Introduction
Insect-borne diseases are a significant public health problem worldwide. The most important vectors of these diseases are mosquitoes and sand flies and, between these, mosquitoes are the best-studied vectors. Much is known about mosquito interactions with malaria-causing plasmodia and arboviruses (Saraiva et al., Reference Saraiva, Kang, Simoes, Anglero-Rodriguez and Dimopoulos2016). Many aspects of these interactions, including the mosquito immune responses to pathogenic microbes and the role of resident microbiota on the infection and on immune responses of the insects have been reviewed (Clayton et al., Reference Clayton, Dong and Dimopoulos2014; Saraiva et al., Reference Saraiva, Kang, Simoes, Anglero-Rodriguez and Dimopoulos2016). In this review, we discuss some of these aspects in sand flies (Diptera: Psychodidae: Phlebotominae). We will focus mainly on bacteria found in resident microbiota and also on some aspects of the interactions of sand fly vectors with viruses, bacteria and Leishmania, with special emphasis on sand fly immune responses.
Sand flies are well-known vectors of leishmaniasis, but they also transmit viruses (Depaquit et al., Reference Depaquit, Grandadam, Fouque, Andry and Peyrefitte2010; Alkan et al., Reference Alkan, Bichaud, de Lamballerie, Alten, Gould and Charrel2013) and bacteria (Herrer and Christensen, Reference Herrer and Christensen1975; Maroli et al., Reference Maroli, Feliciangeli, Bichaud, Charrel and Gradoni2013). The presence of viruses in sand flies has been reported since the middle of last century (reviewed in Tesh and Chaniotis, Reference Tesh and Chaniotis1975; Tesh, Reference Tesh1988; Depaquit et al., Reference Depaquit, Grandadam, Fouque, Andry and Peyrefitte2010). Among the viruses transmitted by sand flies, Phleboviruses are considered the most significant, since many of the viruses in this genus are human pathogens capable of causing symptoms varying from short-term fever to meningitis, encephalitis and haemorrhagic fever (Alkan et al., Reference Alkan, Bichaud, de Lamballerie, Alten, Gould and Charrel2013). Sand flies from the Lutzomyia genus have been incriminated as vectors of the bacteria causing bartonellosis, also known as Carrion's disease, Oroya fever or ‘verruga peruana’ (Schultz, Reference Schultz1968; Cohnstaedt et al., Reference Cohnstaedt, Beati, Caceres, Ferro and Munstermann2011; Battisti et al., Reference Battisti, Lawyer and Minnick2015). This disease is characterized by symptoms such as fever and hemolytic anaemia and in a later phase can produce nodular skin lesions (reviewed in Maguina et al., Reference Maguina, Guerra and Ventosilla2009). Little is known about the molecular interactions of sand flies with viruses and bacteria. The following review will present the progress of the field in addressing the responses of the sand fly to the diverse agents it propagates, with a specific emphasis on the expanding understanding of how these responses may be modulated by the insect's microbiota.
Leishmaniases are the most important illnesses transmitted by phlebotomine sand flies. These multi-spectrum diseases present symptoms that vary from ulcerative skin lesions to mucosal deforming lesions (tegumentary leishmaniasis) or liver and spleen hypertrophy (visceral leishmaniasis). Protozoans from the genus Leishmania (Trypanosomatida: Trypanosomatidae) are the aetiological agents of leishmaniases. Around 20 Leishmania species are known to be pathogenic to humans (Maroli et al., Reference Maroli, Feliciangeli, Bichaud, Charrel and Gradoni2013). These digenetic parasites need two hosts to complete their life cycle: one of them a sand fly, while the other can be a human or another mammal. As an exception to sand fly transmission, in Australia biting midges (Diptera: Ceratopogonidae) were implicated in the transmission of the autoctone Leishmania enriettii (Dougall et al., Reference Dougall, Alexander, Holt, Harris, Sultan, Bates, Rose and Walton2011; Seblova et al., Reference Seblova, Sadlova, Vojtkova, Votypka, Carpenter, Bates and Volf2015).
A detailed understanding of how pathogens interact with their vectors and the resident microbes can lead to the discovery of new tools to block disease transmission. New microbe-based blocking tools have been discovered for the mosquitos that transmit malaria (Wang and Jacobs-Lorena, Reference Wang and Jacobs-Lorena2013) and there is an excellent evidence that Wolbachia endosymbionts can be used as a biocontrol measure to block dengue virus (DENV) transmission (Moreira et al., Reference Moreira, Iturbe-Ormaetxe, Jeffery, Lu, Pyke, Hedges, Rocha, Hall-Mendelin, Day, Riegler, Hugo, Johnson, Kay, McGraw, van den Hurk, Ryan and O'Neill2009; Ye et al., Reference Ye, Carrasco, Frentiu, Chenoweth, Beebe, van den Hurk, Simmons, O'Neill and McGraw2015; Joshi et al., Reference Joshi, Pan, McFadden, Bevins, Liang, Lu, Thiem and Xi2017). Deeper knowledge of the interactions among the sand fly, its microbiota and the pathogens these insects transmit could lead to the discovery of new methods to block sand fly-transmitted diseases. Among alternative control strategies is paratransgenesis, where bacteria normally found in a specific insect is engineered to interfere with pathogen transmission (Coutinho-Abreu et al., Reference Coutinho-Abreu, Zhu and Ramalho-Ortigao2010b; Hurwitz et al., Reference Hurwitz, Fieck, Read, Hillesland, Klein, Kang and Durvasula2011a). The first and crucial step in this approach is the identification of suitable commensal microorganisms in the vector. For safety reasons, these microorganisms should be non-pathogenic to man and other animals.
The search for candidates to paratransgenic blockade of Leishmania transmission by the kala-azar vector Phlebotomus argentipes identified two bacteria which met the above requirements, the commensals Bacillus megaterium and Brevibacterium linens (Hillesland et al., Reference Hillesland, Read, Subhadra, Hurwitz, McKelvey, Ghosh, Das and Durvasula2008). More recently, the same group infected P. argentipes with a transgenic GFP expressing bacteria Baccillus subtilis, and demonstrated that the transduced bacteria was stably maintained in the P. argentipes gut (Hurwitz et al., Reference Hurwitz, Hillesland, Fieck, Das and Durvasula2011b).
Along with the fact that the resident microbiota might establish a competitive or mutualistic interaction with acquired pathogens, insects possess an active immune response to balance and protect themselves from diseases and challenges that these microbes may cause. Insect immune responses are triggered through the recognition of evolutionarily conserved pathogen-associated molecular patterns (PAMPs) by host pattern recognition receptors (PRRs). The binding of PAMPs leads to the activation of defense mechanisms and pathways: RNAi, Janus-kinase/signal transducers and activators of transcription (JAK-STAT), Immune deficiency (IMD) and Toll, that will determine the production of effectors molecules such as antimicrobial peptides (AMPs) and reactive oxygen species (ROS) (Lemaitre et al., Reference Lemaitre, Kromer-Metzger, Michaut, Nicolas, Meister, Georgel, Reichhart and Hoffmann1995; Brennan and Anderson, Reference Brennan and Anderson2004; Blair, Reference Blair2011; Zeidler and Bausek, Reference Zeidler and Bausek2013). In addition to combating foreign invaders, components of the insect innate immune system are also involved in stress responses, wound healing and the management of microbial symbiont populations (Welchman et al., Reference Welchman, Aksoy, Jiggins and Lemaitre2009).
In the following text, we will address the complex interactions among sandflies, their microbiota and pathogens they transmit.
Sand fly and viruses
Insects are hosts to a vast variety of viruses. Some viruses are unique to insects (reviewed in Vasilakis and Tesh, Reference Vasilakis and Tesh2015; Roundy et al., Reference Roundy, Azar, Rossi, Weaver and Vasilakis2017), others (Arboviruses) are transmitted to other organisms, including animals and plants (reviewed in Blanc and Gutierrez, Reference Blanc and Gutierrez2015; Ng and Zhou, Reference Ng and Zhou2015). Vector-borne viral diseases such as dengue, chikungunya and Zika are among the most devasting illnesses to afflict humanity.
Viral presence in insects such as mosquitoes and Drosophila elicits an antiviral immune response mediated by different mechanisms (e.g., Toll, IMD, JAK-STAT, etc.). Although each of these signalling pathways plays a specific role in the antiviral response (Kingsolver et al., Reference Kingsolver, Huang and Hardy2013; Merkling and van Rij, Reference Merkling and van Rij2013; Xu and Cherry, Reference Xu and Cherry2014), the RNAi mechanism is reported to be the most active in insect antiviral response (Kemp et al., Reference Kemp, Mueller, Goto, Barbier, Paro, Bonnay, Dostert, Troxler, Hetru, Meignin, Pfeffer, Hoffmann and Imler2013; Nayak et al., Reference Nayak, Tassetto, Kunitomi and Andino2013; Tassetto et al., Reference Tassetto, Kunitomi and Andino2017). RNAi controls virus replication through the small non-coding RNAs called small interfering RNAs (siRNAs) in conjunction with an enzyme complex. These siRNAs associate with Argonaute (Ago) proteins to identify and destroy viral RNAs in a sequence-specific manner. Other eukaryotic small RNAs, such as microRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs), which regulate cellular gene expression (reviewed in Asgari, Reference Asgari2013) and transposon activity (reviewed in Weick and Miska, Reference Weick and Miska2014), have also been implicated in antiviral defense (Vijayendran et al., Reference Vijayendran, Airs, Dolezal and Bonning2013).
