Introduction
The northern tree shrew Tupaia belangeri chinensis Anderson (Mammalia: Scandentia: Tupaiidae) is a novel ideal animal model for human disease, due to its small size, easy breeding, rapid reproduction and close genetic relationship to primates (Xu et al., Reference Xu, Chen, Nie, Jiang and Yao2012, Reference Xu, Zhang, Liang, Lü, Chen, Chen, Zhou and Yao2013; Xiao et al., Reference Xiao, Liu and Chen2017; Tang et al., Reference Tang, Wu, Xiao, Ge, Wei, Li, Wang, Zeng, Fu and Zhang2018; Wang et al., Reference Wang, Ruan, Song, Wang, Tong, Kuang, Lu, Li, Han, Dai and Sun2021). Tupaia belangeri chinensis is omnivorous, eating fruits, seeds, insects and small vertebrates, which is mainly distributed in southwest China (including Yunnan and Sichuan Provinces) (Xiang and Yang, Reference Xiang and Yang2014) and can act as the intermediate and definitive host for some helminth parasites and protozoa (Brack et al., Reference Brack, Naberhaus and Heymann1987; Tian et al., Reference Tian, Pen and Hou1989; Xiang et al., Reference Xiang, Rosenthal, He, Wang, Wang, Song, Shen, Li and Yang2010; Xiang and Yang, Reference Xiang and Yang2014). However, our present knowledge of the species composition of the acanthocephalans of the northern tree shrew is very limited. To date, only Prosthenorchis sp. (Archiacanthocephala: Oligacanthorhynchidae) has been reported from T. belangeri chinensis (Tian et al., Reference Tian, Pen and Hou1989).
In the present study, some acanthocephalan specimens were collected from T. belangeri chinensis in China. In order to accurately identify these acanthocephalan specimens to species level, the detailed morphology of these specimens was studied using light and scanning electron microscopy. Moreover, the nuclear small subunit ribosomal DNA (18S), internal transcribed spacer (ITS) and large subunit ribosomal DNA (28S), and mitochondrial cytochrome c oxidase subunit 1 (cox1) and subunit 2 (cox2) genes were sequenced and analysed. Phylogenetic analyses were also performed based on the 18S + cox1 sequence data using maximum likelihood (ML) and Bayesian inference (BI) methods, to clarify the phylogenetic relationships between this species and its congeners.
Materials and methods
Morphological observation
Acanthocephalans were isolated from the intestine of the northern tree shrew T. belangeri chinensis in Kunming, Yunnan Province, China. Specimens were washed and kept in cold water for several hours until the proboscis everted, and then stored in 80% ethanol until studied. For light microscopical studies, specimens were made in impermanent mount slide and cleared in lactophenol. Photomicrographs were recorded using a Nikon® digital camera coupled to a Nikon® optical microscopy. For scanning electron microscopy (s.e.m.), specimens were post-fixed in 1% OsO4, dehydrated via an ethanol series and acetone, and then critical point dried. The specimens were coated with gold and examined using a Hitachi S-4800 scanning electron microscope at an accelerating voltage of 20 Kv. Measurements (range, followed by the mean in parentheses) are given in micrometres unless otherwise stated.
Molecular procedures
Genomic DNA from the mid-body of one male and one female was extracted using a Column Genomic DNA Isolation Kit (Shanghai Sangon, China) according to the manufacturer's instructions. The partial 18S region was amplified by polymerase chain reaction (PCR) using the forward primer (5′-AGATTAAGCCATGCATGCGTAAG-3′) and the reverse primer (5′-TGATCCTTCTGCAGGTTCACCTAC-3′) (Garey et al., Reference Garey, Near, Nonnemacher and Nadler1996). The partial 28S region was amplified by PCR using 4 overlapping PCR fragments of 700–800 bp. Primers for 28S amplicon 1 were forward 5′-CAAGTACCGTGAGGGAAAGTTGC-3′ and reverse 5′-CAGCTATCCTGAGGGAAAC-3′; amplicon 2, forward 5′-ACCCGAAAGATGGTGAACTATG-3′ and reverse 5′-CTTCTCCAAC(T/G)TCAGTCTTCAA-3′; amplicon 3, forward 5′-CTAAGGAGTGTGTAACAACTCACC-3′ and reverse 5′-AATGACGAGGCATTTGGCTACCTT-3′; amplicon 4, forward 5′-GATCCGTAACTTCGGGAAAAGGAT-3′ and reverse 5′-CTTCGCAATGATAGGAAGAGCC-3′ (García-Varela and Nadler, Reference García-Varela and Nadler2005). The partial ITS region was amplified by PCR using the forward primer (5'-GTCGTAACAAGGTTTCCGTA-3') and the reverse primer (5'-TATGCTTAAATTCAGCGGGT-3') (Král'ová-Hromadová et al., Reference Král'ová-Hromadová, Tietz, Shinn and Spakulová2003). The partial cox1 region was amplified by PCR using the forward primer (5'-GGTCAACAAATCATAAAGATATTGG-3') and the reverse primer (5'-TAAACTTCAGGGTGACCAAAAAATCA-3') (Gómez et al., Reference Gómez, Serra, Carvalho and Lunt2002). The partial cox2 region was amplified by PCR using the forward primer (5'-AATGTTTGATGGGTTTAGAG-3') and the reverse primer (5'-AACACTGACCATATATAACC-3') (designed by the present study). The cycling conditions were as described previously (Li et al., Reference Li, Wayland, Chen and Yang2019). PCR products were checked on GoldView-stained 1.5% agarose gels and purified with Column PCR Product Purification Kit (Shanghai Sangon, China). Sequencing for each amplification product was carried out from both directions. Sequences were aligned using ClustalW2 and adjusted manually. The DNA sequences obtained herein were compared (using the algorithm BLASTn) with that available in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov).
