Introduction
The digenean family Emprostiotrematidae Cianferoni and Ceccolini, 2021 is the smaller of 2 families making up the Haploporoidea Nicoll, 1914, comprising 4 genera and just 12 species. While species of the Haploporidae Nicoll, 1914 are cosmopolitan and occur in marine and freshwater ecosystems, emprostiotrematids have been found only in marine and estuarine systems of the Indo-West Pacific marine region (Overstreet and Curran, Reference Overstreet and Curran2005; Bray et al., Reference Bray, Cribb, Waeschenbach and Littlewood2014; Andres et al., Reference Andres, Pulis and Overstreet2016; Huston et al., Reference Huston, Cutmore and Cribb2018a). Adult emprostiotrematids parasitize herbivorous or detritivorous fishes and are characterized morphologically by their possession of 2 caeca, 2 near-spherical testes that are symmetrically arranged, and vitelline follicles that form lobed aggregations in lateral fields (Overstreet and Curran, Reference Overstreet and Curran2005; Andres et al., Reference Andres, Pulis and Overstreet2016; Huston et al., Reference Huston, Cutmore and Cribb2018a). This study focuses on the type-genus for the family, Emprostiotrema Cianferoni and Ceccolini, 2021, which was recently proposed as a replacement name for the junior homonym Atractotrema Goto and Ozaki, Reference Goto and Ozaki1929 nec Cossmann, 1888 (see Ceccolini and Cianferoni, Reference Ceccolini and Cianferoni2021). Ceccolini and Cianferoni (Reference Ceccolini and Cianferoni2021) proposed new combinations for the 3 recognized species of the genus – Emprostiotrema fusum (Goto and Ozaki, Reference Goto and Ozaki1929) Cianferoni and Ceccolini, 2021; Emprostiotrema kuntzi (Ahmad, Reference Ahmad1985) Cianferoni and Ceccolini, 2021; and Emprostiotrema sigani (Durio and Manter, Reference Durio and Manter1969) Cianferoni and Ceccolini, 2021. Ceccolini and Cianferoni (Reference Ceccolini and Cianferoni2021) also proposed the replacement of the family group name Atractotrematidae with Emprostiotrematidae. All these modifications appear valid and are comprehensively adopted in this study.
All 3 species of Emprostiotrema have been reported from only rabbitfishes (Acanthuriformes: Siganidae: Siganus) and are distinguished from other emprostiotrematids by their possession of testes that are level with, or anterior to, the ventral sucker and an ovary that is nearly-to-distinctly post-testicular (Overstreet and Curran, Reference Overstreet and Curran2005). The first species to be described was E. fusum, from Siganus fuscescens (Houttuyn), collected from off Takamatsu in the Seto Inland Sea of Japan (Goto and Ozaki, Reference Goto and Ozaki1929). Emprostiotrema sigani was described from Siganus lineatus (Valenciennes), collected off Green Island on the central Great Barrier Reef, Australia and an unidentified Siganus sp. collected in New Caledonia (Durio and Manter, Reference Durio and Manter1969). The most recently described species, E. kuntzi, was described from Siganus argenteus Quoy and Gaimard (as Teuthis rostratus), collected in the Arabian Sea, from off the west coast of India (Ahmad, Reference Ahmad1985).
As part of a broad trematode sampling programme over the past 3 decades, a large number of emprostiotrematids were collected from siganids at locations across the Indo-west Pacific region. These new samples enable a re-examination of the Emprostiotrema species reported from the region, based on integrated morphological and molecular data. In this study, we propose 1 new species, synonymize 1 known species and demonstrate that E. fusum parasitizes a wide range of siganids and spans a broad geographic range in the western Pacific Ocean.
Materials and methods
Specimen collection and morphological analysis
Siganid fishes were wild caught or (rarely) purchased from fish markets from 1986 to 2024 from locations across the Indo-West Pacific (Fig. 1). Fishes were euthanized by cranial pithing or overdose of Aqui-S™ anaesthetic and dissected as per the protocols of Cribb and Bray (Reference Cribb and Bray2010). Trematodes were fixed without pressure in near-boiling saline and preserved in either 10% formalin or 70–80% ethanol. The validity of 1 host species, Siganus canaliculatus (Park), is controversial. The species, being sister to S. fuscescens, is extremely difficult to identify, with recent studies indicating minute (0.2%) genetic differences and instances of hybridization between the 2 (Zolkaply et al., Reference Zolkaply, Do, Asaduzzaman, Seah, Hurwood, Mather, Rahman and Wong2021). Iwamoto et al. (Reference Iwamoto, Abdullah, Chang, Yoshino and Imai2015) proposed synonymy of S. canaliculatus with S. fuscescens, though Zolkaply et al. (Reference Zolkaply, Do, Asaduzzaman, Seah, Hurwood, Mather, Rahman and Wong2021) indicate that more work needs to be done to resolve the relationship between the 2 taxa. Here we follow Eschmeyer's Catalog of Fishes (Fricke et al., Reference Fricke, Eschmeyer and Van der Laan2024) in considering it a valid species. Host common names used in the text follow those used by FishBase.

Figure 1. Map of known Indo-West Pacific collection localities for species of Emprostiotrema. Localities where specimens were obtained for the present study are indicated with a star symbol.
Specimens prepared for morphological analyses were washed in fresh water, stained using Mayer's haematoxylin solution, de-stained using a dilute hydrochloric acid solution, neutralized using a dilute ammonium hydroxide solution, dehydrated using a graded ethanol series, cleared in methyl salicylate and mounted on slides in Canada balsam. Measurements were made using cellSens™ version 1.13 imaging software and an Olympus SC50 digital microscope camera mounted on an Olympus BX-53 compound microscope. Measurements, unless otherwise noted, are reported in micrometres (μm), and are given as a range with the mean in parentheses. Additional morphological data were obtained for a syntype of E. fusum (housed in the Meguro Parasitological Museum, Tokyo, Japan; accession number E0502) by analysing photomicrographs graciously provided by Professor Kazuo Ogawa. Measurement data for 37 features (see Supplementary Material) were imported into R (https://www.R-project.org), log-transformed and examined with principal component analyses (PCAs) of the covariance matrix and visualized using the package ggfortify (Tang and Horikoshi, Reference Tang and Horikoshi2016). Only those specimens that had a complete suite of measurements (i.e. included reliable measurements of all 37 selected features) were included in the PCA. (Thus, poor-quality specimens and hologenophores were excluded.) Drawings were made using a camera lucida, mounted on an Olympus BX-53 compound microscope. Drawings were digitized in Adobe Illustrator CS6. Type and voucher specimens are lodged in the Queensland Museum, Brisbane, Australia (QM), Museum National d'Histoire Naturelles, Paris, France (MNHN), Meguro Parasitological Museum, Tokyo, Japan (MPM), Museum Zoologicum Bogoriense, Bogor, Java, Indonesia (MZB); accession numbers are presented in the taxonomic section of this manuscript.
Molecular sequencing and phylogenetic analysis
Molecular data were generated for 3 markers: the large subunit (28S), the second internal transcribed spacer (ITS2) of ribosomal RNA and the mitochondrial cytochrome c oxidase I (cox1) gene. Molecular specimens used were processed as hologenophores or whole paragenophores (sensu Pleijel et al., Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008). Genomic DNA were extracted using a modified phenol/chloroform technique (Sambrook and Russell, Reference Sambrook and Russell2001). Primers, amplification and sequencing of the ITS2 and 28S regions follow Yong et al. (Reference Yong, Cutmore, Bray, Miller, Semarariana, Palm and Cribb2016) and of the cox1 region followed Wee et al. (Reference Wee, Cribb, Bray and Cutmore2017). Dual direction Sanger sequencing was performed at the Australian Genome Research Facility, Brisbane. Contiguous sequences were assembled and edited with Geneious® version 11.0.5.
