INTRODUCTION
The study of ancient parasites, known as palaeoparasitology, is important for understanding the health status of past populations, their lifestyle and associated medical conditions. The investigation of the life cycles of parasites through the analysis of the markers preserved in archaeological samples can bring to light invaluable anthropological and ecological information (Dutour, Reference Dutour2013; Reinhard et al. Reference Reinhard, Ferreira, Bouchet, Sianto, Dutra, Iniguez, Leles, Le Bailly, Fugassa, Pucu and Araújo2013). For instance, it can highlight food practices when species associated with the consumption of undercooked or raw meat or fish are identified (for example, Tænia sp. or Diphyllobothrium sp.). Living conditions, waste and sanitation management can be evaluated when geohelminth remains such as Ascaris sp. or Trichuris sp. are retrieved in ancient human settlements. Proximity with animals can also be a source of parasite transmission to humans, such as for flukes Fasciola sp. or Dicrocoelium sp. (Le Bailly and Araujo, Reference Le Bailly and Araujo2016). Since the first histological observation of Schistosoma haematobium by Sir Mark A. Ruffer in Egyptian mummies dated to the 20th dynasty (Ruffer, Reference Ruffer1910), the microscopic analysis of eggs is the most usual approach to diagnose helminths preserved in historical contexts. Examination of the shape, size, the presence of opercula, caps and ornamental features of the egg shell, allows for a relatively accurate determination of the residues to the family or genus level. Identification at the species level is possible (for example, Enterobius vermicularis, Clonorchis sinensis, Schistosoma mansoni and S. haematobium) but uncommon as eggs from many species from the same genus can present similar morphological characteristics (for example, the eggs from the family Tæniidae: Tænia sp. and Echinococcus sp.). However, the specific determination would result in a more far-reaching understanding of the health status of the ancient populations under study and the host–parasite relationship.
To overcome this limitation, it can be appropriate to use immunological or molecular tools to identify the species precisely. Immunological diagnosis was performed for the first time in the late 1980s with the detection of Giardia intestinalis oocysts in a coprolite from the American site Big Bone Cave, dated to 2200 Before Present (BP) (Faulkner et al. Reference Faulkner, Patton and Johnson1989), using immunofluorescence. Since the 2000s, G. intestinalis and other intestinal protozoa of medical importance, such as the human pathogenic amoeba Entamoeba histolytica or the coccidia Cryptosporidium parvum, have been identified several times in remains from the Americas, Europe and the Middle East using Enzyme-Linked Immunosorbent Assay (ELISA) (Gonçalves et al. Reference Gonçalves, Araújo, Duarte, da Silva, Reinhard, Bouchet and Ferreira2002, Reference Gonçalves, da Silva, de Andrade, Reinhard, da Rocha, Le Bailly, Bouchet, Ferreira and Araujo2004; Le Bailly et al. Reference Le Bailly, Gonçalves, Lefèvre, Roper, Pye, Araujo and Bouchet2006, Reference Le Bailly, Romon and Kacki2014; Mitchell et al. Reference Mitchell, Stern and Tepper2008; Le Bailly and Araujo, Reference Le Bailly and Araujo2016). Using immunochromatography, such as the ParaSight™-F test, the detection of Plasmodium sp. (Miller et al. Reference Miller, Ikram, Armelagos, Walker, Harer, Shiff, Baggett, Carrigan and Maret1994; Cerutti et al. Reference Cerutti, Marin, Massa and Savoia1999) in Egyptian and Sudanese mummies (4150 years BP) has been reported with up to 35–40% positivity for malaria. In these cases, however, unspecific reactions of the antibodies used in the test have raised questions about the reality of the recovered frequencies of the disease (Bouchaud et al. Reference Bouchaud, Houzé, Longuet, di Piazza, Ruggieri, Sécardin, Coulaud and Bras2000). The sole detection of a helminth using immunological techniques was reported by Deelder et al. (Reference Deelder, Miller, De Jonge and Krijger1990), with the detection using ELISA of S. haematobium from an Egyptian mummy dated to 5150 years BP. PCR combined with molecular hybridization experiments have been performed on Chilean mummified tissues dated to 2000 BP, leading to the identification of Trypanosoma cruzi (Ferreira et al. Reference Ferreira, Britto, Cardoso, Fernandes, Reinhard and Araujo2000).
