Introduction
Schistosomiasis, a debilitating, often fatal, disease, caused by trematode blood fluke parasites of the genus Schistosoma, afflicts over 230 million people in 78 countries (Colley et al., Reference Colley, Bustinduy, Secor and King2014). Three species of schistosomes, Schistosoma mansoni, S. haematobium and S. japonicum, are the most clinically relevant. The zoonotic S. japonicum is currently endemic in P.R. China, the Philippines with small foci occurring in Indonesia (Gordon et al., Reference Gordon, Kurscheid, Williams, Clements, Li, Zhou, Utzinger, McManus and Gray2019). In the Philippines, S. japonicum is prevalent in 28 provinces in 12 regions of the country, with an estimated 28 million individuals at risk of infection (Olveda and Gray, Reference Olveda and Gray2019). Currently, schistosomiasis control relies predominantly on mass praziquantel (PZQ) drug administration (MDA) programs (Olveda and Gray, Reference Olveda and Gray2019). However, MDA on its own is insufficient to provide long term sustainable control of the disease if no additional integrated interventions are implemented (Ross et al., Reference Ross, Olveda, Chy, Olveda, Li, Harn, Gray, McManus, Tallo, Chau and Williams2015; Mutapi et al., Reference Mutapi, Maizels, Fenwick and Woolhouse2017). Accurate diagnostic tools are required in the context of integrated schistosomiasis control programs in the Philippines and other endemic areas.
Currently, three major types of diagnostic methods are available for schistosomiasis: parasitological detection (e.g. the Kato-Katz (KK) test and urine filtration)); serology, including antibody-detection (AbD) and antigen-detection (AgD); and molecular methods (e.g. circulating nucleic acids detection) (Cavalcanti et al., Reference Cavalcanti, Silva, Peralta, Barreto and Peralta2013; Weerakoon et al., Reference Weerakoon, Gobert, Cai and McManus2015). The different methods have different advantages and disadvantages. For example, the traditional KK parasitological technique shows high specificity but an insufficient level of sensitivity, particularly in areas with reduced disease burden (Weerakoon et al., Reference Weerakoon, Gobert, Cai and McManus2015; Reference Weerakoon, Gordon, Williams, Cai, Gobert, Olveda, Ross, Olveda and McManus2017b; Oliveira et al., Reference Oliveira, Magalhaes, Elias, de Castro, Favero, Lindholz, Oliveira, Barbosa, Gil, Gomes, Graeff-Teixeira, Enk, Coelho, Carneiro, Negrao-Correa and Geiger2018; Cai et al., Reference Cai, Weerakoon, Mu, Olveda, Ross, Olveda and McManus2019). AbD-based methods are usually cost-effective and have considerable accuracy yet they have limited ability to discriminate past from active infections. Compared with Ab-based detection assays, AgD-based methods, in the format of lateral flow assays targeting urine samples, provide a rapid and non-invasive diagnostic, but suffer from limited sensitivity in low endemic settings, a relatively high false-positive rate and high cost. Molecular techniques, notably PCR-based methods (Weerakoon et al., Reference Weerakoon, Gordon, Gobert, Cai and McManus2016, Reference Weerakoon, Gordon, Cai, Gobert, Duke, Williams and McManus2017a, Reference Weerakoon, Gordon, Williams, Cai, Gobert, Olveda, Ross, Olveda and McManusb)), exhibit high accuracy for the detection of schistosome infections, but the current costs remain high.