Although much is known about viral infections of mosquito vectors and the model organism Drosophila, much less is known about viral infections of sand flies, in which case, viruses can basically be considered as neglected pathogens (Depaquit et al., Reference Depaquit, Grandadam, Fouque, Andry and Peyrefitte2010). Phleboviruses transmitted by sand flies have a relevant role as human pathogens (Alkan et al., Reference Alkan, Bichaud, de Lamballerie, Alten, Gould and Charrel2013). This genus comprises approximately 70 named viruses that are classified into two broad groups according to their antigenic, genomic and/or vectorial relationships: the sand fly fever virus group and the Uukuniemi-like virus group. The sand fly fever group includes Rift Valley fever virus (RVF) transmitted by mosquitoes and Toscana viruses (TOSV) transmitted by phlebotomine sand flies (Depaquit et al., Reference Depaquit, Grandadam, Fouque, Andry and Peyrefitte2010). A large number of new sand fly-borne phleboviruses were recently described based on phlebovirus phylogeny reconstruction (Moriconi et al., Reference Moriconi, Rugna, Calzolari, Bellini, Albieri, Angelini, Cagarelli, Landini, Charrel and Varani2017).
In the Old World, at least 250 million people are exposed to Phlebovirus infections (Moriconi et al., Reference Moriconi, Rugna, Calzolari, Bellini, Albieri, Angelini, Cagarelli, Landini, Charrel and Varani2017). Sand fly fever Sicilian Viruses (SFSV) and TOSV, both transmitted by sand flies, are prominent human pathogens (Ayhan et al., Reference Ayhan, Sherifi, Taraku, Berxholi and Charrel2017). TOSV is an emerging pathogen and the cause of summer meningitis in the Mediterranean region, for which defined reservoirs were not identified. It is unlikely that humans are the reservoir for TOSV because human viremia is too short-lived (Depaquit et al., Reference Depaquit, Grandadam, Fouque, Andry and Peyrefitte2010). Competent sand fly species might act as reservoirs in the viral cycle through transovarial transmission (Maroli et al., Reference Maroli, Ciufolini and Verani1993, Reference Maroli, Feliciangeli, Bichaud, Charrel and Gradoni2013) since male sand flies were found to be infected by TOSV in nature.
Despite the fact that sand flies are proven vectors of leishmaniasis there are only a few reports describing the phlebotomine midgut infection by both Leishmania and a virus. Interestingly, one study showed that wild-caught and laboratory-reared P. papatasi infected with cytoplasmic polyhedrosis virus (CPVs) were refractory to experimental Leishmania major infection (Warburg and Ostrovska, Reference Warburg and Ostrovska1987). CPVs cause a chronic pathology in the sand fly mid-gut that is characterized by structural abnormalities in the epithelium and the peritrophic matrix (PM) that interferes with blood digestion (Warburg and Ostrovska, Reference Warburg and Ostrovska1987). These gut anomalies might hinder attachment to destroyed midgut epithelial cells and also lead to an early exposure of the parasites to sand fly digestive process and immune effector molecules, thus affecting Leishmania development.
In another study, sand flies trapped in an urban area of Marseille, France were infected with either Leishmania or phlebovirus. Curiously dual infections were not detected in this study, despite the local co-circulation of both pathogens (Faucher et al., Reference Faucher, Bichaud, Charrel, Mary, Izri, de Lamballerie and Piarroux2014). These two publications suggest an incompatibility between Leishmania and concomitant virus infection in the sand fly midgut. The complexity of Leishmania-sand fly-virus relationship is illustrated in another work where sand flies from Eastern Thrace and Northern Cyprus were analysed for presence of a virus and/or Leishmania. A pool of Phlebotomus tobbi was found co-infected by Toscana virus and Leishmania infantum (Ergunay et al., Reference Ergunay, Kasap, Orsten, Oter, Gunay, Yoldar, Dincer, Alten and Ozkul2014), implying different levels of coexistence events between various Leishmania and viruses. Further experiments should be performed with individual insects to confirm the coinfection hypothesis. These results reveal the complexity of the relationship between the Leishmania and viruses inside the sand fly mid-gut. More studies should be carried out to identify whether some of the different sand fly virus species could be used to control the Leishmania transmission. Much more is known about sand fly-transmitted viruses in the Old World than in the New World. The importance given to viruses transmitted by sand flies in Europe is due to the gravity of the disease they cause and high incidence in the local population (Depaquit et al., Reference Depaquit, Grandadam, Fouque, Andry and Peyrefitte2010). On the other hand, there is little information about viruses transmitted by sand flies, or the diseases they cause, in the New World. Approximately 500 Phlebotominae species are known in the Americas, of which at least 56 are known to transmit leishmaniasis (Maroli et al., Reference Maroli, Feliciangeli, Bichaud, Charrel and Gradoni2013; Bates et al., Reference Bates, Depaquit, Galati, Kamhawi, Maroli, McDowell, Picado, Ready, Salomon, Shaw, Traub-Cseko and Warburg2015). Comer et al. (Reference Comer, Stallknecht, Corn and Nettles1991) studied a New Jersey serotype (VSV-NJ) virus of the genus Vesiculovirus (family Rhabdoviridae), a causative agent of vesicular stomatitis in cattle, horses, and pigs (Comer et al., Reference Comer, Stallknecht, Corn and Nettles1991) on Ossabaw Island (Georgia, USA). In this study, the authors suggest that the vector for this virus was the phlebotomine sand fly Lutzomyia shannoni. Nunes-Neto et al. (Reference Nunes-Neto, Souza, Acrani, Romeiro, Fumagalli, Vieira, Medeiros, Lima, Lima, Cardoso, Figueiredo, Silva, Tesh, Nunes and Vasconcelos2017) provided insights into the genetic diversity, classification and evolution of phleboviruses by characterizing six previously unclassified phleboviruses isolated in Brazil (Ambe, Anhanga, Joa, Uriurana, Urucuri and Tapara viruses) (Nunes-Neto et al., Reference Nunes-Neto, Souza, Acrani, Romeiro, Fumagalli, Vieira, Medeiros, Lima, Lima, Cardoso, Figueiredo, Silva, Tesh, Nunes and Vasconcelos2017). Aguiar et al. (Reference Aguiar, Olmo, Paro, Ferreira, de Faria, Todjro, Lobo, Kroon, Meignin, Gatherer, Imler and Marques2015) developed an interesting approach to identify viral infections in different insects based in the production of viral small RNAs produced by host responses as exemplified by the RNA interference pathway (Aguiar et al., Reference Aguiar, Olmo, Paro, Ferreira, de Faria, Todjro, Lobo, Kroon, Meignin, Gatherer, Imler and Marques2015). The authors used the small RNA size profile unique signature to identify novel viruses. Using this method six novel viruses were identified in fruit flies, mosquitoes and sand flies. Among these, viruses named Lutzomyia Piaui reovirus 1 (LPRV1) and Lutzomyia Piaui reovirus 2 (LPRV2) and Lutzomyia Piaui nodavirus (LPNV) were found in L. longipalpis.
JAK-STAT is the classical pathway that responds to viral infections in mammals and this is also true for Drosophila (Arbouzova and Zeidler, Reference Arbouzova and Zeidler2006). In mosquitoes, the JAK-STAT pathway is also active against viruses, but the IMD and Toll pathways also play an important role against the infection in some specific mosquito–virus pairs (Ruckert et al., Reference Ruckert, Bell-Sakyi, Fazakerley and Fragkoudis2014; Saraiva et al., Reference Saraiva, Kang, Simoes, Anglero-Rodriguez and Dimopoulos2016).