In 2001, the first ancient DNA (aDNA) sequences from the helminth Ascaris sp. and the protist T. cruzi were published (Loreille et al. Reference Loreille, Roumat, Verneau, Bouchet and Hänni2001; Madden et al. Reference Madden, Salo, Streitz, Aufderheide, Fornaciari, Jaramillo, Vallejo, Yockteng, Arriaza, CARDENAS-ARROYO and Guhl2001). Since then, approximately 20 publications concerning palaeoparasitology have reported the detection of parasite aDNA. The vast majority of them targeted one to three taxa by PCR, followed by Sanger sequencing. Molecular hybridization, random amplified polymorphic DNA (RAPD) and polymerase chain reaction (PCR) have also been used to identify parasites such as Trichuris sp., Ascaris sp., E. vermicularis and T. cruzi (Guhl et al. Reference Guhl, Jaramillo, Vallejo, Yockteng, Cardenas-Arroyo, Fornaciari, Arriaza and Aufderheide1999; Iñiguez et al. Reference Iñiguez, Araújo, Ferreira and Vicente2003; Jaeger and Iñiguez, Reference Jaeger and Iñiguez2014). Visualization of the PCR or RAPD products with electrophoresis gives interesting and quick information, but amplicon sequencing needs to be performed to assure the specificity of the results. Indeed, archaeological samples can be considered as environmental samples, colonized by numerous species of bacteria, fungi, viruses, etc. Public genetic databases used to verify the specificity of DNA primers and probes, do not list all the genomes of these organisms. These uncharacterized organisms may present similar electrophoresis profiles to the targeted species. If they are present in the sample, they can lead to false-positive conclusions. DNA from these untargeted organisms amplified by the enzymatic reaction can cause chimerical molecules due to jumping PCR, promoted by template damage, a characteristic of aDNA (Pääbo et al. Reference Pääbo, Irwin and Wilson1990). This can also lead to false-positive results when sequencing is not performed.
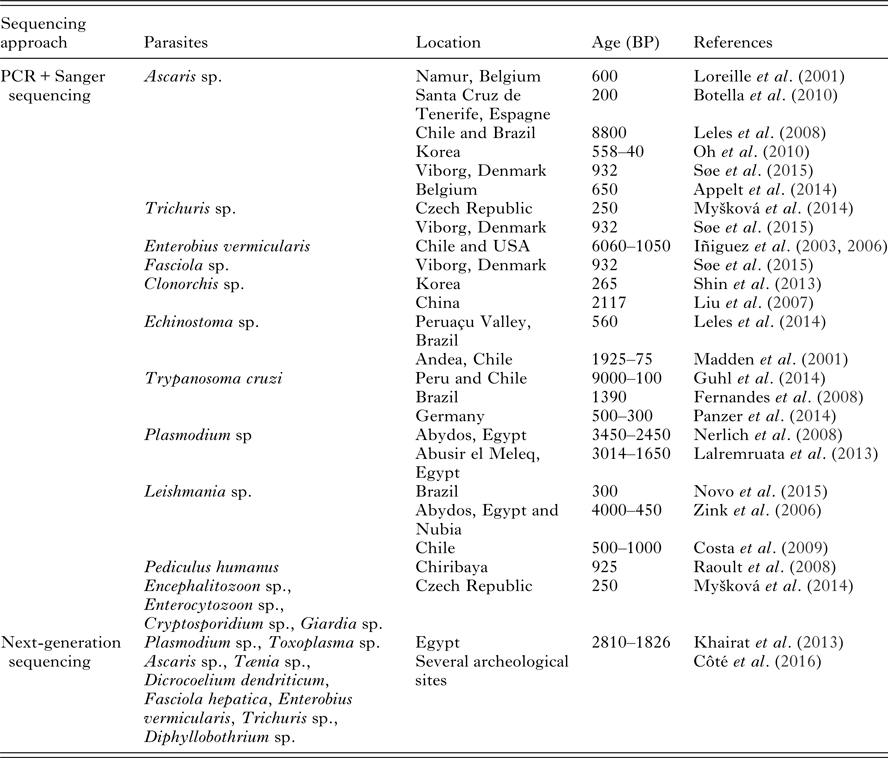
Next-generation sequencing is not commonly used for the analysis of ancient human parasites since only three published works propose this approach. Our paper presents molecular palaeoparasitological works from the past 15 years. Table 1 summarizes the articles that we discuss in this review. We will also expose the benefits of high-throughput approaches and the perspectives for palaeogenomics and parasites.