Phylogenetic analyses
Phylogenetic analyses were performed based on the 18S + cox1 sequence data using maximum likelihood (ML) inference with IQ-TREE and Bayesian inference (BI) with Mrbayes 3.2 (Ronquist et al., Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Hӧhna, Larget, Liu, Suchard and Huelsenbeck2012; Nguyen et al., Reference Nguyen, Schmidt, Haeseler and Minh2015), respectively. Polyacanthorhynchus caballeroi Diaz-Ungria & Rodrigo, 1960 (Polyacanthocephala: Polyacanthorhynchida) was treated as the out-group. The in-group included 15 species of the class Archiacanthocephala representing 6 different genera belonging to 3 orders Gigantorhynchida, Moniliformida and Oligacanthorhynchida. The detailed information of acanthocephalan species included in the present phylogenetic analyses is provided in Table 1.
Table 1. Species of Archiacanthocephala with their detailed information of genetic data included in the phylogenetic analyses

We used a built-in function in IQTREE to select a best-fitting substitution model for the sequences according to the Bayesian information criterion (Posada and Crandall, Reference Posada and Crandall2001). The GTR + F + I + G4 model was identified as optimal nucleotide substitution model. Reliabilities for ML tree were tested using 1000 bootstrap replications and BI tree was tested using 10 million generations. In the ML tree, bootstrap support (BS) values ⩾90 were considered as fully supported; whereas BS values ⩾70 and <90 were considered as generally supported. In the BI tree, Bayesian posterior probabilities (BPP) ⩾0.90 were considered as fully supported, whereas BPP values ⩾0.70 and <0.90 were considered as generally supported.
Results
Description of Moniliformis tupaia n. sp. (×Figs 1–3)
General
Medium-sized worms with small proboscis (Figs 1A; 3A, D). Trunk aspinose, nearly cylindrical and slender, showing pseudosegmentation characteristic of the genus Moniliformis (Fig. 3A). Anterior trunk tapering to gourd-shaped. Proboscis small compared to the trunk, cylindrical, with two apical sensory pores and 14 spiral longitudinal rows of 7–8 hooks each (Figs 1A, B, E; 2A, C; 3B). Proboscis hooks small, with simple roots (Figs 1E, F; 2A–C). Proboscis receptacle double-walled, cerebral ganglion at base of proboscis receptacle (Figs 1A, B; 3D). Neck short. Lemnisci very long, unequal, distinctly longer than proboscis receptacle (Figs 1A; 3D). Gonopore terminal in both sexes (Figs 1C, D; 2D).