Surprisingly, there is almost no information about sand fly responses to viral infections. The first report on sand fly immune responses to a virus infection was published by our group in 2008 when a non-specific antiviral response was identified in the L. longipalpis embryonic cell lineage LL5 (Pitaluga et al., Reference Pitaluga, Mason and Traub-Cseko2008). When these cells were transfected with any double-stranded RNAs, including the mimetic poly(I:C), they became resistant to infection with a West Nile Virus-Like Particle (VLP). A similar non-specific antiviral response elicited by dsRNA was also later identified in a shrimp, the honey bee and a bumble bee (Flenniken and Andino, Reference Flenniken and Andino2013; Piot et al., Reference Piot, Snoeck, Vanlede, Smagghe and Meeus2015; Brutscher et al., Reference Brutscher, Daughenbaugh and Flenniken2017). Recently our group carried out an exoproteomics analysis of LL5 cells after transfection with dsRNA (Martins-da-Silva et al., Reference Martins-da-Silva, Telleria, Batista, Marchini, Traub-Cseko and Tempone2018). Among the secreted proteins positively modulated by dsRNA transfection, several were related to immunity and/or anti-viral response. Of special interest was the increased abundance of a phospholipid scramblase, which in mammals acts as an interferon-induced protein mediating antiviral activity.
The knowledge of the immune mechanisms involved in insect responses to the presence of viruses and other pathogens can provide important tools for the control of arboviral diseases that affect millions of people worldwide. In the case of sand flies, the role of resident microbiota in insect viral infections is still an open question. The identification of molecular agents that make an insect refractory to a specific virus is crucial for the development of novel control strategies.
Sand fly microbiota
Phlebotomine sand flies lay their eggs in the soil, animal burrows or tree trunk niches, and larvae develop feeding on organic matter available in these sites (reviewed in Killick-Kendrick, Reference Killick-Kendrick1999; Feliciangeli, Reference Feliciangeli2004; Ready, Reference Ready2013). The ingested food, together with environmental microbes, gives rise to the larval gut resident microbiota. The interest in the study of this microbiota is multifold, from obtaining basic information on how the vector responds to the presence of different microorganisms to how these interact with other pathogens, such as Leishmania. This knowledge may lead to the development of new strategies to control the spread of diseases, such as paratransgenesis. The importance of this subject led to the production of a large number of publications that are listed in Table 1. This list includes studies that describe microbiota obtained from multiple sources of sand fly samples (e.g. nature or laboratory), that use various technical approaches (e.g. culture or direct sequencing) to identify the resident microbes. In a recent publication focusing on the identification of resident microbiota of Phlebotomus perniciosus from the Western Mediterranean region, the authors performed a network analysis which suggests a pattern of interactions between sand flies and their microbiota (Fig. 1) (Fraihi et al., Reference Fraihi, Fares, Perrin, Dorkeld, Sereno, Barhoumi, Sbissi, Cherni, Chelbi, Durvasula, Ramalho-Ortigao, Gtari and Zhioua2017). The knowledge of the presence of a given bacterial species on various sand fly species might help in the development of paratransgenic bacteria targeting multiple vectors.
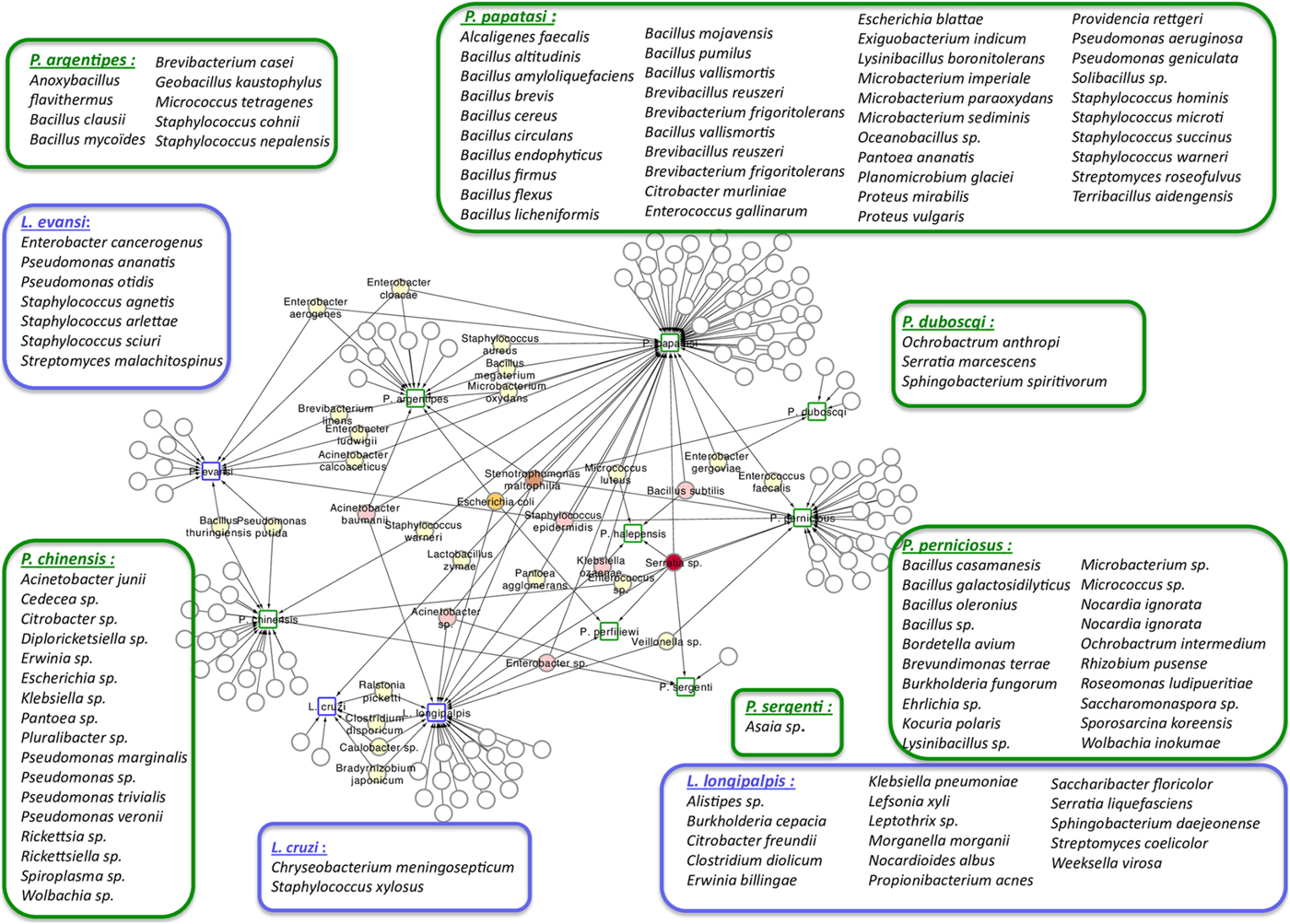
Fig. 1. Network analysis showing the shared bacteria species found in sand flies. Phlebotomus sand flies are identified by squares surrounded by green and bacteria found in Lutzomyia sand flies identified with squares surrounded by blue. Coloured circles represent bacteria species that are shared between sand flies species. White circles represent bacteria species that are unique to each of the sand flies species and are listed inside large rectangles. The network representation suggests some relationships between the 11 studied New World and Old World sand fly species and the bacteria inhabiting their gut. Bacillus thuringiensis was isolated from L. evansi and P. chinensis, two sand fly species belonging to the New World and Old World, respectively (Fraihi et al., Reference Fraihi, Fares, Perrin, Dorkeld, Sereno, Barhoumi, Sbissi, Cherni, Chelbi, Durvasula, Ramalho-Ortigao, Gtari and Zhioua2017).