The cox1 and ITS2 datasets generated during this study were each aligned in MEGA 7 (Kumar et al., Reference Kumar, Stecher and Tamura2016), with UPGMA clustering for iterations 1 and 2; there are no previously published sequence data for either gene region for species of Emprostiotrema available on GenBank. The cox1 alignment was transferred to Mesquite v.3.31 (Maddison and Maddison, Reference Maddison and Maddison2024), translated (echinoderm/flatworm mitochondrial code) and inspected for internal stop codons. All trimmed cox1 sequences were 474 base positions (bp). All codon positions in the cox1 dataset were evaluated for substitution saturation using the ‘Test of substitution saturation by Xia et al.’ function (Xia et al., Reference Xia, Xie, Salemi, Chen and Wang2003; Xia and Lemey, Reference Xia, Lemey, Lemey, Salemi and Vandamm2009) as implemented in DAMBE v. 7.2 (Xia, Reference Xia2018); no significant substitution saturation was detected. Neighbour-joining analyses were conducted for each dataset using MEGA 7, with the following parameters: ‘Model/Method = No. of differences’, ‘Substitutions to Include = d: Transitions + Transversions’, ‘Rates among Sites = Gamma Distributed’ and ‘Gaps/Missing Data Treatment = Pairwise deletion’. Nodal support for the neighbour-joining analyses was estimated by performing 10 000 bootstrap replicates.
The partial 28S rDNA data generated during this study were aligned with emprostiotrematid sequence data available on GenBank (Table 1) using MUSCLE version 3.7 (Edgar, Reference Edgar2004) run on the CIPRES portal, with ClustalW sequence weighting and UPGMA clustering for iterations 1 and 2. The resultant alignment was refined by eye using Mesquite v.3.31; the ends of the alignment were trimmed, and indels constituting more than 3 bp and present in greater than 5% of the sequences in the dataset were removed. Bayesian inference analysis was performed using MrBayes version 3.2.7 (Ronquist et al., Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012) and maximum likelihood analysis using RAxML version 8.2.12 (Stamatakis, Reference Stamatakis2014), both run on the CIPRES portal. The best nucleotide substitution model was estimated using jModelTest version 2.1.10 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012). Both the Akaike information criterion and Bayesian information criterion predicted the GTR + I + G as the best estimator; Bayesian inference and maximum likelihood analyses were conducted using the closest approximation to this model. Nodal support in the maximum likelihood analysis was estimated by performing 1000 bootstrap pseudoreplicates. Bayesian inference analysis was run over 10 000 000 generations (ngen = 10 000 000) with 2 runs each containing 4 simultaneous Markov Chain Monte Carlo (MCMC) chains (nchains = 4) and every 1000th tree saved. Bayesian inference analysis used the following parameters: ‘nst = 6’, ‘rates = invgamma’, ‘ngammacat = 4’, and the prior parameters of the combined dataset were set to ‘ratepr = variable’. Samples of substitution model parameters, and tree and branch lengths were summarized using the parameters ‘sump burnin = 3000’ and ‘sumt burnin = 3000’. Species of the Haploporidae were designated as functional outgroup taxa.
Table 1. Sequence data from GenBank included in this study

Species recognition criteria
Species were distinguished using, as a starting point, the criteria for species delineation proposed by Bray et al. (Reference Bray, Cutmore and Cribb2022).
Results
Collections
Specimens consistent with Emprostiotrema were collected from siganid fishes from 8 localities: Lizard Island on the northern Great Barrier Reef; off Bali in central Indonesia; off Nouméa, New Caledonia; from the main lagoon of Palau atoll; Okinawa Island in southern Japan; and from locations in 3 archipelagos in French Polynesia (Rangiroa, Tuamotu Archipelago; Moorea, Society Archipelago; Mangareva, Gambiers Archipelago). No emprostiotrematids were found in substantial samples of siganids from Heron Island on the southern Great Barrier Reef (n = 250), Moreton Bay in southeastern Queensland (n = 110) or Ningaloo Reef, Western Australia (n = 47). Table 2 summarizes the collections.
Table 2. Siganids examined at Indo-Pacific localities together with the prevalence of 2 species of Emprostiotrema

Number of fish followed by number of infected fish in parentheses. Legend: BA, Bali; NI, Ningaloo; HI, Heron Island; LI, Lizard Island; MB, Moreton Bay; NC, New Caledonia; OK, Okinawa; PA, Palau; GA, Gambiers; MA, Marquesas; MO, Moorea; RA, Rangiroa. The single Bali and all eastern Pacific records relate to E. gotozakiorum n. sp. All Central Pacific records relate to E. fusum except for in S. argenteus where ‘a’ denotes the prevalence of E. fusum and ‘b’ denotes E. gotozakiorum n. sp.
Molecular results
A total of 78 cox1 sequences were generated for samples from all 8 localities, representing 13 host/locality combinations. Neighbour-joining analysis of these data (Fig. 2) resolved 9 lineages that differ by a minimum P-distance of 2.11% (10 bp), which we treat as 9 initial operational taxonomic units (OTUs A–I). All but 2 lineages were represented by multiple sequences, with OTU replication varying from 2 to 38 sequences. Inter-OTU P-distance differences ranged from 2.32 to 17.10% (11–81 bp) and intra-OTU variation ranged from 0 to 1.26% (0–6 bp). All 9 OTUs were restricted to single geographical localities with the exception of OTU I, which was found at sites in 3 French Polynesia archipelagos (Gambiers, Tuamotu and Society). The OTUs formed 2 major and deeply divided clades, clade 1 comprising 6 OTUs and clade 2 comprising 3 OTUs; these 2 clades differ by 13.10–17.10% (62–81 bp). Clade 1 corresponds to sequences of most specimens from Lizard Island (OTUs A, C and E), and all specimens from Palau (OTU B), New Caledonia (OTU D) and Japan (OTU F); inter-OTU P-distance differences within clade 1 range from 3.37 to 7.59% (16–36 bp). Clade 2 corresponds to all sequences of French Polynesian specimens (OTU I), 2 specimens from Lizard Island (OTU G) and the single sequenced specimen from Bali (OTU H); inter-OTU P-distance differences within clade 2 range from 2.32 to 7.38% (11–35 bp). Lizard Island was the only site from which multiple OTUs were found, with samples from this site resolving in 3 OTUs in clade 1 (OTUs A, C and E) and 1 OTU from clade 2 (OTU G). Notably, combinations of specimens relating to the 4 Lizard Island OTUs were found in total sympatry (same individual fish host).

Figure 2. Phylogram from the unrooted neighbour-joining analysis of the cytochrome c oxidase subunit 1 (cox1) mtDNA dataset. Strongly supported nodes (>80) are indicated by a filled circle. The scale bar indicates the number of base differences.
A total of 34 ITS2 sequences were generated, representing 8 of the 9 cox1 OTUs; no ITS2 data could be generated for OTU G (Bali). Neighbour-joining analysis of these data (Fig. 3) resolved the same 2 major clades as in the cox1 analysis with P-distance differences 1.40–2.10% (6–9 bp). These clades had less geographic structuring than was seen from the cox1 data. Sequences relating to clade 1 formed genotypes which differed by 0.23–1.40% (1–6 bp). Of 13 samples from Lizard Island, 11 were identical including multiple samples for each of OTUs A and E, which differed by 4.43–5.27% (21–25 bp) for the cox1 dataset. The remaining Lizard Island sequences were identical and differed from all others by 0.23% (1 bp). Four sequences from New Caledonia (OTU D) were identical and differed from the Lizard Island sequences by 0.23–0.47% (1–2 bp). Samples from Palau and Japan (OTUs B and F) were identical and differed from all other clade 1 sequences by 0.93–1.40% (4–6 bp). All sequences relating to clade 2, 10 sequences from French Polynesia (OTU I) and 2 sequences from Lizard Island (OTU G), were identical.