MicroRNAs (miRNAs) are small non-coding RNAs (~22 nt), which are dysregulated in a wide array of biological processes including carcinogenesis (Peng and Croce, Reference Peng and Croce2016; Anvarnia et al., Reference Anvarnia, Mohaddes-Gharamaleki, Asadi, Akbari, Yousefi and Shanehbandi2019). As potential targets for novel diagnosis (Li and Kowdley, Reference Li and Kowdley2012), circulating and/or extracellular vesicle (EV)-derived miRNA signatures have been tested as biomarkers for different types of diseases and disorders including cancers, infectious and inflammatory diseases (Correia et al., Reference Correia, Nalpas, McLoughlin, Browne, Gordon, MacHugh and Shaughnessy2017; Jia et al., Reference Jia, Liu, Zhang, Xu, Gao, Bai, Xu, Xu and Zhang2017; Meningher et al., Reference Meningher, Lerman, Regev-Rudzki, Gold, Ben-Dov, Sidi, Avni and Schwartz2017; Jamali et al., Reference Jamali, Tofigh, Tutunchi, Panahi, Borhani, Akhavan, Nourmohammadi, Ghaderian, Rasouli and Mirzaei2018; Schonauen et al., Reference Schonauen, Le, von Arnim, Schulz, Malfertheiner and Link2018; Tengda et al., Reference Tengda, Shuping, Mingli, Jie, Yun, Weiwei and Anmei2018; Filipów and Łaczmański, Reference Filipów and Łaczmański2019; Xue et al., Reference Xue, Zhao, Wang, Qin and Hu2019). Circulating miRNAs also have been proposed as having potential to detect parasitic helminth infections (Hoy et al., Reference Hoy, Lundie, Ivens, Quintana, Nausch, Forster, Jones, Kabatereine, Dunne, Mutapi, Macdonald and Buck2014; Tritten et al., Reference Tritten, Burkman, Moorhead, Satti, Geary, Mackenzie and Geary2014; Cai et al., Reference Cai, Gobert, You, Duke and McManus2015; Dong et al., Reference Dong, Gao, Peng, Sun, Han, Zhao, Liu, Wang, Song, Wu and Yang2017; Guo and Zheng, Reference Guo and Zheng2017). To date, a number of parasite-derived miRNAs in plasma/serum have been validated for the purpose of schistosomiasis diagnosis (Hoy et al., Reference Hoy, Lundie, Ivens, Quintana, Nausch, Forster, Jones, Kabatereine, Dunne, Mutapi, Macdonald and Buck2014; Meningher et al., Reference Meningher, Lerman, Regev-Rudzki, Gold, Ben-Dov, Sidi, Avni and Schwartz2017). However, these investigations focused on the diagnosis of S. mansoni and S. haematobium infection by testing a limited number of patient samples; currently, there are no data on the potential of detecting circulating miRNAs in individuals infected with S. japonicum. Recent advances in characterizing miRNA profiles in extracellular vesicles secreted by Schistosoma species (Nowacki et al., Reference Nowacki, Swain, Klychnikov, Niazi, Ivens, Quintana, Hensbergen, Hokke, Buck and Hoffmann2015; Zhu et al., Reference Zhu, Liu, Dao, Lu, Li, Gu, Liu, Feng and Cheng2016a, Reference Zhu, Wang, Lin, Jiang, Cui, Wang, Zhang and Panb; Samoil et al., Reference Samoil, Dagenais, Ganapathy, Aldridge, Glebov, Jardim and Ribeiro2018) have raised the possibility for validating more parasite-derived miRNAs as potentially novel biomarkers for schistosomiasis detection.
In this study, we evaluated the potential of detecting circulating parasite-derived miRNAs in S. japonicum infected human subjects. Initially, we employed the BALB/c mouse as a schistosomiasis model to validate 21 parasite-derived miRNA candidates in serum during S. japonicum infection. Then, following another step of screening, six candidate miRNAs were selected for further validation, individually or in combination, using human sera from a cohort of residents in an area in the rural Philippines endemic for schistosomiasis japonica. We presented the diagnostic performance of parasite-derived miRNA signatures in a S. japonicum-endemic setting with a low-intensity infection.
Materials and methods
Parasites
S. japonicum-infected Oncomelania hupensis hupensis snails were purchased from Nanjing Municipal Center for Disease Control and Prevention, China, and transported to QIMRB, Brisbane, Australia. Cercariae were shed from the infected snails under light stimulation.
Mouse infection and serum collection
Three 8-week-old female BALB/c mice were percutaneously infected with a low-dose challenge of S. japonicum cercariae (16 ± 2). Mice were sacrificed at 9 weeks post infection (pi) and ~1 ml blood was collected by cardiac puncture. Blood samples were then allowed to stand at room temperature for 2 h. After centrifugation at 3000 g for 10 min, the serum samples were collected and stored at − 80°C. Serum samples from three naive mice were used as controls.
Study cohort and human sample collection
The human subjects were recruited from schistosomiasis-endemic areas in Laoang and Palapag, Northern Samar, the Philippines. Additional information on the study population is available in previous reports (Ross et al., Reference Ross, Olveda, Chy, Olveda, Li, Harn, Gray, McManus, Tallo, Chau and Williams2015; Olveda et al., Reference Olveda, Inobaya, Olveda, Vinluan, Ng, Weerakoon, McManus, Ramm, Harn, Li, Lam, Guevarra and Ross2017). For each participant, ~10 ml blood was drawn and serum was then collected after centrifugation and stored at 2–8°C. The serum samples were transported to RITM and stored at −80°C. Subsequently, a subset of samples was shipped to QIMRB, Australia, on dry ice.