Table 1. Palaeoparasitological findings confirmed with sequencing approaches. The review has been established based on publications available in 2016
PALAEOPARASITOLOGY AND SAMPLES
Whether they are colonized with bacteria, viruses, or fungi, the majority of the remains studied with palaeogenetics (bones, teeth or coprolites, for example) belong to a single-living organism. On the other hand, samples studied in palaeoparasitology are more complex, since the analyses are often performed on sediment sampled from cesspits, occupational layers, ditches or latrines, and are potentially made up of a mixture of organic matter. This mixture can originate from several individuals, humans and animals, all living together in villages or settlements. Palaeoparasitological remains associated with individuals, such as coprolites or mummified organs, are scarce, especially in Europe (Bouchet et al. Reference Bouchet, Harter and Le Bailly2003; Appelt et al. Reference Appelt, Drancourt and Le Bailly2016). Accuracy issues in the diagnosis of ancient parasites with microscopic approaches can occur due to the difficulty in distinguishing human parasites from animal parasites in many cases. For example, with Taeniidae, it is not possible to distinguish eggs from the genus Tænia sp. (T. saginata, T. solium and T. asiatica, which may infest humans) from eggs from the Echinococcus sp. genus (eggs from E. multilocularis and E. granulosus can be found in canidae or felidae feces). This is also the case for many nematodes belonging to the Strongylidea order (Le Bailly et al. Reference Le Bailly, Lepetz, Samashev, Francfort and Bouchet2008). Moreover, if various strains of a species are present within a mixed sample, it is not possible to assess this situation solely with microscopic analysis.
MOLECULAR PALAEOPARASITOLOGY
Molecular palaeoparasitology, i.e. the genetic determination of ancient parasites, can overcome the problem of mixed samples and the diagnostic limitations of microscopy. It is possible to target genes that will allow for the discrimination of taxa with similar egg characteristics. Moreover, with the careful choice of genetic targets, it is possible to distinguish taxa at the species level. Accurate taxonomic determination would be more precise and highlight important parasitological, anthropological, ecological and archaeological information. Indeed, helminths present complex life cycles involving various intermediate hosts. A better understanding of interactions between the final and intermediate hosts could be possible with specific identification. For instance, discrimination between the Tænia saginata and T. solium tapeworms would indicate the human consumption of undercooked beef or pork, respectively.
The molecular approach can also identify parasites even when their eggs are destroyed or damaged due to taphonomic processes (Leles et al. Reference Leles, Araújo, Ferreira, Vicente and Iñiguez2008; Côté et al. Reference Côté, Daligault, Pruvost, Bennett, Gorgé, Guimaraes, Capelli, Bailly, Geigl and Grange2016). Discrepancies between microscopic and palaeogenetic analyses due to the heterogeneous distribution of eggs and DNA within the sediment samples can be expected. If different subsamples from the archaeological remains are independently processed with both approaches, the likelihood of identifying taxa with the genetic approach only or with microscopy only is high. Moreover, the DNA and the eggs in the sediment are subject to various biological, chemical and physical factors that affect their preservation (Levy-Booth et al. Reference Levy-Booth, Campbell, Gulden, Hart, Powell, Klironomos, Pauls, Swanton, Trevors and Dunfield2007; Morrow et al. Reference Morrow, Newby, Piombino-Mascali and Reinhard2016). For these reasons, it has been suggested that palaeoparasitological samples should be analysed with a complementary approach combining microscopy and genetics (Cleeland et al. Reference Cleeland, Reichard, Tito, Reinhard and Lewis2013; Appelt et al. Reference Appelt, Armougom, Le Bailly, Robert and Drancourt2014; Côté et al. Reference Côté, Daligault, Pruvost, Bennett, Gorgé, Guimaraes, Capelli, Bailly, Geigl and Grange2016).
The determination of new genetic variants is also possible with the molecular approach. Phylogenetic analysis would then provide precious information on past diversity and cross-transmission events between humans and animals, such as during the domestication process of animals. Reference DNA sequences are required to proceed with PCR and sequencing leading to species identification. Thus, palaeogenetic analyses are confined to species for which genetic references are available. This strongly limits the number of taxa that can be targeted, in particular for animal parasites. Moreover, the morphological identification of modern specimens prior to genetic characterization must be accurate. An example of the impact of taxonomic misassignment has been exposed by Cleeland et al. (Reference Cleeland, Reichard, Tito, Reinhard and Lewis2013). The misidentification of Physaloptera sp. as Contracaecum spiculigerum due to the immature stage of development of the reference specimen was reported in that paper. Finally, phylogenetic analysis can also be limited by the poor representativeness of modern intraspecific variants. In 2013, most of the mitochondrial sequences from Ascaris sp. in Genebank were obtained from specimens from China or the USA. In that case, discrimination between ancient single-nucleotide polymorphisms (SNP) and modern biogeographic SNP can be laborious.