Figure 3. Phylogram from the unrooted neighbour-joining analysis of the internal transcribed spacer (ITS2) rDNA dataset. Strongly supported nodes (>80) are indicated by a filled circle. The scale bar indicates the number of base differences.
28S data were generated from specimens from 7 of the 9 cox1 OTUs; no sequence data could be generated for OTU D (New Caledonia) or OTU H (Bali). Maximum likelihood and Bayesian inference analyses of the 28S dataset (Fig. 4) resolve Emprostiotrema as monophyletic, with sequences representing clades 1 and 2 forming well-supported clades that differ by 1.42–1.74% (18–22 bp). Sequences of OTUs A and E (both Lizard Island) were largely identical in the 28S dataset, with most differing inconsistently at a maximum of a P-distance of 0.08% (1 bp) and 2 sequences differing by 0.32–0.40% (4–5 bp). Sequences of OTUs B and F (Palau and Japan) formed a well-supported clade, differing from those from Lizard Island by 0.40–0.79% (5–10) bp. Within clade 2, there was no variation among sequences relating to OTU I (3 French Polynesian sites); the 2 sequences of OTU G (Lizard Island) differ from the French Polynesian data by 0.08% (1 bp).

Figure 4. Relationships between species of the Emprostiotrematidae based on maximum likelihood phylogenetic analysis of the 28S dataset. Strongly supported nodes (Bayesian posterior probabilities >0.8 and maximum likelihood bootstrap values >80) are indicated by a filled circle. The scale-bar indicates expected number of substitutions per site.
Morphological results
Specimens from Bali (OTU H) were excluded from morphological analysis as they had clearly been dead for some time at the point of fixation and had a distinctly distorted form. No hologenophore specimens were available for the OTUs C and G (both Lizard Island). Thus, morphological analyses were based on specimens representing 6 of the 9 OTUs (A, B, D, E, F and I), with a total of 222 specimens considered. Based on the overwhelmingly weighted molecular data (of the 52 Lizard Island specimens, just 2 specimens, not represented by hologenophores, belonged to clade 2), all Great Barrier Reef specimens were interpreted as relating to clade 1 (OTUs A, C or E).
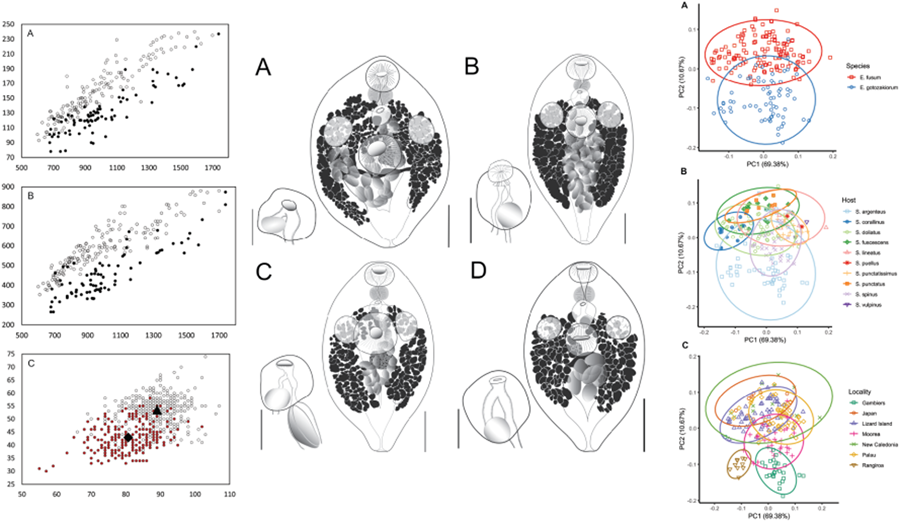
All specimens examined were broadly comparable, with no immediate distinctions suggesting the presence of multiple species. In the light of the molecular results, however, we considered a range of characters for samples from French Polynesia (OTU I) and all other sites (OTU A–F). Plotting of characters relative to body length and identified as relating to the 2 major clades suggest some morphological distinction; Fig. 5A shows body length vs width, Fig. 5B shows body length vs oral sucker width, and Fig. 5C shows egg size. In each there is a clear though incomplete separation between specimens relating to the 2 clades. Inspection of samples and plotting of measurements did not suggest any further morphological distinctions for subdivisions of the 2 clades.

Figure 5. Morphometric comparison of Emprostiotrema fusum and E. gotozakiorum n. sp. (A) body length vs body width; (B) body length vs oral sucker width; (C) egg length vs width. Filled circle data points represent E. fusum; open circle data points represent E. gotozakiorum n. sp. The triangle in B represents mean egg size for E. fusum; the diamond represents mean egg size for E. gotozakiorum n. sp.
PCA analysis
PCA analysis of morphometric data was considered relative to the 2 major clades as identified by molecular analyses (Fig. 6A), host (Fig. 6B) and collection locality (Fig. 6C). The 95% confidence ellipses based on host suggested no informative break-up of the data. Ellipses based on locality suggested no distinction between localities in the western Pacific (Japan, Lizard Island, New Caledonia, Palau), some distinction between specimens from 2 French Polynesian localities (Rangiroa and Gambiers), and distinction between specimens from these 2 localities and all other more western Pacific localities. However, the Rangiroa and Gambiers ellipses overlapped with the Moorea ellipse, and in turn, the Moorea ellipse overlapped with all other western Pacific localities. The analysis based on just the 2 major clades (clade 1: Japan, Palau, Lizard Island and New Caledonia; clade 2: French Polynesia – 3 localities) produced a separation with overlap of ellipses between only 26% of the data points.

Figure 6. Principal component analysis (PCA) of morphometric data obtained from adult specimens of Emprostiotrema from multiple localities in the Indo-West Pacific. (A) Data points coded by species clade as informed by molecular data; (B) data points coded by host; (C) data points coded by collection locality. Ellipses represent 95% confidence levels.
Synthesis
Integrated consideration of host, molecular, morphological and morphometric analyses leads us to interpret the new specimens as representing 2 species. We conclude that host identity is uninformative in this system. Notably, S. argenteus is infected by specimens of both species at Lizard Island. PCA analysis also suggested no distinctions on the basis of host identity. Molecular data suggested a consistent division between clade 1 specimens (from Japan, Palau, New Caledonia and all but 2 from the Great Barrier Reef) relative to clade 2 specimens (from French Polynesia, Bali and 2 from Lizard Island). Sequences belonging to clade 1 have variable geographic division, with strong division in the cox1 dataset and limited division in the ITS2 and 28S datasets. Sequences belonging to clade 2 had strong geographic divisions in the cox1 dataset, and none or almost none in the ITS2 and 28S datasets, respectively. These levels of distinction within the 2 major clades, in the absence of correlation with morphological or host differences, are best interpreted as intra-specific variation. Morphology, in the form of 3 individual characters (body length vs body width; body length vs oral sucker width; egg length vs egg width), suggests distinction between specimens from Japan, Palau, New Caledonia and the Great Barrier Reef (consistent with clade 1) and those from the 3 French Polynesian sites (consistent with clade 2). PCA analysis suggested some division on the basis of geographical origin within French Polynesia. However, we infer that these distinctions cannot be interpreted as correlating with species distinctions, because separation within French Polynesian localities is not supported by any other data. Notably, cox1 sequences are now well-established to frequently show regional population differences in Indo-Pacific fish-infecting trematodes (Huston et al., Reference Huston, Cutmore, Miller, Sasal, Smit and Cribb2021; Bray et al., Reference Bray, Cutmore and Cribb2022; Cutmore and Cribb, Reference Cutmore and Cribb2022; Cutmore et al., Reference Cutmore, Corner and Cribb2023; Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Solórzano-García, Huston, Mendoza-Garfias, Cabañas-Granillo, Cutmore and Cribb2024), as seen here among clade 1 samples; the absence of differences between the 3 French Polynesian sites is thus intriguing. We infer that the PCA distinctions found between French Polynesian sites reflect either stochastic variation based on slight differences in handling, age of the parasites or geographical phenotypic plasticity.
