serna-Saldivar, Reference Rooney, Serna-Saldivar, Lorenz and Kulp1991). It is caused by a complex of more than 40 pathogenic and opportunistic fungi including Fusarium thapsinum (Klittich, Leslie, Nelson, & Manasas.), Fusarium semitectum (Berk. & Ravenel), Curvularia lunata (Wakk.) Boedijn, Alternaria alternata (Fr.) Keissler, and C. sublineola which are considered the most important species worldwide (Thakur et al., Reference Thakur, Rao, Reddy, Sanjana Reddy, Thakur, Reddy and Mathur2007). Therefore, the evaluation of grain mould resistance is mostly measured based on the correlation of seed emergence, deterioration and weight (Prom, Reference Prom2004; Erpelding and Prom, Reference Erpelding and Prom2006). The warm climates of tropical and sub-tropical regions are the most favourable environments for the development of the disease (Ackerman et al., Reference Ackerman, Wenndt and Boyles2021) and resistant cultivars provide the most effective method to control grain mould. At present, some research has identified resistance sources and genomic regions associated with grain mould resistance. The evaluation of the sorghum association panel (SAP) conducted in Puerto Rico and Texas identified resistant accessions and genomic regions associated with seed emergence and deterioration (Cuevas et al., Reference Cuevas, Fermin-Perez, Prom, Cooper, Bean and Rooney2019a; Prom et al., Reference Prom, Cuevas, Ahn, Isakeit, Rooney and Magill2020). This multiyear evaluation identified 25 resistant accessions in Puerto Rico and 18 resistant accessions in Texas. Only four accessions (‘SUMAC’, ‘SC623’, ‘Sureño’ and ‘Della’) were resistant across locations. GWAS found three genomic regions associated with seed emergence and deterioration which enclose one R gene and another related to plant immune system (Cuevas et al., Reference Cuevas, Fermin-Perez, Prom, Cooper, Bean and Rooney2019a). A subset of 1425 Ethiopian landraces from EBI were evaluated for grain mould resistant in Ethiopia (Nida et al., Reference Nida, Girma, Mekonen, Lee, Seyoum, Dessalegn, Tadesse, Ayana, Senbetay, Tesso, Ejeta and Mengiste2019). The analysis based on seed deterioration found that most of the landraces (>66%) showed a resistant response. GWAS found that the YELLOW SEED1 (Y1) locus associated with kernel colour (Ibraheem et al., Reference Ibraheem, Gaffoor and Chopra2010; Morris et al., Reference Morris, Ramu, Deshpande, Hash, Shah, Upadhyaya, Riera-Lizarazu, PJ, Acharya, SE, Harriman, JC, ES and Kresovich2013) also decreased seed deterioration caused by mould and unfavourable weather conditions. These studies revealed the complex inheritance of grain mould resistance and how different environments can affect the observed resistance response. In fact, the screening and inheritance studies for resistance to grain mould of sorghum are difficult due to the many pathogens associated with the disease and its low heritability. Additional evaluations and studies of the inheritance of resistance with others sorghum germplasm will provide better insight into the genetic control of grain mould in sorghum.
The identification of grain mould resistant germplasm is required for the development of new resistant cultivars. In this regard, the NPGS sorghum germplasm collection is a major source of genetic diversity and disease resistance genes for breeding programmes. But the lack of passport information in this collection inhibits the selection of germplasm based on geographical regions. In the current study, we evaluated the grain mould resistance response of NPGS Ethiopian core set (Cuevas et al., Reference Cuevas, Rosa-Valentin, Hayes, Rooney and Hoffmann2017) to: (1) identify new grain mould resistant germplasm, (2) determine the association of NPGS Ethiopian population structure with grain mould resistance, and (3) associate genomic regions with grain mould resistance through GWAS.
Material and methods
Germplasm and field experiment
A total of 330 accessions from a core set of the NPGS Ethiopian germplasm collections (Cuevas et al., Reference Cuevas, Rosa-Valentin, Hayes, Rooney and Hoffmann2017) were evaluated for grain mould resistance (online Supplemental Table S1). The core set and susceptible (RTx430 and RTx2536) and resistant (Sureño and PI 267548 [Prom and Erpelding, Reference Prom and Erpelding2009]) controls were planted in research farms at the USDA-ARS Tropical Agriculture Research Station in Isabela (18.471569, −67.043472) and Mayaguez (18.211536, −67.135562), Puerto Rico for two consecutive years (November 2014 and October 2015, respectively). At both locations, a completely randomized design with a single replication was used, with plots measuring 1.8 m in length with 0.9 m between rows.