PALAEOGENETICS AND HUMAN GASTROINTESTINAL HELMINTHS
Between 2001 and 2015, six human gastrointestinal helminths were targeted by PCR after the observation of intact eggs under a light microscope. The soil-transmitted nematode Ascaris sp. (roundworm) was the most intensively analysed. This helminth has a similar life cycle to the whipworms from the genus Trichuris. Co-infestations with these two nematodes are frequent when food and water are contaminated with human feces. Ascaris lumbricoides and Ascaris suum are described as species that infest humans and pigs, respectively. However, genetic discrimination based on mitochondrial genes is not possible. Differences in the mitogenomes of the two species appear to be associated with geographical distribution rather than signifying specific variants (Liu et al. Reference Liu, Wu, Song, Wei, Xu, Lin, Zhao, Huang and Zhu2012). Recent works suggest that the cytochrome c oxidase- (cox 1) haplogroup can cluster Ascaris with human or pig hosts. However, the modern distribution of this nematode is complex. Indeed, specific mitochondrial haplogroups cluster with a specific host in endemic regions but zoonotic infection appears to be frequent in Europe (Betson et al. Reference Betson, Nejsum, Bendall, Deb and Stothard2014). Due to the degradation of aDNA molecules (Pääbo, Reference Pääbo1989), the 450 base pair (bp) fragments used to determine the cox 1 haplogroup may not be appropriate for palaeogenetic studies. aDNA from the roundworm Ascaris has been identified in French, Spanish, Korean and Danish remains sampled from coprolites, from the surface of pelvis or from egg concentrations processed in sediments (Loreille et al. Reference Loreille, Roumat, Verneau, Bouchet and Hänni2001; Leles et al. Reference Leles, Araújo, Ferreira, Vicente and Iñiguez2008; Botella et al. Reference Botella, Vargas, Rosa, Arnay, Leles, Reimers, Vicente and Iñiguez2010; Oh et al. Reference Oh, Seo, Lim, Lee, Lee, Lee and Shin2010; Søe et al. Reference Søe, Nejsum, Fredensborg and Kapel2015). The cytochrome b and/or the 18S rDNA genes were targeted in five studies and do not allow for the identification of the species. The mitochondrial haplogroup 07, typical of parasites from pigs in Europe, has been described from the Danish sediments from which eggs from the human whipworm Trichuris trichiura were also determined. This is indirect evidence of an anthropic infestation, since the sample was a mixture of various structures, including a waste-dumping pit that may have been used for the disposal of organic matter from humans and animals (Søe et al. Reference Søe, Nejsum, Fredensborg and Kapel2015). Ascaris and Trichuris co-infestations have also been described for the microscopic analysis of remains from Korea, Denmark and the Czech Republic and amplification of the 18S rDNA gene confirmed contamination by T. trichiura (Oh et al. Reference Oh, Seo, Lim, Lee, Lee, Lee and Shin2010; Myšková et al. Reference Myšková, Ditrich, Sak, Kváč and Cymbalak2014; Søe et al. Reference Søe, Nejsum, Fredensborg and Kapel2015).
The human pinworm, E. vermicularis, was also studied using palaeogenetics. Infestations by this parasite are frequent in modern populations. The light and fragile eggs are easily transmitted by handling contaminated objects. Preserved eggs were described in archaeological remains from the Americas (Fry and Moore, Reference Fry and Moore1969; Araujo et al. Reference Araujo, Ferreira, Confalonieri, Nunez and Ribeiro Filho1985; Gonçalves et al. Reference Gonçalves, Araujo and Ferreira2003) and the Middle East (Nezamabadi et al. Reference Nezamabadi, Aali, Stöllner, Mashkour and Le Bailly2013; Paknazhad et al. Reference Paknazhad, Mowlavi, Dupouy Camet, Esmaeili Jelodar, Mobedi, Makki, Kia, Rezaeian, Mohebali, Sarlak and Najafi2016). Molecular determination based on the 5S rDNA intergenic region was performed on 27 coprolites from Chile and the USA, dated between 4110 before Christ (BC) to 900 anno Domini (AD) (Iñiguez et al. Reference Iñiguez, Araújo, Ferreira and Vicente2003, Reference Iñiguez, Reinhard, Carvalho Gonçalves, Ferreira, Araújo and Paulo Vicente2006). Strikingly, with the exception of Herrmann (Reference Herrmann1988), who analysed latrines from medieval Germany, no microscopic study has yet characterized pinworm eggs in European samples. Thus, no palaeogenetic diagnosis based on the prior observation of eggs has been performed for this region, raising questions concerning the early distribution of pinworms.