Based on the integrated data, clade 1 specimens are identified as E. fusum. Notably, clade 1 includes new specimens from the type-host and relatively close to the type-locality of E. fusum [described by Goto and Ozaki (Reference Goto and Ozaki1929) from S. fuscescens off Takamatsu, Seto Inland Sea, Japan]; our new specimens from S. fuscescens, collected off Okinawa (approximately 1000 km to the south), conform closely to the original description of this species. Study of photographs of a syntype of E. fusum collected by Goto and Ozaki (MPM accession number E0502) further supports the conclusion that the new specimens from Okinawa are best interpreted as E. fusum. Durio and Manter (Reference Durio and Manter1969) described E. sigani based on specimens from S. lineatus caught off Green Island, central Great Barrier Reef, and from an unidentified Siganus sp. caught off Nouméa, New Caledonia. All new specimens of Emprostiotrema from S. lineatus from the Great Barrier Reef and from New Caledonia belong to clade 1. These new specimens correspond closely to the description of E. sigani of Durio and Manter (Reference Durio and Manter1969). Measurements provided by Durio and Manter (Reference Durio and Manter1969) to distinguish E. sigani completely overlap with those from E. fusum when our expanded dataset is considered, and we recognize E. sigani as a junior synonym of E. fusum.
Clade 2 is identified as a new species, which is recognizable on the basis of clear molecular distinctions and moderately clear morphological distinction. Notably, E. fusum and the new species were both collected from the same host species (and the same host individual) at Lizard Island, but there is as yet no evidence of overlap elsewhere in the range of the 2 species. We have not detected any morphological specimens consistent with the new species among the 58 mounted specimens from Lizard Island and conclude that it is an exceptionally rare species there.
Taxonomy
Family Emprostiotrematidae Cianferoni and Ceccolini, 2021
Genus Emprostiotrema Cianferoni and Ceccolini, 2021
Homonym: Atractotrema Goto and Ozaki, Reference Goto and Ozaki1929
Syn Atractotrema Goto and Ozaki, Reference Goto and Ozaki1929 nec Cossmann, 1888
Emprostiotrema fusum (Goto and Ozaki, Reference Goto and Ozaki1929) Cianferoni and Ceccolini, 2021 (Fig. 7A–D)

Figure 7. Plate showing whole-body ventrally mounted representatives of Emprostiotrema fusum and their terminal genitalia (hermaphroditic sacs), from (A) Japan ex Siganus fuscescens (accession no. MPM25300); (B) Lizard Island ex S. doliatus (QM G241330); (C) New Caledonia ex S. doliatus (MNH HEL 3003/MNH HEL 3001); and (D) Palau ex S. punctatissimus (QM G241418). Whole-mount scale-bars: A, B and D – 200 μm; C – 300 μm. Terminal genitalia scale-bars: A and B – 100 μm; C – 75 μm; D – 50 μm.
Syn: Atractotrema fusum Goto and Ozaki, Reference Goto and Ozaki1929; Atractotrema sigani Durio and Manter, Reference Durio and Manter1969; E. sigani (Durio and Manter, Reference Durio and Manter1969) Cianferoni and Ceccolini, 2021.
Type-host: Siganus fuscescens (Houttuyn), dusky spinefoot (Acanthuriformes: Siganidae).
Type-locality: Takamatsu, Kagawa Prefecture, Seto Inland Sea, Japan.
Reports: Goto and Ozaki (Reference Goto and Ozaki1929); Yamaguti (Reference Yamaguti1939); Durio and Manter (Reference Durio and Manter1969); Machida et al. (Reference Machida, Ichihara and Kamegai1970); Shen (Reference Shen1990); Cribb et al. (Reference Cribb, Bray, Littlewood, Pichelin, Herniou, Littlewood and Bray2001); Olson et al. (Reference Olson, Cribb, Tkach, Bray and Littlewood2003); Bakhoum et al. (Reference Bakhoum, Quilichini, Justine, Bray, Miquel, Feliu, Bâ and Marchand2015); Motson et al. (Reference Motson, Hutson and Hoey2023).
New specimens
New hosts: Siganus argenteus (Quoy and Gaimard), streamlined spinefoot; Siganus corallinus Valenciennes, blue-spotted spinefoot; Siganus puellus (Schlegel), masked spinefoot; Siganus punctatissimus Fowler and Bean, peppered spinefoot; Siganus punctatus (Schneider and Forster), gold-spotted spinefoot; Siganus spinus (Linnaeus), little spinefoot; Siganus vulpinus (Schlegel and Müller), foxface (Acanthuriformes: Siganidae).
Known hosts: Siganus doliatus Guérin-Méneville, barred spinefoot; S. fuscescens, S. lineatus (Valenciennes), golden-lined spinefoot (Acanthuriformes: Siganidae).
New localities: off Nouméa, New Caledonia, Coral Sea (22°20′ S, 166°22′ E); off Lizard Island, northern Great Barrier Reef, Australia (14°40′ S, 145°27′ E); southern Palau lagoon, Palau (7°30′ N, 134°30′ E); Tomari fish market (26°13′ N, 127°40′ E), Naha, Okinawa Prefecture, Japan.
Voucher material: 136 voucher specimens (QM G 241301–440; MNHN 3001–4; MPM 25300).
Prevalence: See Table 2.
Representative DNA sequences: cox1 mtDNA, 59 sequences (34 submitted to GenBank, PQ268408–41); ITS2 rDNA, 23 sequences (10 submitted to GenBank, PQ260955–64); partial 28S rDNA, 13 sequences (10 submitted to GenBank, PQ260973–82).
Description
[Based on 136 whole or partial, dorso-ventrally mounted specimens from Japan, Palau, Australia and New Caledonia. An extended set of measurements and indices covering 15 host/locality combinations are shown in Supplementary Table 1.] Body fusiform to lachrymiform, tapering anteriorly and posteriorly, 600–1699 × 353–880 (979 × 586); 1.4–2.2 (1.7) times longer than wide. Length of body post-point of maximal breadth 256–874 (476) or 38.6–60.5% (48.7%) of total body length. Tegumental spines <1 long, barely visible by light microscopy, arranged in transverse rows which wrap around entirety of body. Tegumental papillae or bristles not detected. Forebody 154–548 (293) long or 17.1–49.3% (30.1%) of total body length. Oral sucker prominent, ovoid, 74–210 × 93–255 (129 × 159), length/width ratio 0.6–1.3 (0.8). Mouth subterminal. Prepharynx not visible posterior to oral sucker (i.e. 0 length) in over half of specimens; when visible, length 3–44 (5); straight distance from mouth to pharynx 24–157 (69). Pharynx prominent, ovoid, 60–148 × 69–176 (91 × 105). Oesophagus variously conspicuous, sometimes barely discernible, straight or sinuous when visible, 0–103 (37) long, occupying 0–8.7% (3.9%) of total body length. Intestine bifurcates in anterior third of body, caeca run parallel to lateral contours of body, subequal in length in most specimens; left caecum 280–1074 (534); right caecum 273–1117 (542); longer caecum occupying 44.0–68.6% (56.1%) of body length. Ventral sucker prominent, ovoid to round, not pedunculate, 115–318 × 115–318 (197 × 193), length/width ratio 0.8–1.3 (1.0). Testes subspherical to spherical, entire, opposite, variously lateral to antero-lateral to ventral sucker, 72–237 × 66–197 (139 × 125), 158–533 (312) from anterior extremity, 331–1078 (580) from posterior extremity. Pre-testicular space 17.5–46.3% (31.3%) of total body length; post-testicular space 44.2–68.1% (58.0%) of total body length. Majority of male genital tract not observable until entering hermaphroditic sac. Hermaphroditic sac subspherical to diamond-shaped, containing seminal vesicle, generally medial, at level intermediate between pharynx and ventral sucker, 63–191 × 56–151 (116 × 102), 37–327 (197) or 6.1–31.7% (20.2%) of total body length from anterior extremity, 137–1178 (649) or 17.8–81.3% (65.4%) of total body length from posterior extremity. Seminal vesicle spherical, positioned in posterior region of hermaphroditic sac, 20–83 × 23–83 (43 × 48). Cirrus or other male intromittent organ absent. Male and female ducts confluent just posterior to common genital pore; no expansion or genital atrial formation apparent. Genital pore simple, medial, 47–357 (217) from lateral margins, 41–387 (206) or 6.7–31.0% (21.1%) of total body length from anterior extremity, 258–1354 (744) or 42.4–88.5% (75.6%) of total body length from posterior extremity. Ovary oblong, submedial, at or just posterior to level of testes and ventral sucker, 38–163 (91) × 37–183 (91 × 86). Oviduct and seminal receptacle not visible in any specimens. Oötype only visible if developing egg present, immediately posterior (and sometimes sinistral) to ovary. Uterus occupying intercaecal region of body, with eggs primarily concentrated posterior to testes, ovary and ventral sucker. Eggs oblong, 1–70 (17) in number, 63–107 × 38–74 (89 × 54), length/breadth ratio 1.3–2.5 (1.7). Vitelline follicles form variably sized clumps in 2 lateral fields, often anteriorly confluent, extending anteriorly to level of testes (just anteriorly exceeding testes in minority of specimens) and posteriorly to level of caecal termini, 110–413 (232) from anterior extremity, 40–366 (187) from posterior extremity, dorsoventrally overlapping and often obscuring caeca, testes and portions of uterus. Vitelline ducts arise from anterior portion of vitelline fields and unite medially, posterior to ovary. Excretory vesicle V-shaped, 319–1326 (663) long, arms pass parallel to lateral margins of body, closely overlapping caeca, terminating variously well anterior to immediately posterior margin of testes.