The data from the two locations were used to select seven accessions with an average seed deterioration of ⩽2.5 (on a 5-point scale) and emergence rate of ⩾75% for two additional replicated trials at Isabela, Puerto Rico. The trials included the seven selected accessions, the susceptible checks (RTx430, RTx2536 and RTx2911) and the resistant checks from the SAP (‘Summac’, ‘Rox Orange’, ‘Red Amber’, ‘Keller’, ‘Kansas Orange’, ‘SC309’, ‘SC609’, ‘Sureño’, ‘SC15’ ‘SC719’ and ‘SC13’; [Cuevas et al., Reference Cuevas, Fermin-Perez, Prom, Cooper, Bean and Rooney2019a]) and the Sudanese tropical accession PI 267548 (Prom and Erpelding, Reference Prom and Erpelding2009) were planted in 2020 and 2021 in randomized block design consisting of four blocks with plots measuring 1.8 m in length with 0.9 m between rows.
The plots and plants in the four experiments were maintained using standard management practices, weed controlled using mechanical tillage and hand hoeing, and aerial watering was supplied in the absence of rainfall. Temperature and relative humidity were recorded throughout the experiments using an Onset HOBO U23 Pro V2 located within research plots.
Grain mould response
The grain mould response was determined based on visual evaluations of seed mould and weathering degradation (hereafter referred to as seed degradation) and seedling emergence. Three panicles from the core set of the NPGS Ethiopian germplasm collections (i.e., experiment 1 and 2) and five panicles from the subset (i.e., experiment 3 and 4) with uniform flowering times were covered with mesh bags for exposure to environmental conditions and to avoid damage by birds. Panicles were harvested at maturity stage (30–40 days after anthesis), dried, and threshed using a single plant thresher (Almaco Single Plant and Head Thresher; Allan Machine Company). Approximately 400 seeds from each panicle were assessed for seed degradation using a 5-point visual scale (Thakur et al., Reference Thakur, Rao, Reddy, Sanjana Reddy, Thakur, Reddy and Mathur2007; Isakeit et al., Reference Isakeit, Collins, Rooney and Prom2008), where 1 indicates seed is bright with no mould and no discoloration resulting from weathering; 2 indicates seed is not as bright and has little or no mould but has some discoloration (1–10% moulded kernels); 3 indicates seed is not bright, with some mould and some discoloration (11–25% moulded kernels); 4 indicates seed is almost entirely covered in mould, and the pericarp is degraded (26–50% moulded kernels); and 5 indicates seed is covered entirely with mould, and the pericarp is degraded and looks dead (>50% moulded kernels). The emergence rate was determined based on the number of seedlings observed after 10 days from 30 planted seeds from each panicle. The seeds were planted in flats containing Metro Mix 200 potting medium and incubated in a greenhouse at room temperature.
Phenotypic analysis
The seed degradation and emergence rate from the first two years were analysed as a randomized complete block design (RCBD) with two blocks. Likewise, the subset of accessions and reference lines evaluated for four years were analysed as a RCBD with four blocks. The seed degradation and emergence rate of both sets of data were subject to analysis of variance using the Proc mixed covtest method type 3 procedure of SAS (SAS Institute, Cary, NC) where years were treated as fixed effects and accessions as random effects. The repeatability of seed degradation and emergence rate was estimated using the following formula:

where σ2g and σ2e refer to the accessions and error variances, respectively, while e is the number of blocks (Bernardo, Reference Bernardo2002). The Pearson correlation coefficients among traits were calculated using the Proc corr procedure of SAS. The seed degradation and emergence rate of accessions and references lines in the subset were compared by using the Tukey–Kramar's post-hoc test at the 5% level of significance. Likewise, the seed degradation and seedling emergence rate among the eleven population present in the NPGS Ethiopian core set (Cuevas et al., Reference Cuevas, Rosa-Valentin, Hayes, Rooney and Hoffmann2017) were compared by using the Tukey–Kramar's post-hoc test at the 5% level of significance.