Parasitological detection (Kato-Katz)
Each individual from the study cohort provided two stool specimens from which six Kato-Katz thick smear slides were prepared. Slides were examined by experienced laboratory technicians under a light microscope. The burden of infection is presented as the number of eggs per gram of faeces (EPG).
RNA extraction, polyadenylation and reverse transcription (RT)
For each mouse, total RNA was extracted from 100 μL serum samples, and for each human subject, total RNA was extracted from 200 μL serum samples, using miRNeasy mini kits (Qiagen, Hilden, Germany) according to manufacturer's instructions. During the RNA extraction procedure, 3.2 fmoles Arabidopsis thaliana ath-miR-159a (IDT, Coralville, IA) was added to each sample as a spike-in control. The total RNA product was eluted with 30 μL nuclease-free water.
A one-step procedure of polyadenylation and RT reaction was performed by the combined use of two kits: a Poly(A) polymerase tailing kit (Epicentre Biotechnologies, Madison, WI) and a TaqMan microRNA reverse transcription kit (Life Technologies, Carlsbad, CA). For mouse samples, the Poly(A) method was used. Briefly, a 10 μL RT reaction comprised: 1 μL 10 × RT buffer, 1 μL ATP (10 mm), 1 μl universal RT primer (1 mm), 0.1 μL dNTPs (25 mm each), 0.13 μL RNase inhibitor, 0.2 μL poly(A) polymerase, 0.5 μL MultiScribe MuLV and 5 μL RNA and 1.07 μL nuclease-free water. RT reactions were carried out using a Veriti 96-well thermal cycler (ABI, Foster City, CA) under the following condition: 37°C for 30 min, 42°C for 30 min, and followed by enzyme inactivation at 85°C for 5 min. For human subjects, polyadenylation and RT reactions were performed using the S-Poly(T) method (Cai et al., Reference Cai, Gobert, You, Duke and McManus2015). The reaction system was the same as that for the Poly(A) method except that it incorporated 1 μL of miRNA-specific primer pool (25 nm of each primer). RT products were stored at −20°C prior to subsequent analysis. The RT primers are listed in Supplementary Table 1.
qRT-PCR for miRNA quantification
Quantification of the serum levels of miRNAs was performed by probe-based qRT-PCR according to essential protocols as described previously (Cai et al., Reference Cai, Gobert, You, Duke and McManus2015, Reference Cai, Mu, Olveda, Ross, Olveda and McManus2018). Briefly, the 5 μL PCR reaction contained 2.5 μL TaqMan Universal Master Mix II (Life Technologies, Carlsbad, CA), 0.5 μL of RT products, 1 μL primer mixture (forward and universal reverse primers) (final concentration: 0.2 μ m), 0.5 μL universal double quenched probe (final concentration: 0.25 μ m) (IDT, Coralville, IA), and 0.5 μL nuclease-free water. The assays were performed on an ABI Quantstudio 5 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA) with the following cycling condition: pre-denaturation at 95°C for 10 min, followed by 50 cycles: 95°C for 15 sec, and 60°C for 1 min. For analyses, a cutoff Ct value of 40 was set as a background for the purpose of calculating signal over noise. The expression levels were determined by the 2−ΔΔCt method with the spiked-in ath-miR-159a used as the normalization control. Three technical replicates were performed for each sample. The primer and probe sequences used are listed in Supplementary Table 1.
Statistical analysis
Unpaired student's t-test (two tails) was used for comparing the serum levels of miRNAs in naïve and S. japonicum-infected BALB/c mice. The Mann–Whitney U-test was used for the analysis of the capability of the serum levels of miRNAs in discriminating the KK (+) group from the control group. The receiver operating characteristic (ROC) curve analyses were performed and the AUC was calculated to assess the potential of using the parasite-derived circulating miRNAs (individually or in combination) as novel biomarkers for schistosomiasis japonica. Cut-off values for determination of sensitivity and specificity were set by maximizing the Youden's index. Pearson's correlation coefficient (r) was used for the assessment of the correlation between the serum levels of miRNAs and infection intensity (egg burden) in the KK (+) subjects. Statistical analysis was performed with GraphPad Prism Version 6.01 for windows.
Results
Detection of parasite-derived miRNAs in the serum of BALB/c mice at 9 weeks post-S. japonicum infection
Twenty-one miRNAs were selected to assess their potential for detection of S. japonicum infection based on prior published studies of schistosome circulating and extracellular vesicles/exosomes associated miRNAs (Supplementary Table 2). The expression of these 21 miRNAs was tested in naïve and S. japonicum-infected (9 weeks post-infection) BALB/c mice by RT-PCR (Fig. 1). A total of 12 miRNAs (sja-miR-277, sja-miR-3479-3p, sja-miR-125a sja-miR-61, sja-miR-2b-5p, sja-miR-2162-3p, sja-miR-36-3p, sja-miR-3489, sja-miR-3487, sja-miR-2c-5p, sja-miR-2a-3p and sja-miR-10) were selected for further investigation based on a fold change cut-off value ⩾4 and a P value cut-off <0.05.