Remarks
The original description of E. fusum by Goto and Ozaki (Reference Goto and Ozaki1929) is somewhat lacking in detail, but the accompanying figure is of high quality and clearly shows the nature of the species. No holotype was designated by Goto and Ozaki (Reference Goto and Ozaki1929) in their original description; instead, 5 specimens were used for a syntype series (K. Ogawa, personal communication). Notably, the overall body size and dimensions of many of the features described by Goto and Ozaki (Reference Goto and Ozaki1929), and those of the syntype examined by us, are much larger than those seen in our specimens. This may be an artefact of flattening of specimens during fixation, a common practice in trematode taxonomy at the time Goto and Ozaki were active (see Huston et al., Reference Huston, Cutmore and Cribb2019). Beyond this general size difference, our specimens conform almost exactly in body plan and in all morphological features.
Durio and Manter (Reference Durio and Manter1969) differentiated E. sigani from E. fusum on the basis of a larger egg size, longer prepharynx and a body length half that of E. fusum. In our expanded concept of E. fusum, we find that the described dimensions of E. sigani, including body length and egg size, fall within the range of those of E. fusum. Indeed, Durio and Manter's description of egg size for E. sigani (80–103 × 50–62 μm) extensively overlapped those for E. fusum (82–88 × 50–58 μm), and merely indicated the presence of (and potential for) slightly larger eggs. When considered in light of the new collections from the type- and other locations, these differences disappear altogether. In addition, we consider prepharynx length a weak feature for species delineation, as it varies widely depending on how each specimen contracts when fixed. Most specimens we collected from the Great Barrier Reef, New Caledonia and Japan did not have a prepharynx visible posterior to the oral sucker. We therefore do not regard this, or any other feature used to differentiate E. sigani from E. fusum, as valid, and see no reason to consider them as separate species.
Emprostiotrema gotozakiorum n. sp. (Figs 8A–B and 9A–D)

Figure 8. Emprostiotrema gotozakiorum n. sp. ex Siganus argenteus. (A) Whole-body ventral mount, holotype (accession no. MNH HEL3011). (B) Terminal genitalia (hermaphroditic sac), paratype (MNH HEL3009). Abbreviations: Cc, caeca; EV, excretory vesicle; GP, genital pore; HS, hermaphroditic sac; Od, oviduct; OS, oral sucker; Ov, ovary; Ph, pharynx; Pr, prepharynx; s.d., seminal duct; SV, seminal vesicle; Tt, testis; VD, vitelline duct; VS, ventral sucker. Scale-bars: A – 200 μm, B – 75 μm.

Figure 9. Plate showing whole-body ventrally mounted representatives of Emprostiotrema gotozakiorum n. sp. and their terminal genitalia (hermaphroditic sacs), from (A) Rangiroa, French Polynesia ex Siganus argenteus (MNH HEL3011/MNH HEL3009); (B) Moorea, French Polynesia ex Siganus spinus (MNH HEL3018/MNH HEL3026); (C) Mangareva, French Polynesia ex S. argenteus (MNH HEL3047); and (D) Bali, Indonesia ex Siganus canaliculatus (MZB Tr. 268). Whole-mount scale-bars: A, C, D – 200 μm, B – 250 μm. Terminal genitalia scale-bars: A, B, C – 75 μm, D – 100 μm.
Type-host: Siganus argenteus (Quoy and Gaimard), streamlined spinefoot (Acanthuriformes: Siganidae).
Additional hosts: Siganus canaliculatus (Park), white-spotted spinefoot; S. spinus (Linnaeus), little spinefoot (Acanthuriformes: Siganidae).
Type-locality: Rangiroa Atoll, Tuamotu Archipelago, French Polynesia (15°06′ S, 147°40′ W).
Additional localities: Moorea, Society Archipelago, French Polynesia (17°32′ S, 149°46′ W); Mangareva, Gambiers Archipelago, French Polynesia (23°07′ S, 134°58′ W); off Lizard Island, northern Great Barrier Reef, Australia (14°40′ S, 145°27′ E); off Kedonganan Beach, Jimbaran, southern Bali, Indonesia (8°46′ S, 115°08′ E).
Type-material: Holotype (MNHN HEL3005) and 12 paratypes (including 3 hologenophores) from Rangiroa, French Polynesia (MNHN 3006–64).
Additional vouchers: 69 vouchers from Gambiers and Moorea, French Polynesia (QM G241441–480); 3 vouchers from Bali, Indonesia (MZB Tr. 268–270).
Prevalence: See Table 2.
Representative DNA sequences: Representative DNA sequences: cox1 mtDNA, 19 sequences (14 submitted to GenBank, PQ268442–55); ITS2 rDNA, 12 sequences (4 submitted to GenBank, PQ260965–68); partial 28S rDNA, 8 sequences (4 submitted to GenBank, PQ260969–72).
ZooBank accession: http://zoobank.org/NomenclaturalActs/6B856044-CFFB-41FD-ABA5-05E96A4DF247
Etymology: The species name is a compound of the names of 2 eminent Japanese parasitologists, Seitaro Goto and Yoshimasa Ozaki. This name honours their contributions to parasite taxonomy and their role as describers of the type-species of the Emprostiotrematidae, Atractotrema (now Emprostiotrema) fusum.