Association and phylogenetic analysis
GWAS was conducted using the genomic characterization for the NPGS Ethiopia core set available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (SRP159248; [Cuevas et al., Reference Cuevas, Rosa-Valentin, Hayes, Rooney and Hoffmann2017]) and the seed degradation and emergence rate of the 333 accessions. The Tassel 5.0 v2 GBS pipeline (Glaubitz et al., Reference Glaubitz, Casstevens, Lu, Harriman, Elshire, Sun and Buckler2014) was used to recall SNPs using the most recent version of the BTx623 sorghum genome (version v.3; www.sorghumbase.com). A total of 151, 210 SNPs with minor allele frequencies (MAF) ⩾ 0.05 were retained for GWAS. The Bayesian-information and Linkage-disequilibrium Interatively Nested Keyway (BLINK) method was implemented for association analysis using the Genome Association and Prediction Integrated Tool (GAPIT) in R (Lipka et al., Reference Lipka, Tian, Wang, Peiffer, Li, Bradbury, MA, ES and Zhang2012). The first three principal components were included as covariates to control population structure and family relatedness. Log quantile-quantile (QQ) P-value plots were visually examined to determine the appropriateness of the linear model. Manhattan and QQ plots were visualized using the R package (Yin, Reference Yin2022). Associated SNPs were determined using the (Benjamini and Hochberg, Reference Benjamini and Hochberg1995) false discovery rate as implemented in GAPIT at the 1% level of significance. The linkage disequilibrium (LD) for associated significance SNPs were delimited using the block function of PLINK (Purcell et al., Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender, Maller, Sklar, De Bakker, Daly and Sham2007). Later, these LD blocks were used to identify candidate genes based on the annotations of BTx623 v3. sorghum genome reference (www.phytozome.com).
The genotypic data were thinned to 2379 unlinked SNPs (r 2 < 0.10) without missing genotypes using PLINK (Purcell et al., Reference Purcell, Neale, Todd-Brown, Thomas, Ferreira, Bender, Maller, Sklar, De Bakker, Daly and Sham2007) and used in phylogenetic analysis. The maximum-likelihood tree was selected using ModelFinder (Kalyaanamoorthy et al., Reference Kalyaanamoorthy, Minh, Wong, Von Haeseler and Jermiin2017) and branch supports were obtained with the ultrafast bootstrap (Hoang et al., Reference Hoang, Chernomor, Von Haeseler, Minh and Vinh2018) both as implemented in the IQ-TREE 2 software (Minh et al., Reference Minh, Schmidt, Chernomor, Schrempf, Woodhams, Von Haeseler and Lanfear2020). The phylogenetic tree was visualized using Interactive Tree of Life (Letunic and Bork, Reference Letunic and Bork2011).
Results
Grain mould resistance in NPGS Ethiopian core set
The seed degradation and seedling emergence rate in the NPGS core set averaged 64.09% ± 19.02 and 3.84 ± 0.62, respectively, indicating that most of the accessions are highly susceptible to grain mould (online Supplementary Fig. S1 and Table S1). A total of 115 accessions had seedling emergence rates ⩾75% and 11 accessions showed seed degradation scores ⩽2.5. Both traits were negatively correlated (r 2 = −0.32; P-value < 0.001) indicating seeds with high seed deterioration rates are likely to have lower emergency rate. Only seven accessions (PI 330821, PI 453177, PI 454221, PI 455036, PI 455213, PI 455248 and PI 457867) exhibited both emergence rate ⩾75% and seed degradation score ⩽2.5. The repeatability of seed degradation and emergency rate was 0.47 for both traits, indicating that environmental factors have large effects on these traits.
The subsequent evaluation of these seven accessions led to the identification of grain mould resistance accessions. Two accessions (PI 457867 and PI 454221) showed seedling emergence rates similar to those observed in grain mould resistant control lines (⩾71.30%; Table 1). Likewise, three accessions (PI 455036, PI 455213, and PI 330821) showed minimal seed degradation similar to the grain mould resistant lines Summac, Rox Orange, Red Amber, Keller, SC13 and SC15 (⩽1.95, Table 2). None of the Ethiopian accession exhibited neither high emergence rate nor low seed degradation as observed in Summac, Rox Orange, Red Amber and Keller, indicating that Ethiopian germplasm includes a low frequency of grain mould resistant genes.