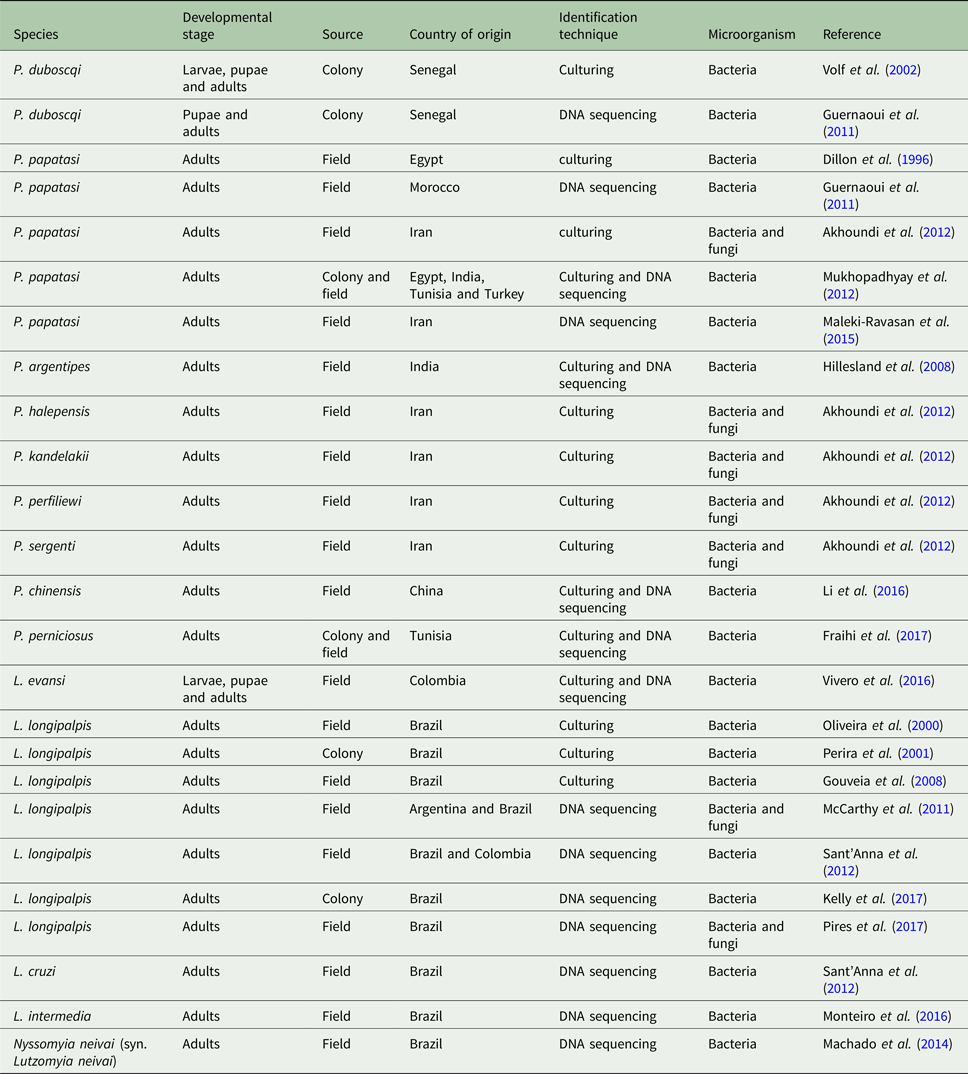
Table 1. Sand fly species with published gut microbiota data
The microbial gut contents of colony-reared Phlebotomus duboscqi were investigated by using standard bacteriological methods to evaluate larvae, pupae and newly emerged insects. In the majority of analysed samples, Ochrobactrum anthropi was the dominant bacterium in all developmental stages, indicating the occurrence of bacterial transtadial passage (Volf et al., Reference Volf, Kiewegova and Nemec2002). Another study on colony-reared P. duboscqi used polymerase chain reaction-temperature gradient gel electrophoresis (TGGE) of the 16S rDNA gene fragment sequences obtained from different developmental stages (Guernaoui et al., Reference Guernaoui, Garcia, Gazanion, Ouhdouch, Boumezzough, Pesson, Fontenille and Sereno2011). In this study, Microbaterium sp. was identified in immature and adult stages. This bacterium was previously identified by a soil bacterial consortium (Zhang et al., Reference Zhang, Shi, Yang, Sha and Zhao2007) indicating that the gut microbiota of immature sand fly stages can be influenced by external microbial populations. This leads to the idea that dispersion of a given microorganism in the larval environment in order to influence or manipulate these insects gut microbiota is a potential strategy for biological control of sand fly-vectored diseases. Indeed, sand fly larvae seem to prefer feeding on a nutrient- and microbe-rich food source mixed with the soil. In the laboratory, when different diets were offered to Lutzomyia intermedia and L. longipalpis, larvae from both species developed better when fed on nutrient-rich food composed of aged rabbit feces (Wermelinger and Zanuncio, Reference Wermelinger and Zanuncio2001). Additionally, it has been suggested that dietary fungi are important to the development of L. intermedia based on pupation ratio (Wermelinger and Zanuncio, Reference Wermelinger and Zanuncio2001).
The gut bacterial content of different developmental stages of Lutzomyia evansi from Central America was studied through culturing in different media and DNA sequencing of the resulting cultures (Vivero et al., Reference Vivero, Jaramillo, Cadavid-Restrepo, Soto and Herrera2016). Identified bacteria across larvae, pupae and adults collected from the same locality included Enterobacter, Pseudomonas, Bacillus and Lysobacter genera (Vivero et al., Reference Vivero, Jaramillo, Cadavid-Restrepo, Soto and Herrera2016). Interestingly, these bacterial genera are abundant in soil (Manfredi et al., Reference Manfredi, Perotti and Martinez2015; Thapa et al., Reference Thapa, Ranjan, Ramakrishnan, Velmourougane and Prasanna2018). The presence of microbial strains in both larvae and adult sand flies will likely increase its utility in developing biological tools to control diseases transmitted by New World sand fly species.
In the same way that ingested food can influence gut microbiota in larvae, it can influence gut microbial content of adults as well. In nature male and female adult sand flies feed on carbohydrate-rich sources such as plant sap and aphid secretions (Wallbanks et al., Reference Wallbanks, Moore, Bennett, Soren, Molyneux, Carlin and Perez1991; Anez et al., Reference Anez, Lugo, Loaiza, Nieves and Orozco1994; Cameron et al., Reference Cameron, Milligan, Llanos-Cuentas and Davies1995; Muller and Schlein, Reference Muller and Schlein2004), while females also feed on blood from birds and mammals, and in some cases, other vertebrates (Ghosh et al., Reference Ghosh, Bhattacharya and Ghosh1990; Mukhopadhyay and Ghosh, Reference Mukhopadhyay and Ghosh1999; Afonso et al., Reference Afonso, Duarte, Miranda, Caranha and Rangel2012; Brito et al., Reference Brito, Almeida Ado, Nakazato, Duarte, Souza Cde and Sousa2014). Several reports focused on the gut microbial content of adult sand flies. Some of them used collected insect in the field, therefore submitted to a diverse diet, while other studies used insect from colonies that were fed on artificial defined diets.
The first report that identified the gut microbial content of New World adult sand flies used L. longipalpis collected from different localities in Brazil, or used insects obtained from a laboratory colony that had been artificially fed on blood or blood followed by sucrose solution (Oliveira et al., Reference Oliveira, Moraes, Goncalves, Giordano-Dias, D'Almeida, Asensi, Mello and Brazil2000; Perira de Oliveira et al., Reference Peterkova-Koci, Robles-Murguia, Ramalho-Ortigao and Zurek2001; Gouveia et al., Reference Gouveia, Asensi, Zahner, Rangel and Oliveira2008). From these studies environmental, gut-associated and opportunistic pathogenic species were identified including Pantoea agglomerans, Stenotrophomonas maltophilia, Enterobacter cloacae, Pseudomonas sp. and Serratia marcescens. These studies indicated that some genera are commonly shared between field and laboratory colony flies.
In addition to the applied culturing methods, L. longipalpis gut microbiota was investigated using denaturing gradient gel electrophoresis (DGGE) of 16S rDNA gene fragments amplified from insects collected at different localities in Brazil and Colombia (Sant'Anna et al., Reference Sant'Anna, Darby, Brazil, Montoya-Lerma, Dillon, Bates and Dillon2012) or high-throughput metatranscriptome analysis using insects collected in Argentina and Brazil (McCarthy et al., Reference McCarthy, Diambra and Rivera Pomar2011). The culture independent-technique using DNA sequencing analysis considerably increased the microbiota detection range, therefore a larger number of bacterial species was identified. Below we will discuss the bacteria that are shared across several sand fly species.
