Published online by Cambridge University Press: 04 August 2022
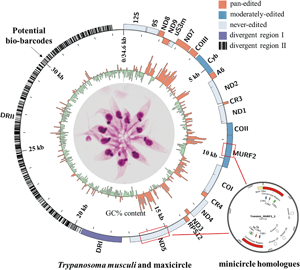
Trypanosoma musculi is a, globally distributed, mouse-specific haemoflagellate, of the family Trypanosomatidae, which shares similar characteristics in morphology with Trypanosoma lewisi. The kinetoplast (mitochondrial) DNA of Trypanosomatidae flagellates is comprised of catenated maxicircles and minicircles. However, genetic information on the T. musculi kinetoplast remains largely unknown. In this study, the T. musculi maxicircle genome was completely assembled, with PacBio and Illumina sequencing, and the size was confirmed at 34 606 bp. It consisted of 2 distinct parts: the coding region and the divergent regions (DRs, DRI and II). In comparison with other trypanosome maxicircles (Trypanosoma brucei, Trypanosoma cruzi and T. lewisi), the T. musculi maxicircle has a syntenic distribution of genes and shares 73.9, 78.0 and 92.7% sequence identity, respectively, over the whole coding region. Moreover, novel insertions in MURF2 (630 bp) and in ND5 (1278 bp) were found, respectively, which are homologous to minicircles. These findings support an evolutionary scenario similar to the one proposed for insertions in Trypanosoma cruzi, the pathogen of American trypanosomiasis. These novel insertions, together with a deletion (281 bp) in ND4, question the role of Complex I in T. musculi. A detailed analysis of DRII indicated that it contains numerous repeat motifs and palindromes, the latter of which are highly conservative and contain A5C elements. The comprehensively annotated kinetoplast maxicircle of T. musculi reveals a high degree of similarity between this parasite and the maxicircle of T. lewisi and suggests that the DRII could be a valuable marker for distinguishing these evolutionarily related species.
Contributed equally.