Table 1. Mean seedling emergence rates for seven NPGS Ethiopian tropical accessions and fifteen temperate adapted references lines evaluated in Puerto Rico for four years

a Different letters indicate significant differences based on Tukey test (minimum significance difference = 20.0; P-value < 0.05).
Table 2. Mean seed deterioration scores for seven NPGS Ethiopian tropical accessions and fifteen temperate adapted references lines evaluated in Puerto Rico for four years.
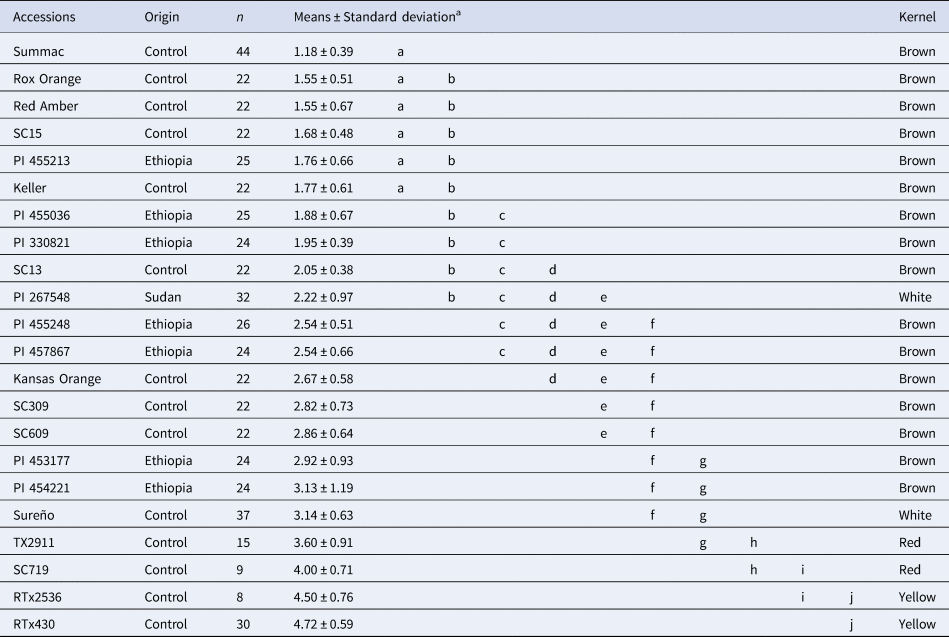
a Different letters indicate significant differences based on Tukey test (minimum significance difference = 0.76; P-value < 0.05).
The association of high emergence rate and low seed degradation with the population structure of the NPGS Ethiopian germplasm can enable the identification of resistance accessions. Population structure analysis divided this germplasm into eleven populations of which population 5 showed the highest seedling emergence rate (86.95%; Table 3) and the lowest seed degradation (3.33). However, these populations show susceptibility to grain mould indicating Ethiopian population structure is not linked to seed quality. In fact, the five accessions with high emergence rate and low seed degradation ratings identified in the subset belong to population 1 and the admixture group. Phylogenetic analysis between these accessions and references lines revealed that PI 457867 is genetically related to grain mould resistance accessions belonging to the Kafir race (Fig. 1). Moreover, this accession cluster nearby to the grain mould resistant line Kansas Orange (PI 641824). The other four accessions (PI 330821, PI 454221, PI 455036 and PI 455213) form one cluster distant from references lines being the grain mould resistance accessions from East Africa/India the closer group (Fig. 1).
Table 3. Populations means analysis for grain mould resistance in NPGS Ethiopian germplasm collection

a Population structure analysis according to Cuevas et al., Reference Cuevas, Rosa-Valentin, Hayes, Rooney and Hoffmann2017.
b Difference letters indicate significance difference based on Tukey test (P-value < 0.05).