Fasciola sp. and Clonorchis sp. are flukes that cause distomatosis in animals and humans. Fasciola hepatica and F. gigantica are distributed worldwide, while most of the infections by C. sinensis are in China (Wu et al. Reference Wu, Qian, Huang and Hong2012). Palaeogenetic analyses have been performed on Danish remains, identifying F. hepatica, in addition to T. trichiura and Ascaris sp. (Søe et al. Reference Søe, Nejsum, Fredensborg and Kapel2015). The environmental origin of the remains does not allow for clear conclusions concerning animal or human infestation by the fluke. Internal Transcribed Spacer (ITS) 1–2, and cox 1 sequences of C. sinensis have been obtained on a Chinese corpse buried in 167 BC (Liu et al. Reference Liu, Liu, Zhang, Long, Lei and Li2007) and in a medieval Korean mummy (Shin et al. Reference Shin, Oh, Lee, Chai, Lee, Hong, Lee and Seo2013). While sequences of ITS1 in the latter showed that 100% identify with modern references, 15 SNP have been identified in the Chinese remains. Combined with previous modern genetic studies (Lee and Huh, Reference Lee and Huh2004), ITS1 appears to be a good marker to study the phylogeny of this Asian fluke. Echinostoma sp., a fluke with worldwide distribution, has also been described in a Brazilian human coprolite (560 BP) by the sequencing of a fragment of cox 1 gene and ITS1 region. Resolution at the species level identified the species E. paraensi (Leles et al. Reference Leles, Cascardo, Freire, Maldonado, Sianto and Araújo2014).
PALAEOGENETICS, PROTISTS AND OTHER PARASITES
The vector-borne unicellular parasite Trypanosoma sp. is widely distributed throughout the world. The causal agent of Chagas disease, T. cruzi, is confined to Central and South America and affects ~10 million people (CDC-parasites). With a reservoir made up of over 100 species of animals and with 12 species of insects acting as vectors, colonization by this parasite appears to predate the arrival of humans in the region.
Morphological indications of infestation reported on mummies, such as the mega visceral syndrome (Rothhammer et al. Reference Rothhammer, Allison, Núñez, Standen and Arriaza1985), and molecular analysis, suggest that humans have been infected by T. cruzi for a long time. The sequencing of kinetoplastic DNA from four Chilean mummies dated between 25 and 1875 AD provides the first ancient sequences of T. cruzi (Madden et al. Reference Madden, Salo, Streitz, Aufderheide, Fornaciari, Jaramillo, Vallejo, Yockteng, Arriaza, CARDENAS-ARROYO and Guhl2001). Subsequently, 283 samples from Peru and Chile were analysed by hybridization of PCR products without sequence analyses. The infection rate reached over 40% of the samples, for a period of 9000 years (7050 BC–1850 AD) (Aufderheide et al. Reference Aufderheide, Salo, Madden, Streitz, Buikstra, Guhl, Arriaza, Renier, Wittmers and Fornaciari2004). Phylogenetic analyses were performed on a set of 17 mummies from the same location using mitochondrial and nuclear gene datasets. Six different genotypes were described (TcI, TcII, TcIV, TcV, TcVI and TcBat), corresponding to oral-transmission strains (Guhl et al. Reference Guhl, Auderheide and Ramírez2014). TcI was also identified in a Brazilian mummy (560 AD) (Fernandes et al. Reference Fernandes, Iñiguez, Lima, Souza, Ferreira, Vicente and Jansen2008). These results highlight the high level of genetic diversity of T. cruzi and raise questions about vectorial and/or oral routes of infestation in the past. A multidisciplinary study, including palaeoradiology, palaeogenetics, forensics, and isotope analysis, was performed on an unidentified mummy conserved in a German museum, suggesting a South American origin for the corpse. Palaeopathological investigations by radiology showed evidence of Chagas disease, confirmed by the palaeogenetic identification of the parasite (Panzer et al. Reference Panzer, Peschel, Haas-Gebhard, Bachmeier, Pusch and Nerlich2014).