More recently, two other studies were carried using colony-reared (Kelly et al., Reference Kelly, Bahr, Serafim, Ajami, Petrosino, Meneses, Kirby, Valenzuela, Kamhawi and Wilson2017) or field (Pires et al., Reference Pires, Villegas, Campolina, Orfano, Pimenta and Secundino2017) L. longipalpis fed under laboratory conditions. Insects were fed on sucrose, blood or artificially infected by Leishmania, and microbial diversity was analysed by 16S or 18S rDNA sequencing. These studies demonstrated that microbial diversity decreased after blood feeding and that after blood digestion contents were eliminated the bacterial diversity recovered to previous sugar fed insect levels. A similar microbial ressurgence was found in L. intermedia (Monteiro et al., Reference Monteiro, Villegas, Campolina, Pires, Miranda, Pimenta and Secundino2016). The suggested idea of a quiescent resident microbiota present in non-fed females being altered by blood feeding and then returning in gravid females to a profile similar to non-fed females adds an interesting aspect to the microbial dynamics in the sand fly gut. It is important to mention that although bacterial diversity decreases after a blood meal, bacterial numbers actually increase. This might be due to some bacteria overgrowing others in a nutrient-rich environment (Volf et al., Reference Volf, Kiewegova and Nemec2002). Bacteria that are shared among L. longipalpis field and laboratory-reared insects (fed on sucrose, blood or Leishmania infected) (Fig. 2) belong mostly to the Proteobacteria phylum including Pantoea, Serratia, Stenotrophomonas and Erwinia genera. These are known to have an impact on L. longipalpis or other insects immunity (Boulanger et al., Reference Boulanger, Lowenberger, Volf, Ursic, Sigutova, Sabatier, Svobodova, Beverley, Spath, Brun, Pesson and Bulet2004; Telleria et al., Reference Telleria, Sant'Anna, Alkurbi, Pitaluga, Dillon and Traub-Cseko2013; Booth et al., Reference Booth, Cambron, Fisher and Greenlee2015; Heerman et al., Reference Heerman, Weng, Hurwitz, Durvasula and Ramalho-Ortigao2015; Husseneder et al., Reference Husseneder, Park, Howells, Tikhe and Davis2017; Keita et al., Reference Keita, Masuzzo, Royet and Kurz2017). Other identified genera including Acinetobacter, Burkolderia, Citrobacter, Enterobacter, Pseudomonas and Ralstonia are commonly associated with Phlebotomus, mosquitoes, other insects or plants (Warburg, Reference Warburg1991; Dillon et al., Reference Dillon, el Kordy, Shehata and Lane1996; Eilmus and Heil, Reference Eilmus and Heil2009; Akhoundi et al., Reference Akhoundi, Bakhtiari, Guillard, Baghaei, Tolouei, Sereno, Toubas, Depaquit and Abyaneh2012; Maleki-Ravasan et al., Reference Maleki-Ravasan, Oshaghi, Afshar, Arandian, Hajikhani, Akhavan, Yakhchali, Shirazi, Rassi, Jafari, Aminian, Fazeli-Varzaneh and Durvasula2015; Lalithambika and Vani, Reference Lalithambika and Vani2016; Sun et al., Reference Sun, Lu, Zhang, Kumar, Liu, Gong, Zhu, Zhu, Liang, Kuang, Chen, Hu, Cao, Xue and Gong2016; Husseneder et al., Reference Husseneder, Park, Howells, Tikhe and Davis2017; Takeshita and Kikuchi, Reference Takeshita and Kikuchi2017; Thapa et al., Reference Thapa, Pant, Shrestha, Gc, Shrestha, Pandey and Gautam2017; Osimani et al., Reference Osimani, Milanovic, Cardinali, Garofalo, Clementi, Pasquini, Riolo, Ruschioni, Isidoro, Loreto, Franciosi, Tuohy, Petruzzelli, Foglini, Gabucci, Tonucci and Aquilanti2018; Song et al., Reference Song, Wang, Dong, Zhu and Wang2018; Ventura et al., Reference Ventura, Briones-Roblero, Hernandez, Rivera-Orduna and Zuniga2018). Interestingly, Caulobacter genus that comprises environmental associated microbes was found in L. longipalpis as well as in glassy-winged sharpshooter Homalodisca vitripennis (Rogers and Backus, Reference Rogers and Backus2014) but very little is known about this bacterium. The Firmicutes phylum, represented by the Staphylococcus, Clostridium and Bacillus genera, was found in L. longipalpis colony fed and field captured insects (Gupta et al., Reference Gupta, Rastogi, Nayduch, Sawant, Bhonde and Shouche2014; Ngo et al., Reference Ngo, Romano-Bertrand, Manguin and Jumas-Bilak2016; Garofalo et al., Reference Garofalo, Osimani, Milanovic, Taccari, Cardinali, Aquilanti, Riolo, Ruschioni, Isidoro and Clementi2017). Bacteria from these genera are pathogenic to several organisms. There is a special interest in Bacillus species for potential sand fly biological control (Robert et al., Reference Robert, Perich, Schlein and Jacobson1998; Wahba et al., Reference Wahba, Labib and el Hamshary1999). More specifically, Bacillus subtilis can colonize L. longipalpis larvae gut under laboratory conditions (Heerman et al., Reference Heerman, Weng, Hurwitz, Durvasula and Ramalho-Ortigao2015). Curiously the Geobacillus genus, that can form biofilms on food industry surfaces (Seale et al., Reference Seale, Dhakal, Chauhan, Craven, Deeth, Pillidge, Powell and Turner2012; Al-Beloshei et al., Reference Al-Beloshei, Al-Awadhi, Al-Khalaf and Afzal2015) is present in the L. longipalpis studied samples and has been rarely studied in insects. The sand fly bacterial diversity and distribution indicates that although laboratory feeding systems can interfere in the L. longipalpis natural microbial diversity, some bacteria species can persist through the adult fly feeding and environmental differences (field and laboratory conditions). Moreover, the sand fly gut bacteria that are found in common in all the analysed conditions mentioned above are tightly associated with the insect environment, including plants. The sand fly plant-visiting habit for sap consumption (Petts et al., Reference Petts, Tang and Ward1997; Muller and Schlein, Reference Muller and Schlein2004) could be considered as a strategy to deliver microbial contents to manipulate the resident gut microbiota.
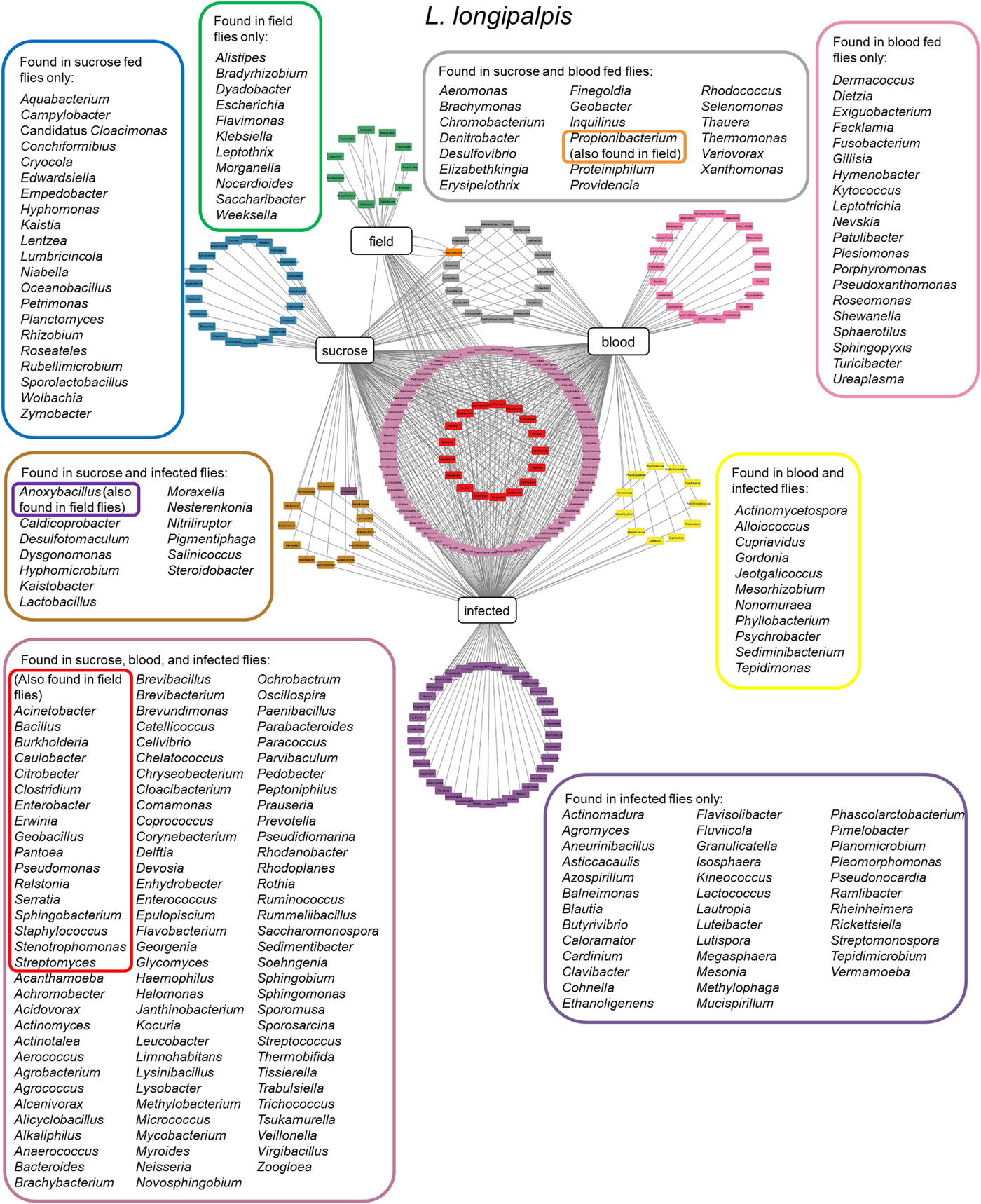
Fig. 2. Lutzomyia longipalpis gut microbiota. Network analysis showing bacteria genera found in L. longipalpis obtained from field collections or laboratory-reared colonies. Feeding regimens are indicated: field (unknown feeding conditions), sucrose, blood, or infected by Leishmania (laboratory artificial feeding). Coloured border rectangles indicate bacteria genera found under each feeding regimen or shared between them. References used: Oliveira et al. (Reference Oliveira, Moraes, Goncalves, Giordano-Dias, D'Almeida, Asensi, Mello and Brazil2000); Perira et al. (Reference Perira de Oliveira, de Morais, Goncalves, Giordano-Dias, Vilela, Brazil, D'Almeida, Asensi and Mello2001); Gouveia et al. (Reference Gouveia, Asensi, Zahner, Rangel and Oliveira2008); McCarthy et al. (Reference McCarthy, Diambra and Rivera Pomar2011); Sant'Anna et al. (Reference Sant'Anna, Darby, Brazil, Montoya-Lerma, Dillon, Bates and Dillon2012); Kelly et al. (Reference Kelly, Bahr, Serafim, Ajami, Petrosino, Meneses, Kirby, Valenzuela, Kamhawi and Wilson2017); Pires et al. (Reference Pires, Villegas, Campolina, Orfano, Pimenta and Secundino2017).