Figure 1. Maximum-likelihood tree based on 2379 SNPs between 15 grain mould resistance accessions presents in the sorghum association panel and 7 NPGS Ethiopian accessions evaluated for grain mould resistance across four years in Isabela, Puerto Rico. Red and green stars represent NPGS Ethiopian accessions accession with low seed deterioration and high emergence rates, respectively.
GWAS and candidate genes
The genome-wide association scan for emergence rate detected two loci on chromosomes 1 (Chr.1: 9,343,797; −log[P-value] = 7.10) and 8 (Chr.8: 7,329,131; −log[P-value] = 6.75) (Table 4; Fig. 2). The LD block for the locus in chromosome 1 is located within the putative gene Sobic.001G120400 which encodes a Phospholipase A2 protein. The associated SNP in chromosome 8 is in an intergenic region of 19 Kb (Sobic.08G065400 – Sobic.08G065600). However, the associated LD block in this intergenic region has homology to the rice putative gene Loc_Os0Gg30220.1 which encodes the Recognition of Peronospora Parasitica 13-like (RPP13-like) protein associated to resistance to downy mildew (Peronospora parasitica). The synergistic allelic effect of these two loci is a 13.84% increase in the seed emergency rate. In a genome-wide association scan for seed deterioration was detected a locus in chromosome 3 (Chr.3: 68,822,874; −log P[value] = 9.97). This LD block for this locus expands 48.41 Kb and encloses seven putative genes (Sobic.03G372600, Sobic.03G372700, Sobic.03G372750, Sobic.03G372800, Sobic.03G372850, Sobic.03G372900, Sobic.03G372950). Candidate gene analysis found that Sobic.03G372900 encodes a Deoxycytidine kinase protein and Sobic.03G372600 is highly expressed in mature dry seeds. The estimated allelic effect of this locus was a 0.31 decrease in the 5-point score rate for the seed deterioration.
Table 4. Genomic regions associated with grain mould resistance in NPGS Ethiopian germplasm collection.