Description
[Based on 82 whole or partial, dorso-ventrally mounted specimens from Rangiroa, Gambiers and Moorea, French Polynesia (specimens from Bali, Indonesia were excluded from consideration). An extended set of measurements and indices covering 5 host/locality combinations are shown in Supplementary Table 1]. Body fusiform, tapering gently anteriorly and distinctly posteriorly, 677–1738 × 265–873 (1019 × 472), 1.5–2.8 (2.3) times longer than wide. Length of body post-point of maximal breadth 300–1060 (585) or 37.0–69.4% (57.3%) of total body length. Tegumental spines <1 long, barely visible by light microscopy, arranged in transverse rows which wrap around entirety of body. Tegumental papillae or bristles not detected. Forebody 210–594 (322) long or 24.8–41.7% (30.9%) of total body length. Oral sucker prominent, 65–202 × 78–237 (113 × 135), length/width ratio 0.6–1.3 (0.8). Mouth subterminal. Prepharynx not visible posterior to oral sucker (i.e. 0 length) in 7 of 82 (8.5%) specimens; when visible, length 2–67 (21); straight distance from mouth to pharynx 14–95 (66). Pharynx prominent, ovoid, 67–199 × 68–200 (104 × 108). Oesophagus variously conspicuous, straight or sinuous when visible, 19–93 (49) long, occupying 2.7–7.5% (4.7%) of total body length. Intestine bifurcates in anterior third of body, caeca run parallel to lateral contours of body, subequal in length in most specimens; left caecum 336–984 (581), right caecum 372–1036 (587); longer caecum occupying 45.9–70.3% (57.6%) of body length. Ventral sucker prominent, ovoid to round, not pedunculate, 124–352 × 117–366 (194 × 192), length/width ratio 0.8–1.3 (1.0). Testes subspherical to spherical, entire, paired, lateral, at level of ventral sucker, 65–262 × 47–207 (132 × 106), 236–740 (371) from anterior extremity, 262–1010 (584) from posterior extremity. Pre-testicular space 28.7–42.3% (34.6%) of total body length; post-testicular space 33.8–63.9% (55.2%) of total body length. Majority of male genital tract not observable until entering hermaphroditic sac. Hermaphroditic sac subspherical to diamond-shaped, containing seminal vesicle, generally medial, at level intermediate between pharynx and ventral sucker, 78–275 × 61–246 (137 × 117), 144–425 (241) or 17.2–30.6% (23.2%) of total body length from anterior extremity, 405–1189 (665) or 55.5–77.7% (64.6%) of total body length from posterior extremity. Seminal vesicle spherical, positioned in posterior region of hermaphroditic sac, 32–89 × 32–111 (54 × 52). Cirrus or other male intromittent organ absent. Male and female ducts confluent just posterior to common genital pore; no expansion or atrial formation apparent. Genital pore simple, medial, 165–459 (259) or 19.2–33.0% (24.9%) of total body length from anterior extremity, 460–1314 (769) or 64.7–83.3% (74.9%) of total body length from posterior extremity. Ovary oblong, submedial, at or just posterior to level of testes and ventral sucker, 49–245 × 38–152 (97 × 72). Oviduct and seminal receptacle not visible in any specimens. Oötype not visible in any specimens. Uterus occupying intercaecal region of body, with eggs primarily concentrated posterior to testes, ovary and ventral sucker. Eggs oblong, 1–77 (13) in number, 55–102 × 30–58 (81 × 43), length/breadth ratio 1.4–2.5 (1.9). Vitelline follicles form variably sized clumps in 2 lateral fields, occasionally anteriorly confluent, extending anteriorly to level of testes (just anteriorly exceeding testes in minority of specimens) and posteriorly to level of caecal termini, 137–666 (313) from anterior extremity, 93–412 (215) from posterior extremity, dorsoventrally overlapping and often obscuring caeca, testes and portions of uterus. Vitelline ducts arise from anterior portion of vitelline fields and unite medially, posterior to ovary. Excretory vesicle V-shaped, arms pass parallel to lateral margins of body, closely overlapping caeca, terminating immediately posterior to posterior margin of testes, 635–997 (822) long.
Remarks
Emprostiotrema gotozakiorum n. sp. is distinguished from its congeners by its body shape. Although all species of Emprostiotrema are generally spindle-to-teardrop-shaped, A. gotozakiorum typically has a slenderer body overall and a distinctly tapering hindbody, which leads to the highest mean length/maximum width ratio of any Emprostiotrema species [2.3 vs 1.7 for E. fusum (this study) and ~2.0 for E. kuntzi]. Correspondingly, A. gotozakiorum has a greater body length post-maximum breadth (both in absolute terms and as a proportion of total body length) and a greater distance of forebody features (ventral sucker, genital pore, hermaphroditic sac) to the posterior extremity than all other Emprostiotrema species.
Discussion
Species recognition
The incorporation of DNA sequence data in taxonomic studies of trematodes has enabled a nuanced interpretation of species delimitation over wide geographical ranges. Widespread but geographically structured species have now been characterized for multiple trematode families in the Indo-west Pacific (Huston et al., Reference Huston, Cutmore, Miller, Sasal, Smit and Cribb2021; Bray et al., Reference Bray, Cutmore and Cribb2022; Cutmore and Cribb, Reference Cutmore and Cribb2022; Cutmore et al., Reference Cutmore, Corner and Cribb2023). However, in some cases, different markers have suggested conflicting interpretations, complicating species delimitation (e.g. Cutmore et al., Reference Cutmore, Yong, Reimer, Shirakashi, Nolan and Cribb2021; Cribb et al., Reference Cribb, Bray, Justine, Reimer, Sasal, Shirakashi and Cutmore2022; Wee et al., Reference Wee, Cribb, Shirakashi and Cutmore2022). Overall, it is now clear that variability in the highly discriminant markers itself varies among families, genera and even species of single genera; there is seemingly no reliable yardstick that can presently be applied to delineate otherwise cryptic trematode species (e.g. Cutmore et al., Reference Cutmore, Yong, Reimer, Shirakashi, Nolan and Cribb2021, Reference Cutmore, Corner and Cribb2023; Huston et al., Reference Huston, Cutmore, Miller, Sasal, Smit and Cribb2021). In the current study, samples relating to E. fusum showed incongruence between the genes analysed. For the ribosomal regions, samples from Australia + New Caledonia and samples from Japan + Palau formed well-supported clades which differed by a P-distance of 0.93–1.40% (4–6 bp) in the ITS2 dataset and by 0.40–0.79% (5–10 bp) in the 28S dataset. Differences less than these are routinely found between morphologically distinct trematode species in the region (e.g. Trieu et al., Reference Trieu, Cutmore, Miller and Cribb2015; Bray et al., Reference Bray, Cutmore and Cribb2018; Huston et al., Reference Huston, Cutmore and Cribb2018b; Huston et al., Reference Huston, Cutmore, Miller, Sasal, Smit and Cribb2021); notably, Bray et al. (Reference Bray, Cutmore and Cribb2023) used mitochondrial data to describe 2 species pairs (each distinct in morphology and host range) that had identical ribosomal data.
In contrast to ribosomal data, the cox1 data for specimens of E. fusum from the 4 Pacific localities form 6 well-supported clades (3 from Australia and 1 from each of New Caledonia, Japan and Palau), with geographical structuring different from that suggested by the ribosomal data; samples from Palau and New Caledonia formed a clade with 3 lineages of Lizard Island samples, sister to samples from Japan. Based on the cox1 data, the specimens of E. fusum could have been interpreted as 4 species (1 from Japan, 2 from Lizard Island and 1 from New Caledonia + Palau + Lizard Island). Only a handful of specimens of E. gotozakiorum were collected outside of French Polynesia, but, based on these samples, this new species demonstrates a topology that would have been predicted over such a range. In the cox1 analyses, samples from French Polynesia formed a well-supported clade, sister to a clade of specimens from Bali and Lizard Island; samples from French Polynesia and Lizard Island were almost identical in the ribosomal analyses. Although the level of intraspecific variation in the cox1 dataset, up to 7.38% (35 bp) and 7.59% (36 bp) within each of the 2 major clades, is high compared to a range of recent studies in the region (e.g. Cutmore and Cribb, Reference Cutmore and Cribb2022; Bray et al., Reference Bray, Cutmore and Cribb2023; Duong et al., Reference Duong, Cribb and Cutmore2023), it is lower than differences interpreted as intraspecific variation for several other species in the Indo-west Pacific. Magro et al. (Reference Magro, Cutmore, Carrassón and Cribb2023) found variation of up to 8.9% (42 bp) for samples of Paraschistorchis stenosoma (Hanson, 1953) Blend, Karar & Dronen, 2017 in sympatry on the Great Barrier Reef; Cribb et al. (Reference Cribb, Cutmore, Wee, Browne, Diaz and Pitt2024) found variation up to 11.2% (53 bp) for samples of Prodistomum orientale (Layman, 1930) Bray and Gibson, 1990 between Australia and Japan; and Cutmore et al. (Reference Cutmore, Corner and Cribb2023) found variation of 12.6% (60 bp) for samples of Transversotrema enceladi Cutmore, Corner & Cribb, Reference Cutmore, Corner and Cribb2023 between Heron and Lizard Islands, both on the Great Barrier Reef. Overall, the final identification of all samples of clade 1 as representing E. fusum and all samples of clade 2 representing E. gotozakiorum was only possible by combining mitochondrial and ribosomal data interpretations, with a systematic consideration of the context of the data within each individual clade.