Although the microbiota of field-collected and laboratory-reared L. longipalpis can have some bacteria species in common, the microbial diversity shared with other Lutzomyia species collected in the field can be quite reduced. To date, five New World sand fly species exclusively collected in the field had their microbial gut content investigated. Two of them are L. evansi (Vivero et al., Reference Vivero, Jaramillo, Cadavid-Restrepo, Soto and Herrera2016) and L. longipalpis (Oliveira et al., Reference Oliveira, Moraes, Goncalves, Giordano-Dias, D'Almeida, Asensi, Mello and Brazil2000; Gouveia et al., Reference Gouveia, Asensi, Zahner, Rangel and Oliveira2008; McCarthy et al., Reference McCarthy, Diambra and Rivera Pomar2011; Sant'Anna et al., Reference Sant'Anna, Darby, Brazil, Montoya-Lerma, Dillon, Bates and Dillon2012) mentioned above. Three other species were investigated using 16S rDNA sequencing or metatranscriptome: Lutzomyia cruzi (Sant'Anna et al., Reference Sant'Anna, Darby, Brazil, Montoya-Lerma, Dillon, Bates and Dillon2012), L. intermedia (Monteiro et al., Reference Monteiro, Villegas, Campolina, Pires, Miranda, Pimenta and Secundino2016) and Nyssomyia neivai (synonymous Lutzomyia neivai) (Machado et al., Reference Machado, Martins, Ferreira, Ferro, Bacci and Pinto2014). These field-collected sand fly species have very few shared bacteria (Fig. 3), most probably because they are exposed to diverse environments leading to diverse and distinct microbiota. Nevertheless, some shared species can be pointed out. Pelomonas sp., also found in other insects (Montoya-Porras et al., Reference Montoya-Porras, Omar, Alzate, Moreno-Herrera and Cadavid-Restrepo2018), was found in N. neivai and L. intermedia. Only Ralstonia sp. and Bradyrhizobium japonicum, present in other insects (Klimaszewski et al., Reference Klimaszewski, Morency, Labrie, Seguin, Langor, Work, Bourdon, Thiffault, Pare, Newton and Thayer2013; Rogers and Backus, Reference Rogers and Backus2014), were found in L. cruzi, L. intermedia and L. longipalpis. Among L. evansi, L. intermedia and L. longipalpis only three bacteria species were found in common: Acinetobacter calcoaceticus (found in other insect species, known for triggering a detectable immune response in tsetse flies) (Kaaya et al., Reference Kaaya, Otieno, Darji and Alemu1986; Hernandez-Flores et al., Reference Hernandez-Flores, Llanderal-Cazares, Guzman-Franco and Aranda-Ocampo2015), Enterobacter aerogenes (found in other insects and potential pathogen to humans) (Memona et al., Reference Memona, Manzoor and Anjum2017) and Pseudomonas putida (associated with soil and water) (Nicoletti et al., Reference Nicoletti, Corbella, Jaber, Marone, Scevola and Faga2015; Colauto et al., Reference Colauto, Fermor, Eira and Linde2016). Staphylococcus agnetis potentially pathogenic to poultry (Poulsen et al., Reference Poulsen, Thofner, Bisgaard, Olsen, Christensen and Christensen2017) and associated to bovine mastitis (Lange et al., Reference Lange, Brito, Reis, Machado, Guimaraes, Azevedo, Salles, Alvim, Silva and Meurer2015) was found only in L. cruzi, L. evansi and L. longipalpis.
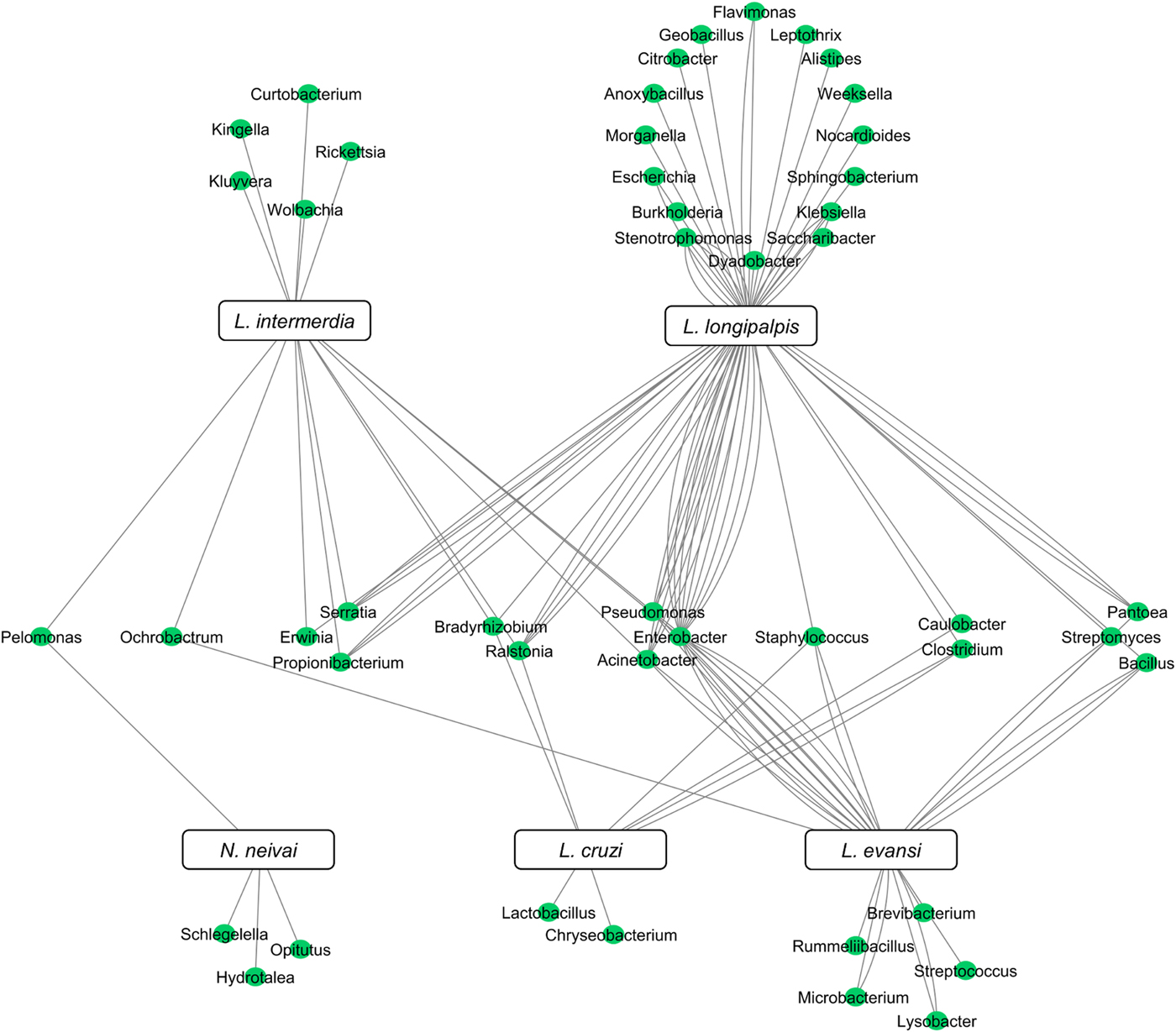
Fig. 3. New World field sand flies microbiota. Network analysis showing bacteria genera found in L. intermedia, L. longipalpis, L. evansi, L. cruzi, and N. neivai (syn. L. neivai) obtained exclusively from field collection studies. References used: Oliveira et al. (Reference Oliveira, Moraes, Goncalves, Giordano-Dias, D'Almeida, Asensi, Mello and Brazil2000); Gouveia et al. (Reference Gouveia, Asensi, Zahner, Rangel and Oliveira2008); McCarthy et al. (Reference McCarthy, Diambra and Rivera Pomar2011); Sant'Anna et al. (Reference Sant'Anna, Darby, Brazil, Montoya-Lerma, Dillon, Bates and Dillon2012); Machado et al. (Reference Machado, Martins, Ferreira, Ferro, Bacci and Pinto2014); Monteiro et al. (Reference Monteiro, Villegas, Campolina, Pires, Miranda, Pimenta and Secundino2016); and Vivero et al. (Reference Vivero, Jaramillo, Cadavid-Restrepo, Soto and Herrera2016).