a SNPs refers to single nucleotide polymorphism (SNPs).
b Chr refers to chromosome.
c MAF refers to minor allele frequency.
d LD refers to linkage disequilibrium.
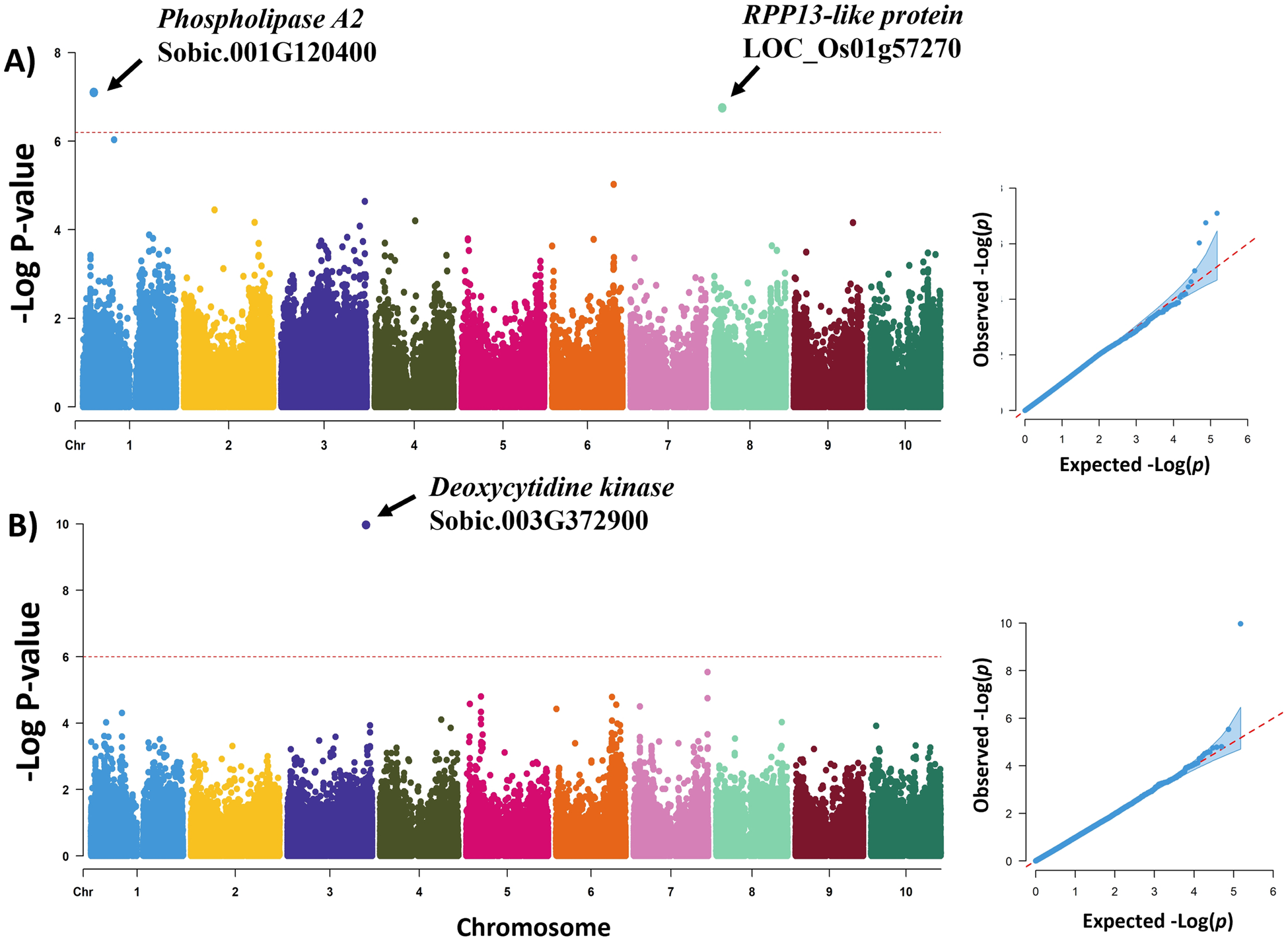
Figure 2. Manhattan and quantile–quantile plots for the GWAS of grain mould resistance in NPGS Ethiopian germplasm collection. (a) GWAS for seed emergence rate. (b) GWAS for seed deterioration.
Discussion
The narrow genetic diversity amongst temperate adapted sorghum lines encourages the utilization of tropical germplasm collection to identify the most valuable accessions. Nevertheless, the large number of tropical accessions in NPGS germplasm collection has limited its phenotypic analysis. Today, core sets for the NPGS Ethiopian, Sudanese, Nigeria and Senegal germplasm collections are available to enable phenotypic evaluations, perform genome-wide association analyses and to establish allele deployment strategies (Cuevas et al., Reference Cuevas, Rosa-Valentin, Hayes, Rooney and Hoffmann2017; Olatoye et al., Reference Olatoye, Hu, Maina and Morris2018; Faye et al., Reference Faye, Maina, Hu, Fonceka, Cisse and Morris2019; Cuevas and Prom, Reference Cuevas and Prom2020). In this study, the evaluation of the grain mould resistance response of the NPGS Ethiopian core set found five accessions with resistance alleles, three novel genetic loci and found that most of this germplasm set is highly susceptible to grain mould. Moreover, our results suggest that the further evaluation of this core set for other seed quality traits (e.g. grain hardness, starch, fat, etc.) and yield should be directed to dry tropical regions with low grain mould incidence. Although the grain deterioration due to mould is not always correlated to the chemical and nutritional quality of the grain (Ratnavathi and Komala, Reference Ratnavathi, Komala, Ratnavathi, Patil and Chava2016).