Fig. 1. The expression levels of 21 miRNAs in the sera of naïve and infected (9 wks p.i.) BALB/c mice determined using qRT-PCR (Control, n = 3; 9 weeks pi, n = 3). P values were calculated using the unpaired student's t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Initial screening of 12 miRNAs for the diagnosis of human S. japonicum infection
In the next stage of screening, qRT-PCR was used to determine the expression levels of the 12 miRNAs selected in serum samples from KK-positive (KK (+)) patients (n = 5) and control individuals (KK and SjSAP4 + Sj23-LHD-ELISA negative (Cai et al., Reference Cai, Weerakoon, Mu, Olveda, Piao, Liu, Olveda, Chen, Ross and McManus2017)) (n = 5) (Supplementary Fig 1). ROC curve analysis was performed and the AUC levels were calculated to evaluate the diagnostic potential of each miRNA (Supplementary Fig 1). As a result, six miRNAs (sja-miRNA-277, sja-miR-125a, sja-miR-2b-5p, sja-miR-36-3p, sja-miR-2c-5p and sja-miR-2a-3p) with an AUC value ⩾0.80 were selected for further validation.
The potential value of serum levels of six miRNAs by singleplex qRT-PCR for the diagnosis of human schistosomiasis
The expression levels of sja-miRNA-277, sja-miR-125a, sja-miR-2b-5p, sja-miR-36-3p, sja-miR-2c-5p and sja-miR-2a-3p were further probed using sera from a human cohort of low-intensity infected individuals from a schistosomiasis-endemic area, Northern Samar, the Philippines (Table 1) by qRT-PCR. The cohort included 53 KK (+) individuals and 25 KK and SjSAP4 + Sj23-LHD-ELISA negatives as controls. The levels of two miRNAs, sja-miR-2b-5p, and sja-miR-2c-5p, were significantly higher in patients than in control individuals (P = 0.0251 and P = 0.0114, respectively), while the serum abundance of the other four miRNAs, sja-miRNA-277, sja-miR-125a, sja-miR-36-3p and sja-miR-2a-3p failed to differentiate the two groups (P < 0.05) (Fig. 2A). Using optimal cut-off points, sja-miR-2b-5p and sja-miR-2c-5p could detect S. japonicum infected individuals with a specificity/sensitivity of 66.0%/68.0% and 54.7%/80.0%, respectively (Fig. 2A). The ROC curve