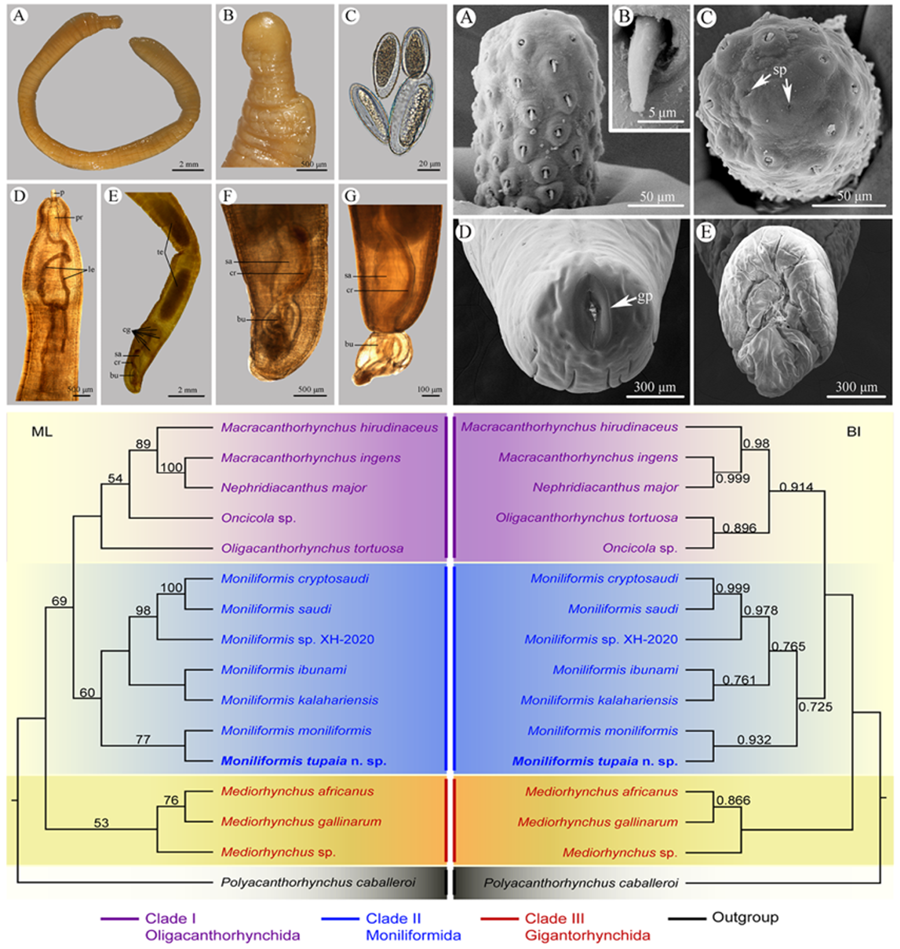
Figure 1. Moniliformis tupaia n. sp. collected from Tupaia belangeri chinensis (Scandentia: Tupaiidae) in China. (A) anterior part of male. (B) anterior end of male. (C) posterior end of female. (D) posterior part of male. (E) proboscis. (F) longitudinal row of hooks. (G) posterior end of male. (H) egg. Scale bars: A, D, G = 1000 μm; B, C = 500 μm; E, F = 100 μm; H = 30 μm.

Figure 2. Scanning electron micrographs of Moniliformis tupaia n. sp. collected from Tupaia belangeri chinensis (Scandentia: Tupaiidae) in China. (A) Proboscis of male, lateral view. (B) Magnified image of proboscis hook. (C) Proboscis of male (sensory pores arrowed), apical view. (D) Posterior end of female (gonopore arrowed). (E) Copulatory bursa. Abbreviations: sp, sensory pores; gp, gonopore.
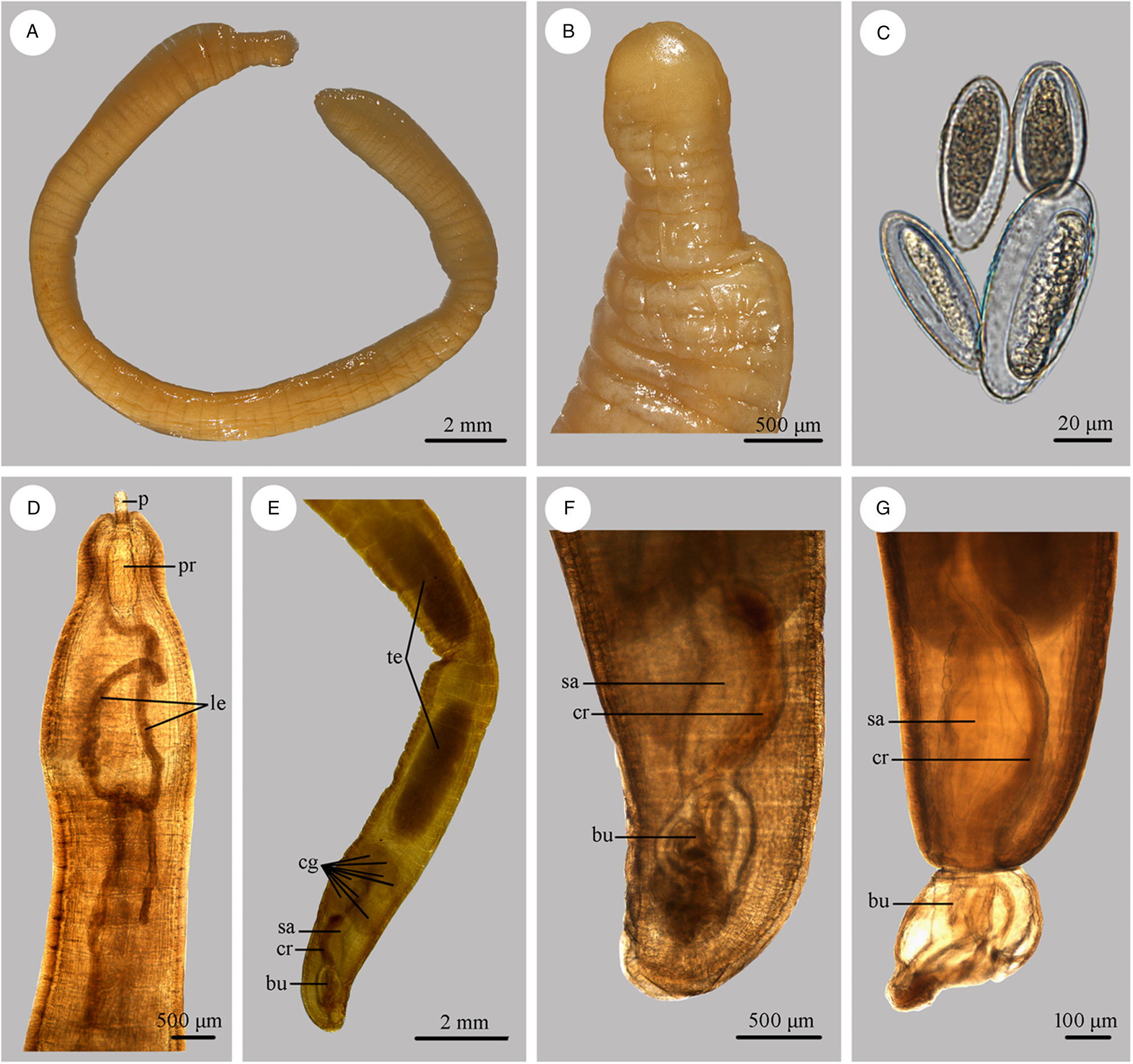
Figure 3. Photomicrographs of Moniliformis tupaia n. sp. collected from Tupaia belangeri chinensis (Scandentia: Tupaiidae) in China. (A) Mature female. (B) Tail of female. (C) Eggs. (D) Anterior part of male. (E) Posterior part of male. (F) Posterior end of male (copulatory bursa not everted). (G) Posterior end of male (copulatory bursa evaginabled). Abbreviations: bu, bursa; cg, cement glands; cr, cement reservoir; le, lemnisci; p, proboscis; pr, proboscis receptacle; sa, saeftigen's pouch; te, testis.