In the Old World, Phlebotomus papatasi females from different collection sites had their microbial gut content investigated. Initial studies identified Enterobacter cloaceae, pathogentic to humans (Nagy et al., Reference Nagy, Pragai, Koczian, Hajdu and Fodor1998), from sand flies collected in Egypt (Dillon et al., Reference Dillon, el Kordy, Shehata and Lane1996) and Microbacterium sp., pathogenic to insects (Thakur et al., Reference Thakur, Dhammi, Saini and Kaur2015), from insects caught in Morocco (Guernaoui et al., Reference Guernaoui, Garcia, Gazanion, Ouhdouch, Boumezzough, Pesson, Fontenille and Sereno2011). A larger number of bacteria species were identified from P. papatasi gut contents collected from Tunisia, Turkey and India, using culture brain heart infusion (BHI) medium followed by 16S rDNA sequencing. The majority of identified sequences belong to the Bacillus genus (Mukhopadhyay et al., Reference Mukhopadhyay, Braig, Rowton and Ghosh2012) and depending on the culture media choice different bacteria could be identified. When comparing the microbial diversity of P. papatasi mentioned above with the data obtained through 16S ribosomal DNA sequencing collected in Iran (Maleki-Ravasan et al., Reference Maleki-Ravasan, Oshaghi, Afshar, Arandian, Hajikhani, Akhavan, Yakhchali, Shirazi, Rassi, Jafari, Aminian, Fazeli-Varzaneh and Durvasula2015) it is possible to point out some similar isolates such as Acinetobacter, Enterobacter, Microbacterium, Staphylococcus and Terribacillus genera. Additionally, other bacteria were identified at the species level such as Bacillus cereus, Bacillus flexus, Bacillus licheniformis, Bacillus pumilus, B. subtilis, Pseudomonas aeruginosa and S. marcescens.
Since sand fly environment and feeding habits can influence the sand fly microbial gut contents, one interesting study used 16S ribosomal DNA sequencing to investigate the bacteria present in the rodent Rhombomys opimus burrows where larvae feed on, R. opimus skin and intestinal track, as well as P. papatasi larvae and adult guts (Maleki-Ravasan et al., Reference Maleki-Ravasan, Oshaghi, Afshar, Arandian, Hajikhani, Akhavan, Yakhchali, Shirazi, Rassi, Jafari, Aminian, Fazeli-Varzaneh and Durvasula2015). B. subtilis and Enterobacter cloacae were identified in all analysed samples indicating that these two bacteria species can survive in P. papatasi environment, vertebrate host skin and gut, and sand fly gut. Moreover, these two bacteria are good candidates to be used in paratransgenic methods (Maleki-Ravasan et al., Reference Maleki-Ravasan, Oshaghi, Afshar, Arandian, Hajikhani, Akhavan, Yakhchali, Shirazi, Rassi, Jafari, Aminian, Fazeli-Varzaneh and Durvasula2015).
Other Phlebotomus species were investigated and their microbial content identified by microbiological methods. Phlebotomus sergenti, Phlebotomus kandelakii, Phlebotomus perfiliewi and Phlebotomus halepensis collected in Iran harbour several bacteria that were also found in P. papatasi such as Acinetobacter, Enterobacter and Pseudomonas genera, and more specifically B. subtilis and S. marcescens (Akhoundi et al., Reference Akhoundi, Bakhtiari, Guillard, Baghaei, Tolouei, Sereno, Toubas, Depaquit and Abyaneh2012). Acinetobacter and Bacillus genera were also identified in P. argentipes collected in India through culturing and 16S rDNA sequencing (Hillesland et al., Reference Hillesland, Read, Subhadra, Hurwitz, McKelvey, Ghosh, Das and Durvasula2008) supporting the potential use of bacteria from these genera in biological control strategies.
In China, the Phlebotomus chinensis associated microbial community presents interesting characteristics. The majority of bacteria identified by sequencing a 16S rDNA clone library obtained from the gut contents of adult females of this species were found to be from the families Coxiellaceae, Enterobacteriaceae mostly from the genus Enterococcus, and Pseudomonadaceae mostly from the genus Pseudomonas. Additionally, the intracellular Diplorickettsia, Rickettsia, Rickettsiella, Spiroplasma and Wolbachia were identified with these methods in the same study (Li et al., Reference Li, Chen, Jiang, Li, Xu and Ma2016). Curiously, Rickettsia and Wolbachia were found only in sand flies collected from a locality where anthroponotic visceral leishmaniasis occurs and dogs and other animals are the reservoirs (Li et al., Reference Li, Chen, Jiang, Li, Xu and Ma2016). Diplorickettsia, Rickettsiella and Spiroplasma were found only in sand flies collected from a locality where zoonotic visceral leishmaniasis occurs and humans are the identified reservoirs. It would be of interest to experimentally show the connection between these different microbiota profiles with vetorial capacity.
A study of the microbial gut content of Phlebotomus perniciosus collected in Tunis used culture-dependent and -independent techniques for bacteria identification (Fraihi et al., Reference Fraihi, Fares, Perrin, Dorkeld, Sereno, Barhoumi, Sbissi, Cherni, Chelbi, Durvasula, Ramalho-Ortigao, Gtari and Zhioua2017). When authors compared gut contents from field-caught and colony-reared insects they found that Stenotrophomonas maltophilia, Bacillus sp. and Lysinibacillus sp. are common to both groups of insects, suggesting that control strategies developed under laboratory conditions might be useful in the field (Fraihi et al., Reference Fraihi, Fares, Perrin, Dorkeld, Sereno, Barhoumi, Sbissi, Cherni, Chelbi, Durvasula, Ramalho-Ortigao, Gtari and Zhioua2017). In the same report, the authors show that variations in the resident microbiota are influenced by the sand fly species and particularities of niches where these insects live. Yet, an extensive meta-analysis showed an intricate network of several bacteria species associated with each of the sand fly species investigated and a relative small number of bacteria being shared among two or more sand fly species independent of the environment of harvest (Fig. 1). Acinetobacter baumanii, E. coli, Stenotrophamonas maltophila, B. subtilis, Staphylococcus epidermidis, Acinetobacter sp., Enterobacter sp., Klebsiella ozaenae and Serratia sp. are shared among at least three phlebotomine insects from Old and New World (Fraihi et al., Reference Fraihi, Fares, Perrin, Dorkeld, Sereno, Barhoumi, Sbissi, Cherni, Chelbi, Durvasula, Ramalho-Ortigao, Gtari and Zhioua2017).
While the gut resident bacteria of several sand fly species have been reported in multiple publication, there are limited reports on the fungal content of the sand fly gut. The presence of fungi species was reported in L. longipalpis guts collected from a non-endemic area for leishmaniasis. The fungal species identified by a high-throughput sequencing analysis were Cunninghamella bertholletiae, Peronospora conglomerata, Mortierella verticillata and Toxicocladosporium irritans, while no fungi sequences were found in the samples collected from an endemic area (McCarthy et al., Reference McCarthy, Diambra and Rivera Pomar2011) suggesting an excluding effect of fungi over Leishmania occurrence. On the other hand, when culturing techniques were used for isolating fungal contents of P. papatasi, P. sergenti, P. kandelakii, P. perfiliewi and P. halepensis collected in endemic areas of northern Iran, species belonging to Penicillium, Aspergillus, Acremonium, Fusarium, Geotrichum and Candida genera were identified (Schlein et al., Reference Schlein, Polacheck and Yuval1985; Akhoundi et al., Reference Akhoundi, Bakhtiari, Guillard, Baghaei, Tolouei, Sereno, Toubas, Depaquit and Abyaneh2012), which are different fungi genus from those identified in L. longipalpis. Whether L. longipalpis resident fungi species have the potentials for controlling Leishmania presence is yet to be explored.
One of the main motivating forces for the study of vector microbiota is the possibility of identifying microorganisms potentially useful for the development of control strategies, as mentioned before. The data discussed above show the high complexity of microbiota in different sand flies and the difficulties of discussing the available data, considering the various and diverse methods of obtaining and analysing samples. In Fig. 2, we condensed these results using field or laboratory feeding parameters into a more comprehensible and friendly figurative display. What we can conclude is that some bacteria genera are regularly found across different sand flies, but at the species level there is less diversity. Thus, identifying a single bacteria that can be applied to control strategies targetted to a majority of sand fly vectors will be quite challenging. Additionally, when sand flies are kept in laboratory colonies they are able to host additional bacteria suggesting that these insects immune response is efficiently tunned to protect the insect integrity.