analysis for the six individual miRNAs in discriminating the KK (+) from the controls showed AUC values of 0.6340, 0.6279, 0.6574, 0.5906, 0.6770 and 0.5804 for sja-miRNA-277, sja-miR-125a, sja-miR-2b-5p, sja-miR-36-3p, sja-miR-2c-5p and sja-miR-2a-3p, respectively (P = 0.0574, 0.0696, 0.0256, 0.1989, 0.0121 and 0.2542, respectively) (Fig. 2B).
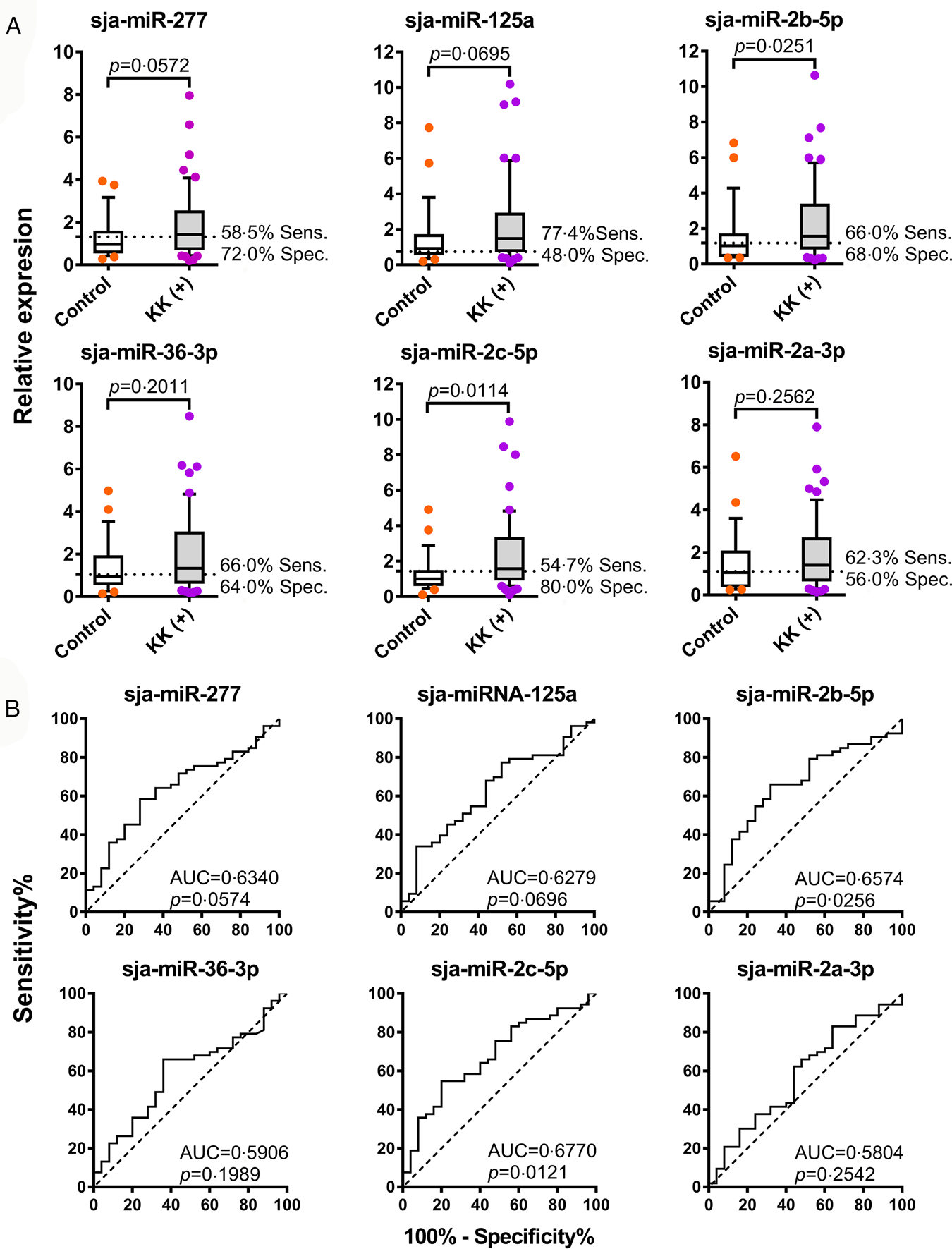
Fig. 2. Discrimination of S. japonicum-infected and non-infected individuals by detection of parasite-derived miRNAs in serum. (A) The serum levels of the six candidate miRNAs in the KK (+) and control subjects. Boxes represent the interquartile range of the data with lines across the boxes indicate the median values. The hash marks below and above the boxes indicate the 10th and 90th percentiles for each group, respectively. (B) ROC curve analysis for the six candidate miRNAs was performed to evaluate the capabilities in differentiating the KK (+) and control participants.
Table 1. Basic information of study participants in the control and KK (+) groups

*ELISA assay detecting IgG antibodies against a combination of recombinant S. japonicum proteins (rSjSAP4 and rSj23-LHD) (Cai et al., Reference Cai, Weerakoon, Mu, Olveda, Piao, Liu, Olveda, Chen, Ross and McManus2017).
EPG: Eggs per gram of faeces.
ROC curve analysis was performed to evaluate the ability of combinations of the miRNAs to distinguish the KK (+) from the control participants (Table 2). Using the combined data for six miRNAs, the combination of sja-miR-2b-5p and sja-miR-2c-5p was best able to differentiate between the two groups with an AUC value of 0.6906 (95% CI 0.5645-0.8166; P = 0.0069; sensitivity 77.4%, specificity 60.0%), followed by the combination of sja-miR-125a, sja-miR-2b-5p and sja-miR-2c-5p (AUC 0.6792; 95% CI 0.5541-0.8044; P = 0.0110; sensitivity 55.8%, specificity 80.0%).
Table 2. Discrimination of S. japonicum infected individuals from controls using serum levels of combined miRNAs

# Sja-miR-36-3p and sja-miR-2a-3p were excluded from analysis for the combinations of 2, 3, and 4 miRNAs.
CI, confidence interval.