Male [based on 5 mature specimens]
Trunk 34.0–47.5 (40.0) mm long, maximum width 1.83–2.07 (1.93) mm. Proboscis 366–439 (395) long, 146–171 (162) wide. Proboscis hooks similar in shape, 27–31 (30), 28–31 (29), 26–29 (28), 24–27 (26), 21–25 (24), 18–23 (21), 18–22 (20), 17–22 (19) in length from anteriorly to posteriorly. Neck 49–100 (68) long, 180–244 (204) wide. Proboscis receptacle 854–1195 (1000) long, 341–390 (368) wide. Shorter lemniscus 5.00–6.10 (5.72) mm long, longer lemniscus 7.68–9.32 (8.61) mm long. Testes two, oval, nearly equal in size; anterior testis 2.44–3.49 (3.05) mm long, 732–1024 (888) wide; posterior testis 2.44–3.54 (2.99) mm long, 585–1000 (849) wide (Fig. 1D). Cement glands eight, ovoid, clustered together; a short distance from posterior testis, 854–1829 (1256) long, 659–854 (761) wide (Figs 1D, G; 3E). Saefftigen's pouch 927–1512 (1317) long, 293–463 (378) wide. Copulatory bursa evaginabled or not everted, 780–1171 (971) long, 366–854 (644) wide (Figs 1G, 2E; 3F, G). Gonopore nearly terminal (Fig. 1D, G).