Plasmodium sp. is a blood parasite that infects vertebrates and four species can cause malaria in humans. Due to its vector-borne transmission cycle involving the insect Anopheles, this disease is restricted to tropical and subtropical regions below an altitude of 1500 m (CDC-parasites). While classic molecular markers for Plasmodium in ancient remains have led to unspecific sequences, the Plasmodium falciparum chloroquine resistant transporter gene was identified in two Egyptian samples (Nerlich et al. Reference Nerlich, Schraut, Dittrich, Jelinek and Zink2008). These remains, dating from the New Kingdom to the Late Period in West-Thebes, were associated with osteopathological signs of anaemia, which supported the genetic results. Palaeopathological analyses were also performed on mummies from the Egyptian necropolis of Abusir el Meleq (1064 BC–300 AD), with molecular evidence of co-infection with Mycobacterium tuberculosis and P. falciparum (Lalremruata et al. Reference Lalremruata, Ball, Bianucci, Welte, Nerlich, Kun and Pusch2013). A 196-bp fragment of the apical membrane antigen gene was targeted, followed by genotyping of the merozoite surface protein-1 gene. Many cases of co-infection by tuberculosis and malaria were reported at the end of the 19th century/beginning of the 20th century.
More than 20 species of Leishmania can infest humans, causing visceral, cutaneous or mucosal forms of leishmaniasis. Leishmania tropica, L. major and L. aethiopica, L. infantum and L. donovani are species that affect populations from the Old World. Leishmania mexicana and L. (V.) braziliensis are complex species commonly affecting populations from the New World (CDC-parasites). In 2009, DNA from skulls with unspecific signs of infection have been analysed using PCR and sequencing. The identification of L. donovani in this area of San Pedro de Atacama (Chile) was unexpected. This arid desert at high altitude is unlikely to have been endemic and the presence of Leishmania can be associated with the migration of people from an endemic neighbourhood (Costa et al. Reference Costa, Matheson, Iachetta, Llagostera and Appenzeller2009). Palaeogenetic analysis reported the infection of a Brazilian mummy dating to the Colonial Period by L. tarentolae (Novo et al. Reference Novo, Leles, Bianucci and Araujo2015). This species is considered non-pathogenic for humans, but infected geckos from the Old World. This result raises questions as to an ancient occurrence of the disease, but also as regards the ecology of the parasite, since this L. tarentolae was not detected in modern Brazilian reptiles. Leishmania donovani was identified in remains from Abydos (Middle Kingdom, Egypt, 2050–1650 BC) and Upper Nubia (550–1500 AD) (Zink et al. Reference Zink, Spigelman, Schraut, Greenblatt, Nerlich and Donoghue2006). Due to the sandfly vector and woodland distribution, Leishmania is presumed to be non-endemic in ancient Egypt. Since no molecular evidence of Leishmania infection has been retrieved from the Pre-to Early Dynasty (3500–2800 BC) and in the Late Period (2050–500 BC), ancient Egyptians are thought to have been contaminated during trade with Nubians.
Analyses of sediment from archaeological structures preserved in Prague (18th–19th centuries) indicate infestation by several intestinal parasites, including Trichuris sp., Capillaria sp., Ascaris sp., Diphyllobothrium latum, Dicrocoelium dendriticum, F. hepatica, and from the Opisthorchiidae, Taeniidae and Heterophyidae families (Myšková et al. Reference Myšková, Ditrich, Sak, Kváč and Cymbalak2014). Molecular analyses detected the whipworm Trichuris sp., the microsporidia Encephalitozoon sp. and Enterocytozoon bieneusi, the Apicomplexa Cryptosporidium sp., and the flagellate Giardia sp. Cysts from these organisms are rarely preserved over time, thus they do not provide a source or potential protection for ancient biomolecules.
Finally, the human louse Pediculus humanus was collected from hair belonging to a Chiribaya mummy from Peru (1025 AD cal.). Sequence analysis and phylogeny based on cox 1 gene identified the phylotype A of P. humanus, the most widespread phylotype in modern populations (Raoult et al. Reference Raoult, Reed, Dittmar, Kirchman, Rolain, Guillen and Light2008).
NEXT-GENERATION SEQUENCING APPROACHES
Palaeogenetic analyses based on classic Sanger sequencing of PCR products can give an interesting overall picture of the presence of pathogenic agents in individuals from ancient populations. It is also an appropriate approach to support the hypothesis of parasitic infestation when macroscopic signs are observed. However, methodological issues can arise for the analysis of multiple samples and when several taxa are targeted. This would drastically increase the number of PCR amplification experiments, which can be expensive and time-consuming. Moreover, depending on the taxonomic identification level that can be achieved with the primer pairs, infestation with multiple species from the same genus can be difficult to diagnose with cloning techniques and Sanger sequencing. While the sediment collected can be a mixture of organic matter from several individuals and/or animals, then this approach can draw an incomplete palaeopathological picture. Next-generation sequencing technologies can generate a large number of sequences and thus provide better parasitological diagnosis.