The diagnostic performance of serum miRNA levels determined by duplex and multiplex qRT-PCR assays for human schistosomiasis
The serum miRNA levels were also probed using a duplex (designated as 2P, targeting sja-miR-2b-5p and sja-miR-2c-5p) and three multiplex qRT-PCR assays with the same cohort. The multiplex qRT-PCR assays were designated as 3P (targeting sja-miR-277, sja-miR-2b-5p, and sja-miR-2c-5p), 5P (targeting sja-miR-125a, sja-miR-2b-5p, sja-miR-36-3p, sja-miR-2c-5p and sja-miR-2a-3p) and 6P (targeting sja-miRNA-277, sja-miR-125a, sja-miR-2b-5p, sja-miR-36-3p, sja-miR-2c-5p and sja-miR-2a-3p). In the 2P and 5P assays, the serum levels of targeted miRNAs were significantly higher in the KK (+) than control individuals (P = 0.0491 and P = 0.0202, respectively), while no significant difference was observed between the two groups in the 3P and 6P assays (Fig. 3A). The ROC curve analysis for discriminating the KK (+) from the controls yielded AUC values of 0.6385, 0.6302, 0.6630, and 0.6185, for the 2P, 3P, 5P and 6P assays, respectively (P = 0.0495, 0.0648, 0.0208 and 0.0928, respectively) (Fig. 3B).

Fig. 3. The diagnostic performance for detecting human schistosomiasis japonica using serum parasite-derived miRNAs quantified by duplex and multiplex qRT-PCR assays. (A) The serum levels of parasite-derived miRNA combinations in the control and KK (+) individuals. Boxes represent the interquartile range of the data with lines across the boxes indicating the median values. The hash marks below and above the boxes indicate the 10th and 90th percentiles for each group, respectively. (B) ROC curve analysis was performed for the levels of different miRNA combinations determined by the duplex and multiplex qRT-PCR assays to evaluate the capabilities in discriminating the KK (+) from the control individuals.
Correlations of the serum miRNA levels with egg burden in the KK (+) individuals
The associations between the levels of the six miRNA signatures (individually or in combination) in serum and egg burden were then investigated in the KK (+) group. The serum level of miRNA-2c-5p correlated with EPG (r = 0.3222, P = 0.0186), whereas the serum levels of the other 5 miRNAs did not show a significant correlation with infection intensity determined by the KK method (Fig. 4). Also, no significant correlation was observed between the serum miRNA levels determined by the duplex (2P) or multiplex assays (3P, 5P and 6P) and faecal egg burden (Supplementary Fig 2).

Fig 4. Correlations between the serum abundance of six miRNAs and faecal egg burden (EPG) in the KK (+) individuals using Pearson's correlation coefficient.
Discussion
Accurate diagnosis of schistosomiasis, especially in low-intensity areas following MDA and other control programs, remains a great challenge. Nevertheless, the development and deployment of novel diagnostic tools, with the requisite accuracy, for the purpose of monitoring control efforts in endemic areas to ensure schistosomiasis elimination will be critical (Utzinger et al., Reference Utzinger, Becker, van Lieshout, van Dam and Knopp2015; Weerakoon et al., Reference Weerakoon, Gobert, Cai and McManus2015; Cai et al., Reference Cai, Weerakoon, Mu, Olveda, Piao, Liu, Olveda, Chen, Ross and McManus2017; Oliveira et al., Reference Oliveira, Magalhaes, Elias, de Castro, Favero, Lindholz, Oliveira, Barbosa, Gil, Gomes, Graeff-Teixeira, Enk, Coelho, Carneiro, Negrao-Correa and Geiger2018). The realization that detection of parasite-derived miRNAs in the host circulatory system during an infection is possible has generated much interest in their application as diagnostic indicators (Manzano-Roman and Siles-Lucas, Reference Manzano-Roman and Siles-Lucas2012; Cai et al., Reference Cai, Gobert and McManus2016a). The utility of using circulating miRNAs as biomarkers for the detection of schistosome infections has been shown in several recent pioneering investigations using animal models of schistosomiasis and/or with clinical samples (Cheng et al., Reference Cheng, Luo, Hu, Cao and Jin2013; Hoy et al., Reference Hoy, Lundie, Ivens, Quintana, Nausch, Forster, Jones, Kabatereine, Dunne, Mutapi, Macdonald and Buck2014; Cai et al., Reference Cai, Gobert, You, Duke and McManus2015; Meningher et al., Reference Meningher, Lerman, Regev-Rudzki, Gold, Ben-Dov, Sidi, Avni and Schwartz2017). However, there had been no reports hitherto of their use in the clinical diagnosis of schistosomiasis japonica.