Female [Based on 1 mature specimen]. Trunk 41.0 mm long, maximum width 2.15 mm. Proboscis 390 long, 171 wide. Proboscis hooks similar in shape, 28–33 (31), 29–33 (32), 27–31 (29), 26–29 (27), 25–29 (27), 23–26 (24), 22–26 (24), 20–25 (22) in length from anteriorly to posteriorly. Neck 73 long, 195 wide. Proboscis receptacle 927 long, 439 wide. Shorter lemniscus 4.63 mm long, longer lemniscus 8.54 mm long. Uterine bell 350 long, 300 wide. Uterus 680 long, vagina 270 long (Fig. 1C). Eggs ellipsoid, 58–68 (65) × 24–32 (30) in size (n = 20) (Figs 1H; 3C). Gonopore nearly terminal (Figs 1C, 2D).
Type-host: Northern tree shrew Tupaia belangeri chinensis Anderson (Scandentia: Tupaiidae).
Type-locality: Kunming, Yunnan Province, China.
Site in host: Intestine.
Type specimens: Holotype, male (HBNU-A-M20231201CL); allotype, female (HBNU-A-M20231202CL); paratypes: 4 males (HBNU-A-M20231203CL); deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, China.
Etymology: The species name refers to the generic name of the type host.
Molecular characterization
Partial 18s region
Two 18S sequences of M. tupaia n. sp. obtained herein are both 1188 bp in length, with no nucleotide divergence detected. In the genus Moniliformis, there are 6 species with their 18S sequences available in GenBank, including M. cryptosaudi Amin, Heckmann, Sharifdini and Albayati, Reference Amin, Heckmann, Sharifdini and Albayati2019 (MH401043), M. ibunami Lynggaard, García-Prieto, Guzmán-Cornejo and García-Varela, Reference Lynggaard, García-Prieto, Guzmán-Cornejo and García-Varela2021 (MW136271, MW136272), M. kalahariensis Meyer, 1931 (MH401042), M. moniliformis (Bremser, 1811) (HQ536017, Z19562), M. saudi Amin, Heckmann, Mohammed and Evans, Reference Amin, Heckmann, Mohammed and Evans2016 (KU206782) and Moniliformis sp. XH-2020 (OM388438). Pairwise comparison of the 18S sequences of M. tupaia n. sp. obtained herein with that of other Moniliformis species showed no nucleotide divergence (Moniliformis sp. XH-2020) to 0.66% (M. ibunami) nucleotide divergence. The 18S sequences of M. tupaia n. sp. obtained herein were deposited in GenBank database (http://www.ncbi.nlm.nih.gov) (under accession numbers PP002170, PP002171).
Partial 28s region
Two 28S sequences of M. tupaia n. sp. obtained herein are both 2692 bp in length, with no nucleotide divergence detected. In the genus Moniliformis, there are M. ibunami (MW136276, MW136277) and M. moniliformis (AY829086) with 28S sequences available in GenBank. Pairwise comparison of the 28S sequences of M. tupaia n. sp. obtained herein with that of other Moniliformis species showed 1.49% (M. ibunami) to 2.04% (M. moniliformis) nucleotide divergence. The 28S sequences of M. tupaia n. sp. obtained herein were deposited in GenBank database (http://www.ncbi.nlm.nih.gov) (under accession numbers PP002172, PP002173).
Partial ITS region
Two ITS sequences of M. tupaia n. sp. obtained herein are both 671 bp in length, with no nucleotide divergence detected. In the genus Moniliformis, only M. moniliformis (AF416415) has an ITS sequence available in GenBank. Pairwise comparison of the ITS sequences of M. tupaia n. sp. obtained herein with that of M. moniliformis showed 17.2% nucleotide divergence. The ITS sequences of M. tupaia n. sp. obtained herein were deposited in GenBank database (http://www.ncbi.nlm.nih.gov) (under accession numbers PP002174, PP002175).
Partial cox1 region
Two cox1 sequences of M. tupaia n. sp. obtained herein are both 658 bp in length, with no nucleotide divergence detected. In the genus Moniliformis, 7 species have their cox1 sequences available in GenBank, namely M. cryptosaudi (MH401041), M. ibunami (MW115575, MW115576), M. kalahariensis (MH401040), M. moniliformis (AF416998), M. necromysi Gomes, Costa, Gentile, Vilela and Maldonado, Reference Gomes, Costa, Gentile, Vilela and Maldonado2020 (MT803593), M. saudi (KU206783, OQ078755) and Moniliformis sp. XH-2020 (OK415026). Pairwise comparison of the cox1 sequences of M. tupaia n. sp. obtained herein with that of other Moniliformis species showed 24.9% (M. ibunami) to 27.3% (M. moniliformis) nucleotide divergence. The cox1 sequences of M. tupaia n. sp. obtained herein were deposited in GenBank database (http://www.ncbi.nlm.nih.gov) (under accession numbers OR997666, OR997667).