As it is not limited by the selection of species targeted by PCR, shotgun sequencing may offer a more complete diagnosis of the pathogens (parasites and bacteria) preserved in the sample. The study of intestinal microbiota by metagenomic analysis can lead to the identification of gastrointestinal parasites. Metagenomic sequencing was performed on a sample collected from an Egyptian mummy (806 BC–124 AD) and led to the identification of the pathogens Plasmodium sp. and Toxoplasma gondii (Khairat et al. Reference Khairat, Ball, Chang, Bianucci, Nerlich, Trautmann, Ismail, Shanab, Karim, Gad and Pusch2013). Toxoplasma sp. is an intracellular parasite with a complex life cycle, involving the Felidae, and mice or birds as intermediate hosts. Humans can be infected by eating undercooked meat from infected animals or by consuming food or water contaminated with cat feces. Cats were a cult animal in ancient Egypt and the identification of T. gondii in a human mummy indicated proximity with this animal. Metagenomics can thus provide important knowledge on intestinal microbiota and pathogens that affected ancient populations. However, this approach may be not appropriate for genetic studies focusing on parasites. A relatively high throughput is necessary to detect ancient parasitic molecules mixed with high numbers of prokaryotic, viral and fungal DNA molecules. For example, while microscopy and PCR identified Ascaris sp. in a 14th-century coprolite, metagenomic analyses performed on the same sample yielded ~ 107 000 reads, with 32% corresponding to known sequences, and none have been assigned to Ascaris (Appelt et al. Reference Appelt, Armougom, Le Bailly, Robert and Drancourt2014). Again, the association of several approaches, combining microscopic identification with molecular determination, will result in more reliable palaeoparasitological identification.
Depending on the research project, the sequencing of amplicons with medium throughput technology could provide more information than Sanger sequencing of individual PCR products. Even though this approach restricts the diversity of the taxa that can be identified, its low cost can be an alternative to metagenomics. Based on this type of approach, 14 archaeological sites were analysed with a method called aMPlex Torrent. Out of the 17 targeted species of gastrointestinal helminths, nine taxa were identified (Ascaris sp., T. saginata, T. solium, T. asiatica, D. dendriticum, F. hepatica, E. vermicularis, T. trichiura, Diphyllobothrium sp.) (Côté et al. Reference Côté, Daligault, Pruvost, Bennett, Gorgé, Guimaraes, Capelli, Bailly, Geigl and Grange2016). This flexible method appears to be a suitable approach for screening taxa from multiple samples.
PROCEDURES FOR aDNA ANALYSES
Since the first report on the analysis of aDNA fragments from a 140-year-old Quagga, an extinct subspecies of horse (Higuchi et al. Reference Higuchi, Bowman, Freiberger, Ryder and Wilson1984), palaeogenetics raises interest from the scientific community. Due to the characteristics of aDNA [minute amount of short fragments diluted in environmental DNA, and biochemical modifications of nucleotides due to taphonomic processes (Pääbo, Reference Pääbo1989)], authenticity of the DNA sequences also became a focus of interest. In 2000, a list of criteria of authenticity has been published (Cooper and Poinar, Reference Cooper and Poinar2000) and as we consider that this list is still relevant and topical, we briefly present the most important points.
A separated and isolated laboratory dedicated for the processing of ancient samples (preparation of the remains, extraction and purification of DNA and preparation of the reagents for enzymatic reactions) is essential to avoid contamination with modern DNA. Multiple negative controls, such as parallel mock purifications and non-template PCR controls should always be included during the preparation of DNA and PCR. Positive control using modern DNA should be avoided. Positive results should be repeatable using the same DNA extract, and when possible, re-extraction from the same samples should confirm the results. Finally, quantitative characterization of the DNA extract should be performed to assess the starting number of templates and to test the molecular behaviour: due to the nature of the ancient molecules, PCR products longer than 1000 bp are not usual. PCR amplification efficiency should be inversely correlated to the product size to reflect this characteristic. When possible, independent laboratories should proceed to the replication of the complete experiment (from the extraction to the sequencing analysis). In our opinion, two additional criteria need to be mentioned. Prevention of contamination, especially when working with human remains, should start during archaeological excavations. Contaminant DNA can come from the archaeologist or the anthropologist who excavate and handle the remains. Palaeogeneticists and archaeologists should collaborate to establish good field practice. Finally, as commercial reagents are potential sources of modern DNA, we suggest proceeding to the decontamination of buffers, enzymes and primers, especially when targeting ancient human or bacterial DNA (Champlot et al. Reference Champlot, Berthelot, Pruvost, Bennett, Grange and Geigl2010).