Of the 12 initially selected miRNAs, based on results obtained in the animal model of schistosomiasis japonica, the majority were unable to discriminate infected from uninfected individuals in a clinical cohort (Fig. 2 and Supplementary Fig 1), although an increased volume of serum was used for RNA extraction from clinical samples and miRNAs extracted from human samples are subjected to RT with the more sensitive S-Poly(T) method (Kang et al., Reference Kang, Zhang, Liu, Wang, Zhong, Huang, Peng, Zeng, Wang, Yang, Luo and Gou2012). This may have been due to the facts that: (1) the severity of a schistosome infection is far more pronounced in the experimental murine model of schistosomiasis than is found in subjects who are KK positive, since that even a single worm pair in a mouse represents a high infection burden when body weight is taken into consideration; and (2) S. japonicum adult worm pairs digest a considerable number of erythrocytes daily in order to obtain essential amino acids (Cai et al., Reference Cai, Liu, Piao, Hou, Gobert, McManus and Chen2016b), and in so doing this results in the release of a high concentration of small RNA signatures of host origin, which may readily cause non-specific amplification in the samples obtained from individuals with a high burden of infection, as was the case with the BALB/c mouse model utilised here.
Of the six miRNAs tested, any individual miRNA provided only moderate diagnostic power for differentiating the KK (+) and control participants (AUC from 0.5804 to 0.6770); slightly higher to the most powerful singleplex test targeting sja-miR-2c-5p (AUC: 0.6770, P = 0.0121) (Fig. 1), the best diagnostic performance was obtained with a combination of sja-miR-2b-5p and sja-miR-2c-5p (AUC: 0.6906, P = 0.0069), showing a sensitivity of 76% and specificity of 60% (Table 2). In order to improve the diagnostic potential by amplification two or multiple miRNAs simultaneously, duplex/multiplex qRT-PCR assays were developed. The duplex assay 2P targeting the two most powerful miRNA signatures (sja-miR-2b-5p and sja-miR-2c-5p) only marginally discriminated the control and KK (+) individuals with an AUC of 0.6385 (P = 0.0495) and sensitivity/specificity values of 66.0%/60.0%. The multiplex assay 3P failed to differentiate the control and KK (+) subjects (AUC = 0.6302, P = 0.0648), while the accumulative data based on singleplex assay targeting the same miRNAs exhibited moderate diagnostic power with an AUC of 0.6687 (P = 0.0168). Furthermore, both the multiplex assay 6P and the combined data based on singleplex assays targeting all six miRNAs failed to show any discrimination ability in the diagnosis of clinical S. japonicum infections (Fig. 3 and Table 2). However, the multiplex assay 5P exhibited a superior diagnostic power than that obtained by the combination targeting the same miRNAs (AUC 0.6630, P = 0.0208 vs 0.6294, P = 0.0664) (Fig. 3 and Table 2). Nevertheless, the diagnostic power of the 5P assay was inferior to that of the singleplex assay detecting sja-miR-2c-5p. The failure of the duplex and multiplex assays to increase the diagnostic power may be due to: (1) the data obtained with the duplex/multiplex assays still mainly depend on a highly expressed signature(s) within the target miRNAs; (2) a relatively higher noise background may be introduced by targeting two or multiple targets, especially when the samples from a low-intensity infection setting were tested.
Overall, the diagnostic performance of the assays (singleplex, duplex, and multiplex) developed in the current study for detecting S. japonicum miRNAs in serum, was moderate but is consistent with the results obtained by Meningher et al. (Reference Meningher, Lerman, Regev-Rudzki, Gold, Ben-Dov, Sidi, Avni and Schwartz2017) when detecting S. mansoni, S. haematobium and S. mekongi infections in 26 returning travellers with schistosomiasis (based on the detection of eggs or the positive results of serologic tests) returning from either sub-Saharan Africa or Laos by amplification of miRNAs extracted from serum. Furthermore, it has been reported that parasite miRNAs are not present in plasma at a sufficiently high level to be used as a biomarker for Onchocerca volvulus infection or for monitoring treatment using miRCURY Locked Nucleic Acid (LNA) primer-based RT-qPCR (Lagatie et al., Reference Lagatie, Batsa Debrah, Debrah and Stuyver2017). The modest AUC values we obtained in efforts to diagnose schistosomal infections in the human Philippines cohort may be attributable to the following factors: (1) most of the KK (+) individuals tested harboured light schistosome infections (Table 1), a feature which itself poses a challenge for any of the currently available diagnostic tools for schistosomiasis; (2) we have previously shown that the targeted cohort is extensively co-parasitised with intestinal worms and intestinal protozoa (Gordon et al., Reference Gordon, McManus, Acosta, Olveda, Williams, Ross, Gray and Gobert2015; Ross et al., Reference Ross, Olveda, Chy, Olveda, Li, Harn, Gray, McManus, Tallo, Chau and Williams2015; Weerakoon et al., Reference Weerakoon, Gordon, Williams, Cai, Gobert, Olveda, Ross, Olveda and McManus2018). These pathogens are likely to secrete RNA signatures with sequence similarity to the miRNAs detected here, thereby affecting the specificity of the assays we employed; (3) the limited cohort sample number may also have impaired our ability to measure elevated diagnostic scores.