Partial cox2 region
Two cox2 sequences of M. tupaia n. sp. obtained herein are both 514 bp in length, with no nucleotide divergence detected. In the genus Moniliformis, only Moniliformis sp. XH-2020 (OK415026) has a cox2 sequence available in GenBank. Pairwise comparison of the cox2 sequences of M. tupaia n. sp. obtained herein with that of Moniliformis sp. XH-2020 showed 23.6% nucleotide divergence. The cox2 sequences of M. tupaia n. sp. obtained herein were deposited in GenBank database (http://www.ncbi.nlm.nih.gov) (under accession numbers PP002935, PP002936).
Phylogenetic analyses
Phylogenetic trees of the class Archiacanthocephala constructed from the 18S + cox1 sequence data using ML and BI methods have almost identical topology (Fig. 4). The representatives of Archiacanthocephala were divided into three major clades. Clade I included species of Macracanthorhynchus, Nephridiacanthus, Oligacanthorhynchus and Oncicola, representing the order Oligacanthorhynchida. Among them, the phylogenetic results showed N. major (Bremser, 1811) clustered together with M. ingens (Von Linstow, 1879). Clade I contained species of Moniliformis, representing the order Moniliformida. Clade III included species of Mediorhynchus, representing the order Gigantorhynchida. In the genus Moniliformis, M. tupaia n. sp. showed sister relationship with M. moniliformis.
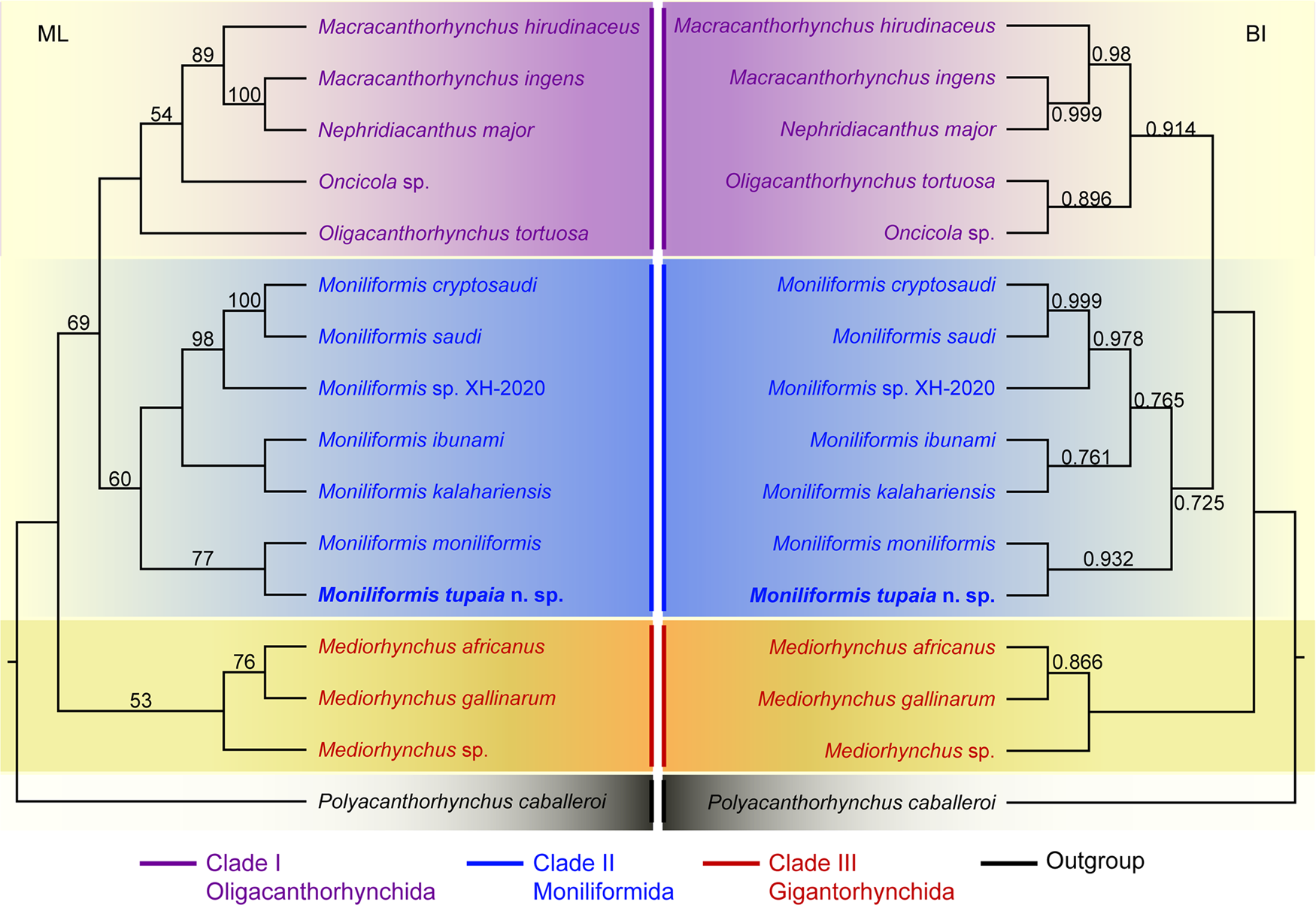
Figure 4. Maximum likelihood (ML) and Bayesian inference (BI) based on the 18S + cox1 sequence data showing the phylogenetic relationships of representatives of Archiacanthocephala. Polyacanthorhynchus caballeroi (Polyacanthocephala: Polyacanthorhynchidae) was chose as the outgroup. Bootstrap support (BS) values ⩾50 in ML tree and Bayesian posterior probabilities (BPP) ⩾ 0.70 in BI tree are shown.
Discussion
The present specimens collected from the northern tree shrew T. belangeri chinensis belong to the genus Moniliformis (Moniliformida: Moniliformidae), due to the pseudosegmented trunk, the very small cylindrical proboscis, the double-walled proboscis receptacle, the very long lemnisci and the presence of 8 spherical cement glands (Travassos, Reference Travassos1917; Van Cleave, Reference Van Cleave1923, Reference Van Cleave1953; Southwell and Macfie, Reference Southwell and Macfie1925; Yamaguti, Reference