PCA analysis provided a general separation between the 2 species recognized here and 3 individual characters enabled a partial distinction between them. However, we consider reliable identification of these forms to be genotype-dependent; that is, molecular data provide a far more reliable basis for separation than do morphological characters. Should samples from new localities or hosts become available, a combined molecular and morphological assessment is desirable, but we predict that molecular data will provide the strongest evidence. In light of the results of this study, it may be possible to partly predict the identity of species in this genus by their geographic origin, but the presence of 2 species at Lizard Island where 1 is seemingly rare makes even this somewhat problematic.
Mitochondrial populations
One striking feature of the cox1 dataset is the multiple lineages of E. fusum at Lizard Island; 3 lineages (OTUs A, C and E) were found at this location, with combinations of these lineages found in total sympatry (same individual fish); none of OTUs A, C and E are each other's sister clade. Analogous occurrences of multiple Lizard Island lineages have recently been reported recently, but few with such a complicated topology. Bray et al. (Reference Bray, Cutmore and Cribb2022) found 2 lineages of Preptetos zebravaranus Bray, Cutmore & Cribb, Reference Bray, Cutmore and Cribb2022, Cutmore et al. (Reference Cutmore, Corner and Cribb2023) found 2 lineages each for Transversotrema fusilieri Hunter and Cribb, 2012 and T. iapeti Cutmore, Corner & Cribb, Reference Cutmore, Corner and Cribb2023 and Magro et al. (Reference Magro, Cutmore, Carrassón and Cribb2023) found 2 lineages for P. stenosoma (Hanson, 1953) Blend, Karar & Dronen, 2017; in all these cases, the 2 lineages were sister to each other. More comparable to the current topology is that of Preptetos laguncula Bray and Cribb, 1996. Bray et al. (Reference Bray, Cutmore and Cribb2022) demonstrated that P. laguncula is represented by 2 clades at Lizard Island, 1 more closely related to samples from French Polynesia (Society and Gambiers Archipelagos) than to the other Lizard Island clade; as in the current study, the ribosomal data for these 2 clades were identical. Bray et al. (Reference Bray, Cutmore and Cribb2022) speculated that the phenomenon may be a result of Lizard Island representing a ‘suture zone’, where previously isolated populations of P. laguncula had merged and the ribosomal DNA recombined while the mitochondrial populations remained distinct; the new data for E. fusum, as well as that reported for P. stenosoma, P. zebravaranus, T. fusilieri and T. iapeti, all support this interpretation.
Another potential key factor in play may be connectivity. It is well understood that host vagility is an important determinant in parasite dispersal ability; the more vagile a host, the higher the ability of the parasite to disperse widely across oceanic regions, while the retained connectivity between populations in turn limits genetic variability. If we take several of our sampling locations (Okinawa, Palau, Bali and the northern Great Barrier Reef) to represent the western boundary of the West Pacific region, French Polynesia represents the near-eastern extent of this region. The distances between these localities therefore represent biogeographical extremes in relation to 1 another, ones that are largely insurmountable for most inshore reef-dwelling species. Siganus argenteus, however, is the most vagile siganid species, being among the most pelagic of siganids and the only species with a long pelagic larval duration (Zolkaply et al., Reference Zolkaply, Do, Asaduzzaman, Seah, Hurwood, Mather, Rahman and Wong2021). Such traits have enabled it to become, along with S. spinus, the most widespread siganid species in the Tropical Indo-West Pacific (TIWP).
The centre of radiation for much of TIWP biodiversity, including of the rabbitfish family Siganidae, is held to be the Coral Triangle, i.e. the region bound by Peninsular Malaysia, the Philippines and the Solomon Islands (Carpenter and Springer, Reference Carpenter and Springer2005). It is therefore likely that species of Emprostiotrema also radiated in this region, dispersed eastwards along with the only hosts able to carry them and ultimately speciated in isolation in French Polynesia. In this interpretation, the presence of genotypes consistent with this Polynesian taxon in the western Pacific represents rare instances of ‘spillback’ of this Polynesian genotype, dispersing westwards in relatively small numbers in highly vagile hosts like S. argenteus. The westerly flowing trans-oceanic currents in the South Pacific split northwards and southwards upon encountering the Australian coastline in the vicinity of the southern Coral Sea. This current pattern may contribute to a biogeographical connectivity barrier, which may explain some of the observed absences and limited genetic variability between parasites from Polynesia and those from the northern and southern GBR.
It is known that disparities in ecology can lead to genetic separation due to both pre- and post-zygotic isolation mechanisms forming barriers to recombination; for instance, Williamson et al. (Reference Williamson, Gyllenhaal, Bauernfeind, Bautista, Baumann, Gadek, Marra, Ricote, Valqui, Bozinovic, Singh and Witt2024) demonstrated that some individuals of a widespread and ecologically versatile species being resident and others being highly migratory effectively form 2 populations which ultimately speciate, though they may remain morphologically cryptic and still often exist in sympatry. In the case of Emprostiotrema spp., genomic-scale analysis of the 2 Emprostiotrema species may reveal even deeper genetic divergences between them than is apparent from our ribosomal and cox1 mitochondrial data, facilitated by the greater dispersal capacity of S. argenteus relative to other Siganus species (Zolkaply et al., Reference Zolkaply, Do, Asaduzzaman, Seah, Hurwood, Mather, Rahman and Wong2021). We speculate that the divergence of E. gotozakiorum closely tracks that of S. argenteus in the late Miocene (approximately 14.3 mya) and subsequent eastward dispersal to Polynesia. These divergences would effectively prevent E. gotozakiorum from recombining with E. fusum in the relatively rare occasions the former re-encounter the latter.
Prevalence, distribution and host-specificity
Although we have examined 689 individual siganids, the distribution of these samples between 12 collection localities and 13 siganid species dilutes the capacity to draw robust inferences relating to prevalence. The sample sizes do seem sufficiently robust to allow the inference that species of Emprostiotrema do not occur on the southern Great Barrier Reef, at Moreton Bay or at Ningaloo Reef (Western Australia). The siganids examined at these 3 sites overlap with species found infected elsewhere, except for S. trispilos and S. virgatus, which have been examined only at Ningaloo Reef. Absence of Emprostiotrema species from sites where suitable hosts are abundant contrasts with the genuine widespread distributions demonstrated for other Indo-Pacific fish trematode species, e.g. Elaphrobates chaetodontis (Yamaguti, 1970) Yong, Cribb & Cutmore, 2021 reported from the Great Barrier Reef, Japan and French Polynesia (Cutmore and Cribb, Reference Cutmore and Cribb2022), Gorgocephalus yaaji Bray and Cribb, 2005 reported from eastern South Africa, Lizard Island and French Polynesia (Huston et al., Reference Huston, Cutmore, Miller, Sasal, Smit and Cribb2021) and Schikhobalotrema acutum (Linton, 1910) Skrjabin and Guschanskaja, 1955 reported from the Gulf of Mexico to Australia (Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Solórzano-García, Huston, Mendoza-Garfias, Cabañas-Granillo, Cutmore and Cribb2024). We speculate that the absences relate to the absence of suitable intermediate hosts; however, as discussed below, these remain completely unknown.