Grain mould resistance is a complex trait that involves the interaction of multiple genes (Ackerman et al., Reference Ackerman, Wenndt and Boyles2021). Resistance germplasm is the result of the fixation of multiple resistance alleles by natural or human selection. The climate of Ethiopia includes eight agro-ecological zones (cool/humid, cool/subhumid, cool/semiarid, cool/arid, warm/humid, warm/subhumid, warm/semiarid and warm/arid) that are associated with the sorghum population structure (Amede et al., Reference Amede, Auricht, Boffa, Dixon, Mallawaarachchi, Rukuni and Teklewold-Deneke2015; Wondimu et al., Reference Wondimu, Dong, Paterson, Worku and Bantte2021). The lack of passport information in NPGS Ethiopian germplasm collection makes it difficult to determine which of the eight agro-ecological zones are represented. The tropical environmental conditions of this research included high relative humidity (77% ± 10) and warm temperature (24°C ± 0.6) that might represent the warm/humid zones of Ethiopia. We hypothesize that most of the NPGS Ethiopia germplasm might have been collected within the arid regions with minimal grain mould incidence. Likewise, the compact panicle shape associated with the Durra germplasm and prevalent in Ethiopian germplasm favours the development of fungi in the seeds. In fact, a study using the sorghum mini-core collection from the International Crops Research Institute for the Semi-arid Tropics (ICRISAT) found that panicle compactness is highly correlated with grain mould susceptibility (Sharma et al., Reference Sharma, Rao, Upadhyaya, Reddy and Thakur2010). In addition, high frequency of grain mould resistance germplasm was found in a subset of the NPGS Senegal collection where the loose panicle of Guinea race was the most prevalent (Cuevas et al., Reference Cuevas, Prom and Rosa-Valentin2018). Our results contrast with the evaluation for Ethiopian landraces from EBI at Bako Agriculture Research Center in Ethiopia where most of the accessions were classified as grain mould resistance germplasm based on seed deterioration (Nida et al., Reference Nida, Girma, Mekonen, Lee, Seyoum, Dessalegn, Tadesse, Ayana, Senbetay, Tesso, Ejeta and Mengiste2019). A recent genetic analysis showed that EBI collections is more genetically diverse than NPGS Ethiopian collections and includes different landraces (Girma et al., Reference Girma, Nida, Tirfessa, Lule, Bejiga, Seyoum, Mekonen, Nega, Dessalegn, Birhanu, Bekele, Gebreyohannes, Ayana, Tesso, Ejeta and Mengiste2020). Moreover, differences in environmental conditions and mycoflora populations (i.e. fungi species and/or strains) between studies, as well as the absence of seed emergence analysis for EBI germplasm could have contributed to different grain mould resistance results. The prior identification of the most likely resistance germplasm in the NPGS Ethiopia collection based on genetic profile could be an adequate approach to identify grain mould resistant germplasm.
The genetic variation for seedling emergence rate and seed deterioration was suitable to identify novel grain mould resistance loci. During pathogen invasion the plant immune system could recognize specific pathogen secreted effectors and microbe-associated molecular pattern (MAMP) molecules by intracellular resistant genes (R) and pattern recognition receptors, respectively. Both induce a signalling pathway that led to a plant defence response, but the MAMP-triggered immunity response render plants resistant to a wider range of pathogens (Boller and Felix, Reference Boller and Felix2009). Grain mould is a complex disease that involves multiple fungi species, and its resistance mechanism is likely to be associated to MAMP-triggered immunity response (Ackerman et al., Reference Ackerman, Wenndt and Boyles2021). The two candidate genes associated with seedling emergence rate are associated with MAMP immunity response system (phospholipase A2) and intracellular resistance protein (RPP13). The involvement of phospholipase activity in response to diverse abiotic and biotic stresses has been widely documented (Hong et al., Reference Hong, Zhao, Guo, Kim, Deng and Wang2016). In fact, phospholipase A2 has been reported to elicit a resistance reaction to Phytophthora infestans in potato (Solanum tuberosum L.) (Senda et al., Reference Senda, Doke and Kawakita1998) and play an important role in the hypersensitive reactions of Arabidopsis thaliana (Reina-Pinto et al., Reference Reina-Pinto, Voisin, Kurdyukov, Faust, Haslam, Michaelson, Efremova, Franke, Schreiber, JA and Yephremov2009). Other isoforms of phospholipase (e.g. phospholipase C4, phospholipase D) are also associated with the resistance response of tomato [Solanum lycopersicum (SI)] to Cladosporium fulvum (Vossen et al., Reference Vossen, Abd-El-Haliem, Fradin, Van Den Berg, Ekengren, Meijer, Seife, Bai, Ten Have, Munnik, Thomma and Joosten2010) and citrus [Citrus sinensis (L.) Osbeck] fruit