Yamaguti1963; Schmidt, Reference Schmidt1972; Amin, Reference Amin1987). The genus Moniliformis currently comprises 19 species mainly parasitic in mammals (Amin, Reference Amin2013; Amin et al., Reference Amin, Heckmann, Mohammed and Evans2016, Reference Amin, Heckmann, Sharifdini and Albayati2019; Martins et al., Reference Martins, Del Rosario Robles and Navone2017; Gomes et al., Reference Gomes, Costa, Gentile, Vilela and Maldonado2020; Lynggaard et al., Reference Lynggaard, García-Prieto, Guzmán-Cornejo and García-Varela2021). Among them, only M. moniliformis and Moniliformis sp. XH-2020 have been reported in China (Chen, Reference Chen1933; Chandler, Reference Chandler1941; Dai et al., Reference Dai, Yan, Li, Zhang, Liu, Gao, Ohiolei, Wu, Guo, Fu and Jia2022).
The proboscis of the new species has 14 spiral longitudinal rows of 7–8 simple rooted hooks each, which is similar to the proboscis of following species M. acomysi Ward and Nelson, Reference Ward and Nelson1967, M. cryptosaudi, M. moniliformis, M. saudi and M. siciliensis Meyer, Reference Meyer1932. Moniliformis tupaia n. sp. can be easily distinguished from M. acomysi by its much longer proboscis and lemnisci (proboscis 0.37–0.44 mm and lemnisci 5.00–9.32 mm long in the male of new species vs proboscis 0.19–0.36 mm and lemnisci 2.73–4.42 mm long in the male of M. acomysi). Moniliformis tupaia n. sp. differs from M. cryptosaudi and M. saudi by having larger cement glands (854–1829 long in the new species vs 312–811 long in the latter two species). Moreover, M. cryptosaudi and M. saudi are both parasitic in hedgehogs (Erinaceomorpha: Erinaceidae) in Saudi Arabia and Iraq, but the new species parasitizes the northern tree shrew T. belangeri chinensis in China. Furthermore, molecular analysis revealed strong genetic divergence (25.9‒26.9% difference in nucleotides in the cox1 region) between the new species and M. cryptosaudi and M. saudi. Moniliformis siciliensis is a poorly known acanthocephalan species only reported from the garden dormouse Eliomys quercinus Linnaeus (Mammalia: Rodentia) in the Italian island of Sicily (Meyer, Reference Meyer1932; Petrochenko, Reference Petrochenko1958). The new species differs from M. siciliensis in having shorter lemnisci (5.00–9.32 mm long in the former vs about 10.0 mm in the latter) and different localities and hosts.