The overall prevalence of E. fusum in all susceptible siganids at sites at which it was found was 70/228 (30.7%) and is remarkably similar to the figure for E. gotozakiorum in French Polynesia (11/36, 30.5%). In contrast to these moderate prevalences, just 2 cox1 sequences were consistent with E. gotozakiorum compared to 50 sequences of E. fusum from Lizard Island, <4% of the specimens of Emprostiotrema at this locality. Both E. gotozakiorum sequences were from specimens of S. argenteus, one of the 2 known hosts from French Polynesia. It remains to be determined whether E. gotozakiorum has a narrower host range than E. fusum on the GBR; in French Polynesia it has been found in the only 2 siganids that occur there (S. argenteus and S. spinus). Emprostiotrema gotozakiorum is thus both nearly morphologically cryptic relative to E. fusum and much less abundant. Discovery of such rare species is clearly problematic as it is dependent on time-consuming and moderately expensive sequencing. Almost by definition, we have little understanding of the distribution and significance of rarity of marine fish parasites. However, as discussed above, such occurrences are likely at least partly explained by host population connectivity and dispersal ability.
The variation in the prevalence of E. fusum is interesting. Eight of 10 species of Siganus examined at Lizard Island (by far the best sampled site at which it is present) were infected by E. fusum; it was not found in robust samples of S. puellus (n = 17), or S. spinus (n = 13). However, both of those species were found infected in small fish samples at Palau (just 3 examined fishes of each species). Overall, every siganid species examined at sites where infections were present was infected in at least 1 locality.
The broad definitive host range of E. fusum (11 siganid species) is somewhat surprising in that ecological differences in host biology typically correlate with differences in digenean fauna (Miller et al., Reference Miller, Bray and Cribb2011). Although all species of Siganus form a monophyletic clade, these fishes are far from uniform in their behaviour and diet. Siganid species can be broadly divided into 2 general body types which have phylogenetic support: a ‘fusiform’ body [clades 1 and 2 sensu Kuriiwa et al. (Reference Kuriiwa, Hanzawa, Yoshino, Kimura and Nishida2007), including S. argenteus, S. canaliculatus, S. fuscescens and S. spinus], associated with species that school on reef and algal flats, and a ‘deep’ body [clade 3 sensu Kuriiwa et al. (Reference Kuriiwa, Hanzawa, Yoshino, Kimura and Nishida2007), including S. corallinus, S. doliatus, S. lineatus, S. puellus, S. punctatissimus, S. punctatus and S. vulpinus], associated with species that form pairs or small groups and feed near hard corals and within reef crevices (Kuriiwa et al., Reference Kuriiwa, Hanzawa, Yoshino, Kimura and Nishida2007; Fox and Bellwood, Reference Fox and Bellwood2013; Hoey et al., Reference Hoey, Brandl and Bellwood2013). Diets also differ among siganids, with some feeding primarily on algae, others feeding mostly on detrital aggregates, and S. puellus feeding primarily on sponges (Fox et al., Reference Fox, Sunderland, Hoey and Bellwood2009; Fox and Bellwood, Reference Fox and Bellwood2013; Hoey et al., Reference Hoey, Brandl and Bellwood2013). Emprostiotrema fusum parasitizes siganids of both body types, multiple algal and detrital feeding species, and even the sponge-feeding S. puellus. In contrast, E. gotozakiorum is known only from clades 1 and 2 species (S. argenteus, S. canaliculatus and S. spinus); notably, however S. argenteus and S. spinus are the only species of Siganus known to occur in French Polynesia (Siu et al., Reference Siu, Bacchet, Bernardi, Brooks, Carlot, Causse, Claudet, Clua, Delrieu-Trottin and Espiau2017), where E. gotozakiorum is most commonly found. Emprostiotrema kuntzi is only known from S. argenteus; this species, however, has not been reported since its original description (Ahmad, Reference Ahmad1985) and the lack of additional reports from the Indian Ocean do not allow for commentary of the host-specificity of this species.
Life cycles
The life cycles of Emprostiotrema species remain unknown. However, the ability of species of Emprostiotrema to parasitize siganids that occupy multiple ecological and dietary niches, coupled with a known intermediate host for another emprostiotrematid, Isorchis cannoni Huston, Cutmore & Cribb, 2017, allows inference of the cercarial dispersal and encystment strategy used by these digeneans. The cercariae of I. cannoni emerge from the intertidal gastropod Clypeomorus batillariaeformis Habe and Kosuge (Caenogastropoda: Cerithiidae) and swim for a short period before they encyst in the benthos where they are incidentally consumed by grazing S. fuscescens and S. lineatus (Cannon, Reference Cannon1978; Huston et al., Reference Huston, Cutmore and Cribb2018a). The variety of dietary niches occupied by hosts of E. fusum suggests that the cercariae of this species also encyst in the environment, likely with little site specificity. Although no intermediate hosts are yet known for any species of Emprostiotrema, cerithiids seem likely as indicated by I. cannoni, and Rissooidea or Truncatelloidea gastropods have also been suggested as potential hosts for emprostiotrematids, as these groups serve as intermediate hosts for haploporids (Andres et al., Reference Andres, Pulis and Overstreet2016). As with most digenean lineages, the next step in our understanding of emprostiotrematids is likely to be obtained through discovery of new intermediate host species.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182024001252.
Data availability statement
Data underlying the present work, including R code and raw data used in principle component analysis, and an extended set of measurements of specimens summarized by host/locality combinations, have been uploaded as Supplementary Material.
Acknowledgements
We thank the staff of the Lizard Island (Australian Museum), the Direction of Marine Resources from French Polynesia, and Professor James Reimer (University of the Ryukyus, Japan) for their support of our collecting efforts. We thank Drs Rod Bray, Storm Martin, Nick Wee and all students of the Marine Parasitology Laboratory (University of Queensland) for their assistance in collecting fish and digeneans over the many years of this project, Professor Kazuo Ogawa (Meguro Parasitological Museum) for graciously providing photographs of a syntype of E. fusum, and Jeff Johnson (Queensland Museum) for advising on the status of Siganus canaliculatus.
Author contributions
D. C. H. conceived and designed the study, collected and prepared specimens and performed PCA. S. C. C. performed sequencing, phylogenetic analyses and prepared phylogenetic trees. P. S. arranged work in French Polynesia and collected specimens. T. H. C. explored the morphometric dataset and developed morphometric indices and figures. R. Q.-Y. Y. prepared specimens, performed morphometric analyses and prepared illustrations. All authors contributed to writing the final manuscript.
Financial support
This project represents a contribution to Taxonomy Australia (2020), a national initiative organized under the auspices of the Australian Academy of Science that brings together the taxonomic community to develop approaches that will significantly increase the rate at which new species are discovered, resolved and named, with a view to completely documenting the Australian biota within a generation. This project is supported by funding from the Australian Government's Australian Biological Resources Study National Taxonomy Research Grants Program (4-H04JDSM, awarded to S. C. C. and T. H. C.). Collecting in Japan was supported by an Australian Society for Parasitology (ASP) Network Researcher Exchange, Training and Travel Award grant to S. C. C. R. Q.-Y. Y. acknowledges the current support of a North-West University (NWU) Postdoctoral Research Fellowship. We thank Professor Harry Palm for organising collecting in Indonesia, supported through the Science for the Protection of Indonesian Coastal Marine Ecosystems (SPICE III) programme (BMBF Grant No. 03F0641D). This is contribution number 914 from the NWU Water Research Group.
Competing interests
None.
Ethical standards
This study was conducted in compliance with all institutional, national and international guidelines on the care and use of animals.