to Penicillium digitatum (Lafuente et al., Reference Lafuente, Ballester, Holland, Cerveró and Romero2021). The R gene RPP13 encodes a protein containing a coiled-coil (CC) domain, nucleotide-binding site (NBS) and a leucine-rich repeat (LRR) region. This gene has been associated with resistance to downy mildew in A. thaliana (Bittner-Eddy and Beynon, Reference Bittner-Eddy and Beynon2001) and wheat (Liu et al., Reference Liu, Zhang, Zhang, Huang, Dang, Xie and Wang2020b). Most of the intracellular R gene encoding NBS and LRR domain belong to a superfamily of genes that provide resistance to pathogens through point mutations that led to the recognition of specific effectors proteins. Indeed, the rapid evolution of R genes led to new resistance reactions and the degeneration of susceptible alleles. An analysis of 346 NBS-encoding genes in sorghum found high nucleotide diversity driven by purifying and balance selections (Mace et al., Reference Mace, Tai, Innes, Godwin, Hu, Campbell, Gilding, Cruickshank, Prentis, Wang and Jordan2014). The candidate gene associated with seedling rating (Deoxycytidine kinase) is an intracellular protein involved in the salvage pathway for the nucleotide synthesis in plants (Liu et al., Reference Liu, You, Zhu, Chen, Hu, Gu, Liu, ZY, YH, SJ, LM, Liu, YL, Zhou, Jiang and JM2020a). This pathway plays important roles in seed germination (Jung et al., Reference Jung, Flörchinger, Kunz, Traub, Wartenberg and Jeblick2009) and embryo maturation (Stasolla et al., Reference Stasolla, Loukanina, Ashihara, Yeung and Thorpe2003). Therefore, it is possible that this gene is associated with the MAMP immunity response system. These candidate genes might be validated based on the transcriptome analysis of the resistance and susceptible accessions and through the development of high-throughput SNP markers to be tested in segregating populations. Our GWAS provides insight into the grain mould resistance response in sorghum. The results suggest that both the direct and indirect recognition of multiple pathogens involved in the grain mould diseases are required to activate the whole immunity system and boost its resistance response.
Germplasm improvement for multigenic inheritance traits is based on recurrent selection programmes where superior progenies are selected and recombined for consecutives breeding cycles (Fehr, Reference Fehr1991). The complexity of grain mould disease requires the stacking of multiple alleles to confer a resistant response. Herein, we identified five grain mould resistance Ethiopian tropical accessions that include resistance alleles which can be introduce into recurrent selection breeding programmes. However, the existence of multiple unsuitable agronomic traits (e.g. photoperiod, plant height, open panicle shape, etc.) among these tropical accessions restricts their acceptance in sorghum breeding programmes. Pre-breeding provides a unique opportunity to enhance the use of landraces genetic variability and desirable genes alleles in crop improvement programmes (Sharma et al., Reference Sharma, Upadhyaya, Varshney and Gowda2013; Byrne et al., Reference Byrne, Volk, Gardner, Gore, Simon and Smith2018). In this regard, the four most genetically distance tropical accessions could be initially crossed to temperate adapted breeding line with an excellent combining ability to develop a pre-breeding population. In this regard, the first two segregating generations (i.e. F 2 populations and F 2:3 families) should be focused on the selection of plant height, flowering time and other agronomical traits in fields with high pressure of grain mould disease. Later, these families could be tests for grain mould resistance in replicate trial to select and advance superior families (i.e. F 2:4 to F 2:6). This could lead to an intermediate set of lines with grain mould resistance that can be used in breeding programmes to broaden the genetic diversity of resistance in new varieties.
Conclusion
The NPGS Ethiopian germplasm collection includes grain mould resistant accessions in low frequency. The evaluation of 330 Ethiopian accessions identified two and three accessions with high seedling emergence rates and low seed degradation, respectively. GWAS identified three grain mould resistance loci in chromosome 1, 3 and 8 that explain a limited portion of the observed variation. Candidate gene analysis identified that these three loci include diseases resistance genes involved in pathogen recognition and signalling cascades of the plant immunity system. These five NPGS Ethiopian accessions (PI 457867, PI 454221, PI 455036, PI 455213 and PI 330821) could be used by a pre-breeding germplasm programme to develop new grain mould resistance cultivars.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1479262124000157
Acknowledgements
This research was funded by the USDA-ARS Current Research Information System (Project 6090-21000-053-00-D; HC)