CONCLUSIONS/FUTURE DIRECTIONS
Over the last century, gastrointestinal helminths from archaeological sites have been described based on the preservation of fossilized remains. While their eggs can be preserved for a long time, with evidence of parasitic infestation in palaeontological coprolites (Poinar and Boucot, Reference Poinar and Boucot2006; Dentzien-Dias et al. Reference Dentzien-Dias, Poinar, de Figueiredo, Pacheco, Horn and Schultz2013; Hugot et al. Reference Hugot, Gardner, Borba, Araujo, Leles, Stock Da-Rosa, Dutra, Ferreira and Araújo2014; Silva et al. Reference Silva, Borba, Dutra, Leles, da-Rosa, Ferreira and Araujo2014), taphonomic processes can affect their morphological features, leading to imprecise taxonomic identification. Five taphonomic factors have been described as having an impact on egg preservation: abiotic, contextual, anthropogenic, organismal and ecological factors (Morrow et al. Reference Morrow, Newby, Piombino-Mascali and Reinhard2016). Badly preserved or destroyed eggs can lead to an underestimation of the occurrence of taxa. This was the case for the E. vermicularis pinworm in Europe. Eggs have been described in a single archaeological site in Germany (Herrmann, Reference Herrmann1988), while positive molecular results have been obtained from various European locations since the Neolithic period (Côté et al. Reference Côté, Daligault, Pruvost, Bennett, Gorgé, Guimaraes, Capelli, Bailly, Geigl and Grange2016). Genetics can thus provide an accurate diagnosis of species, even when the eggs are poorly preserved. The biomolecules are also affected by taphonomic processes or can be adsorbed on mineral particles (Blum et al. Reference Blum, Lorenz and Wackernagel1997). This phenomenon could lead to false-negative results, in particular if the molecular methods used for detection are not sensitive enough. Besides, for Apicomplexa parasites, such as Toxoplasma sp. or Plasmodium sp., cysts are rarely preserved. Genetics appears today to be the only approach for the identification of these pathogens.
In Côté et al. (Reference Côté, Daligault, Pruvost, Bennett, Gorgé, Guimaraes, Capelli, Bailly, Geigl and Grange2016), we studied 25 samples from 14 archaeological using microscopic determination and genetic analyses. For each samples, using only one approach would have resulted in a less accurate parasitic diagnose. Complementary studies combining palaeogenetics with microscopic identification are thus the best method to provide the most complete and accurate diagnosis possible.
Genetic determination is still limited by the presence of modern reference sequences. The lack of modern molecular analyses performed on animal parasites or rare human parasites, such as the capillariid group, limits the diversity of species that can be characterized in archaeological contexts. Intraspecific genetic variants, including geographical variants, are impossible to distinguish from ancestral states if few modern sequences are available. This can also cause limited phylogenetic conclusions. Until recently, Ascaris phylogeny was only based on Asian and American samples and did not allow for discrimination between animal and human species, if such differences truly exist. Extensive analyses of mitochondrial and nuclear markers of worms from multiple locations allow for a better understanding of the natural history of this nematode (Betson et al. Reference Betson, Nejsum, Bendall, Deb and Stothard2014). Including ancient sequences in the analyses would increase our knowledge of its evolution and may indicate past cross-transmission events between humans and swine.
As discussed, new sequencing technologies have been underemployed for molecular palaeoparasitology up until now. Recently, target-enrichment capture methods based on DNA hybridization have been used for the reconstruction of ancient prokaryotic genomes (Bos et al. Reference Bos, Schuenemann, Golding, Burbano, Waglechner, Coombes, McPhee, DeWitte, Meyer, Schmedes, Wood, Earn, Herring, Bauer, Poinar and Krause2011; Maixner et al. Reference Maixner, Krause-Kyora, Turaev, Herbig, Hoopmann, Hallows, Kusebauch, Vigl, Malfertheiner, Megraud, O'Sullivan, Cipollini, Coia, Samadelli, Engstrand, Linz, Moritz, Grimm, Krause, Nebel, Moodley, Rattei and Zink2016). Palaeogenomics appears to be the next step for a better understanding of the evolution of parasites. For instance, ancient genomes could highlight genetic adaptations due to the migration of populations, to host shifting, to climatic changes, and to the use of antiparasitic drugs.
FINANCIAL SUPPORT
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.