Moniliformis moniliformis is an important zoonotic acanthocephan species, parasitizing rodents, canines and felines worldwide, including China (Meyer, Reference Meyer1932; Petrochenko, Reference Petrochenko1958; Yamaguti, Reference Yamaguti1963; Ward and Nelson, Reference Ward and Nelson1967; Bhattacharya, Reference Bhattacharya2007; Naidu, Reference Naidu2012). This species has a proboscis with 11–14 (usually 12) rows of 9–14 (usually 10–11) hooks each and much larger trunk (over 50.0 mm long in male), which is different from the new species (vs proboscis with 14 rows of 7–8 hooks each, and male 34.0–47.5 mm long in M. tupaia n. sp.). Additionally, molecular analysis displayed 27.3% and 17.2% nucleotide divergence in the cox1 and ITS regions, between the new species and M. moniliformis, which strongly indicated that they represent 2 distinct species. Dai et al. (Reference Dai, Yan, Li, Zhang, Liu, Gao, Ohiolei, Wu, Guo, Fu and Jia2022) reported Moniliformis sp. XH-2020 from the plateau zokor (Eospalax fontanierii baileyi) in China, but they only provided the mitochondrial genomic data of their specimens (they did not describe the morphology). Pairwise comparison between M. tupaia n. sp. and Moniliformis sp. XH-2020 showed 27.3% and 23.6% nucleotide divergence in the cox1 and cox2 regions. Consequently, they belong to different species.
The class Archiacanthocephala currently includes 4 orders, namely Gigantorhynchida, Moniliformida, Oligacanthorhynchida and Apororhynchida (Amin, Reference Amin2013). However, the phylogenetic relationships of the 4 orders remain unclear, due to a lack of genetic data of some taxa, especially the order Apororhynchida. The previous phylogenetic study using 18S or 18S + cox1 genetic data suggested a close affinity between Moniliformida and Gigantorhynchida (Amin et al., Reference Amin, Evans, Heckmann and El-Naggar2013, Reference Amin, Sharifdini, Heckmann and Zarean2020). However, our phylogenetic results based on the 18S + cox1 data suggested Moniliformida is a sister to Oligacanthorhynchida, rather than Gigantorhynchida, which are consistent with some previous studies based on cox1 or 18S data (Gomes et al., Reference Gomes, Costa, Gentile, Vilela and Maldonado2020; Rodríguez et al., Reference Rodríguez, Amin, Heckmann, Sharifdini and D'Elía2021; Amin et al., Reference Amin, Heckmann, Sist and Basso2021, Reference Amin, Chaudhary, Heckmann, Swenson and Singh2022). In the order Oligacanthorhynchida, the present phylogeny displayed Nephridiacanthus major nested in representatives of Macracanthorhynchus (clustered together with M. ingens), which challenged the current systematic position of this species. The present results agreed well with the previous phylogenetic study based on cox1 data (Rodríguez et al., Reference Rodríguez, Amin, Heckmann, Sharifdini and D'Elía2021). In the genus Moniliformis, our molecular phylogenetic analyses indicate that M. tupaia n. sp. is a sister to M. moniliformis.
Data availability statement
The nuclear and mitochondrial DNA sequences of M. tupaia n. sp. obtained herein were deposited in the GenBank database [http://www.ncbi.nlm.nih.gov, accession numbers: PP002170, PP002171 (18S); PP002172, PP002173 (28S); PP002174, PP002175 (ITS); OR997666, OR997667 (cox1); PP002935, PP002936 (cox2)]. Type specimens of M. tupaia n. sp. (HBNU-A-M20231201-3CL) were deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, China.
Author contributions
HXC and LL contributed to the study design and identification of the acanthocephalan specimens. HXC, CHZ and LL sequenced and analysed genetic data. HXC, ZJY and LL conducted the phylogenetic analyses. JM and FQC provided acanthocephalan specimens. HXC and LL wrote the manuscript. All authors read and approved the final manuscript.
Financial support
This study was supported by the National Natural Science Foundation of China (NSFC31872197) and the National Key R&D Program of China (Grant No. 2022YFC2601200) for Dr Liang Li, the Ecological Postdoctoral Research Mobile Station and the Post-doctoral Research Program of Hebei Normal University for Dr Hui-Xia Chen.
Competing interests
The authors declare that they have no conflict of interest.
Ethical standards
This study was conducted under the protocol of Hebei Normal University. All applicable national and international guidelines for the protection and use of animals were followed.