The two most powerful serum-based signatures identified here, sja-miR-2b-5p and sja-miR-2c-5p, were listed as the top fourth and fourteenth miRNAs associated with S. japonicum adult EVs (Zhu et al., Reference Zhu, Liu, Dao, Lu, Li, Gu, Liu, Feng and Cheng2016a), indicating that serum and serum-exosomal miRNAomes are significantly different in terms of miRNA numbers, types and expression profiles (Zhao et al., Reference Zhao, Liang, Sun and Guan le2016). Although accumulating evidence indicates that extracellular miRNAs are mainly found bound to AGO proteins (Lopez and Granados-Lopez, Reference Lopez and Granados-Lopez2017), an active sorting mechanism of exosomal miRNA may enrich specific miRNA members in extracellular vesicles/exosomes (Villarroya-Beltri et al., Reference Villarroya-Beltri, Gutierrez-Vazquez, Sanchez-Cabo, Perez-Hernandez, Vazquez, Martin-Cofreces, Martinez-Herrera, Pascual-Montano, Mittelbrunn and Sanchez-Madrid2013; Janas et al., Reference Janas, Janas, Sapon and Janas2015; Zhang et al., Reference Zhang, Li, Li, Li, Guo, Yao and Mi2015; Gon et al., Reference Gon, Maruoka, Inoue, Kuroda, Yamagishi, Kozu, Shikano, Soda, Lotvall and Hashimoto2017). Further detection of parasite-derived miRNAs from serum EV composition represents another direction for the diagnosis of human S. japonicum infection.
Previously, we showed that the serum levels of two parasite-derived miRNAs, sja-miR-277 and sja-miR-3479-3p, exhibited a strong correlation with hepatic egg burden (P < 0.0001) during the course of S. japonicum infection in C57BL/6 and BALB/c mice (Cai et al., Reference Cai, Gobert, You, Duke and McManus2015). In the current study, only the serum level of sja-miR-2c-5p weakly correlated with infection intensity based on the KK test, indicating that the infectious status or disease progression in schistosomiasis patients may be complicated. It is worth noting that the accuracy of the KK test may be affected by the uneven distribution of eggs in the fecal samples and the daily fluctuation in the number of eggs discharged; particular when most of the tested KK (+) individuals in the cohort harboured light infections. Also, as the target cohort was located in a medium-high prevalence schistosomiasis-endemic area, reinfection with S. japonicum was considered to be a frequent occurrence. Accordingly, the abundance of parasite-derived miRNAs in the sera of re-infected individuals has less chance to show a significant linear relationship with infection intensity.
Concluding remarks
In summary, we have developed singleplex, duplex and multiplex qRT-PCR assays for the diagnosis of S. japonicum infection by targeting parasite-derived serum miRNAs in a clinical cohort from a medium-high prevalence but low infection burden schistosomiasis-endemic area. The best diagnostic performance was achieved using a combination of sja-miR-2b-5p and sja-miR-2c-5p (AUC: 0.6906, P = 0.0069). The results here shed light on the diagnostic performance of parasite-derived serum miRNAs for the detection of schistosomiasis japonica by probing a human cohort with low infection burden.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182019001690
Acknowledgements
We thank Mary Duke for maintenance of the S. japonicum lifecycle at QIMRB. We also appreciate the efforts of the local field and clinical staff in Palapag, Northern Samar for the collection of the clinical samples.
Financial support
This work was funded by the National Health and Medical Research Council (NHMRC) of Australia (ID APP1160046, APP1037304, APP1102926 and APP1098244). D.P.M. is an NHMRC Senior Principal Research Fellow and Senior Scientist at QIMRB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of interest
None
Ethical standards
Animal experiments were carried out according to the Australian Code for the Care and Use of Animals for Scientific Purposes (8th edition) and with the approval of the Animal Ethics Committee, QIMR Berghofer Medical Research Institute (QIMRB), Brisbane, Australia (Ethics Approval: Project P288). The human cohort study was approved by the Institutional Review Board of the Research Institute for Tropical Medicine (RITM), Department of Health, Manila, the Philippines (Approval No: 2012-13-0) and the Human Ethics Committee, QIMRB (Ethics Approval: Project P524), in accordance with the Declaration of Helsinki. When conducting the human study, written informed consent was obtained from all participants.