Sand flies and bacteria: interdependence and immune responses
Recent research in many fields highlights the interdependence between many animals and their microbiomes. In the case of insect disease vectors, there is significant evidence showing the influence of resident non-pathogenic microorganisms on parasite–vector interactions.
The importance of bacteria in insect vector development had been demonstrated in mosquitoes as diverse as Aedes aegypti, Anopheles gambiae and the autogenous Georgecraigius atropalpus (Coon et al., Reference Coon, Vogel, Brown and Strand2014). In all three mosquitoes, axenic larvae were unable to develop and the reintroduction of one single bacteria species was capable of rescuing development of A. aegypti. On the other hand, the complex interactions between microorganisms and disease-causing agents as viruses and Plasmodium, have been investigated. The review by Caragata and Walker (Reference Caragata and Walker2012) covers the potential use of modified resident bacteria to fight pathogens and emphasizes the recent successful use of Wolbachia in controlling mosquito-borne diseases. The complex interactions of microbiota and mosquito vectors that go as far as influencing vector competence are also discussed (Caragata and Walker, Reference Caragata and Walker2012; Hegde et al., Reference Hegde, Rasgon and Hughes2015).
In the case of sand flies, bacteria seem to be important in many aspects of the flies’ life, starting with early development (Peterkova-Koci et al., Reference Peterkova-Koci, Robles-Murguia, Ramalho-Ortigao and Zurek2012). This study showed that L. longipalpis flies fed a diet containing raw rabbit feces were much more likely to lay eggs than flies fed feces that had been sterilized to remove all rabbit intestinal track-supplied bacteria. In addition, larvae fed on sterile feces had delayed hatching and lower survival rates. When different bacteria were reintroduced into sterile feces, there was a wide difference in hatching time and survival, demonstrating once again the importance of bacteria presence and specificity for insect development (Peterkova-Koci et al., Reference Peterkova-Koci, Robles-Murguia, Ramalho-Ortigao and Zurek2012).
Although breeding habits of sand flies in nature are still not clear, sand flies from tropical regions, like L. longipalpis, apparently breed in soil enriched with decomposed leaves and other detritus, with a preference for tree bases (Alencar et al., Reference Alencar, de Queiroz and Barrett2011). Feeding habits of larvae in this environment are also scantily known, but evidence suggests a participation of microorganisms in the diet. In the laboratory, the direct feeding of larvae on fungi mycelia has been observed (Moraes et al., Reference Moraes, Lucena, Moreira, Brazil, Gontijo and Genta2012). Furthermore, the incorporation of different fluorescent bacteria to the diet and later detection of these fluorescent microorganisms in the gut proved the ingestion of these by the larvae. The presence of enzymes capable of digesting bacterial and fungal walls has also been found in insect guts confirming that environmental microbes could be a nutritional source for insects (Moraes et al., Reference Moraes, Lucena, Moreira, Brazil, Gontijo and Genta2012).
Although in most cases insects live at peace with their resident microbiota, this peaceful coexistence is the result of a complex balance between acceptance and rejection. Insects tend to mount immune responses to keep this balance. The expression of the anti-microbial peptide (AMP) defensin was investigated in L. longipalpis early stages of development (Telleria et al., Reference Telleria, Sant'Anna, Alkurbi, Pitaluga, Dillon and Traub-Cseko2013). AMPs are small effector molecules involved in the innate immune response, composed of 5–100 amino acid residues, and found in plants and animals. AMPs are active against a broad spectrum of targets including viruses, bacteria, fungi and parasites (Bahar and Ren, Reference Bahar and Ren2013). Interestingly an increase of defensin expression was detected in late L4 larva stage, which stops eating in preparation for pupation and in pupae (Telleria et al., Reference Telleria, Sant'Anna, Alkurbi, Pitaluga, Dillon and Traub-Cseko2013). Since it is well known that microbiota is practically abolished in pupal stages, the increased production of this defensin may be helping in clearing the insect gut of bacteria.
Further immunological studies in sand fly larvae involved the artificial introduction of Gram+ (B. subtilis) and Gram− (P. agglomerans) bacteria, normally present in the gut of L. longipalpis (Gouveia et al., Reference Gouveia, Asensi, Zahner, Rangel and Oliveira2008), and the investigation of immune responses. A quite complex response was observed, with different outcomes related to Gram+ or −bacterial infections, and an apparent interplay between different immune pathways and effector molecules (Heerman et al., Reference Heerman, Weng, Hurwitz, Durvasula and Ramalho-Ortigao2015). One of the effectors investigated in this study was the negative IMD regulatory gene named ‘Poor immune response upon knock-in’ or Pirk. The expression of this gene was elevated at early times post infection (PI) with P. agglomerans, and this increased expression was maintained until 36 h PI. This might explain the downregulation of attacin at initial times PI. B. subtilis only affected the expression of Pirk at 24 h. On the other hand, IMD was upregulated only at 24 h PI with P. agglomerans, which might be responsible for an upregulated defensin expression at 24 h PI.
Immune responses to bacteria have also been reported in cultures of L. longipalpis LL5 embryonic cell line (Tesh and Modi, Reference Tesh and Modi1983). Insect cell lines have been widely used as a model to study vector immunity, being extensively exploited as a surrogate for understanding responses of mosquitoes to arbovirus infections (Walker et al., Reference Walker, Jeffries, Mansfield and Johnson2014). Mosquito cells have also been employed in studies of insect immune response against bacteria, revealing the involvement of the Toll and IMD pathways upon exposure to both Gram+ and Gram− heat inactivated bacteria (Barletta et al., Reference Barletta, Silva and Sorgine2012) in a way similar to what was observed previously in adult mosquitoes. With the advent of mosquito control approaches utilizing the bacteria Wolbachia, studies have been carried out to investigate mechanisms involved in the resistance to viral infection in mosquitoes harboring this endosymbiont, using both cell lines and insects (Rances et al., Reference Rances, Ye, Woolfit, McGraw and O'Neill2012).
In the case of L. longipalpis, previous studies utilizing cell lines focused on the interaction of these cells with Leishmania (Rey et al., Reference Rey, Ferro and Bello2000; Cortes et al., Reference Cortes, Silva, Pereira, Guerra, Zapata, Bello, Finkelstein, Madeira, Brazil, Corte-Real and Alves2011). Our group has investigated the response of LL5 cells to various organisms including viruses (Pitaluga et al., Reference Pitaluga, Mason and Traub-Cseko2008) as well as yeast, Leishmania and bacteria (Tinoco-Nunes et al., Reference Tinoco-Nunes, Telleria, da Silva-Neves, Marques, Azevedo-Brito, Pitaluga and Traub-Cseko2016). LL5 challenges with the Gram− Staphylococcus aureus and the Gram+ Escherichia coli and S. marcescens activated the Toll and IMD pathways, with S. aureus and S. marcescens triggering an early response (Tinoco-Nunes et al., Reference Tinoco-Nunes, Telleria, da Silva-Neves, Marques, Azevedo-Brito, Pitaluga and Traub-Cseko2016). AMPs seem to be under the control of different pathways, with cecropin and defensin 2 being under the control of the Toll and IMD pathways, the latter being produced as an early response to all challenges. Cecropin was shown to be produced early only in response to S. marcescens exposure, whereas the AMP attacin was produced at later times (Tinoco-Nunes et al., Reference Tinoco-Nunes, Telleria, da Silva-Neves, Marques, Azevedo-Brito, Pitaluga and Traub-Cseko2016). S. marcescens is known to be pathogenic for L. longipalpis (Diaz-Albiter et al., Reference Diaz-Albiter, Sant'Anna, Genta and Dillon2012), which might explain the early LL5 responses to this specific bacteria. These findings were important to establish LL5 cells as a reliable system to study L. longipalpis immunity.
The first report of putative immune responses of adult sand flies to bacteria came in 1997 when Nimmo et al., detected anti-microbial molecules in the hemolymph of L. longipalpis previously injected with Gram+ or Gram− bacteria. This hemolymph showed lyzing properties against both Micrococcus luteus and E. coli, and specific bands were detected by gel electrophoresis, one of approximately 4kD which is compatible with the AMPs cecropin or defensin (Nimmo et al., Reference Nimmo, Ham, Ward and Maingon1997).
A few years later the presence of AMPs was investigated in the Old World sand fly vector P. duboscqi (Boulanger et al., Reference Boulanger, Lowenberger, Volf, Ursic, Sigutova, Sabatier, Svobodova, Beverley, Spath, Brun, Pesson and Bulet2004). In these studies, insects were exposed to the Gram− bacteria Erwinia carotovora, normally found in plants and in insect guts, initially by intrathoracic injection. The effect of this challenge was investigated by submitting insects’ hemolymph to HPLC where specific peptide peaks were identified. Two fractions harvested from HPLC analyses were found to have an antibacterial activity and after amino acid sequencing, one of them was identified as a defensin with high levels of similarity to mosquito defensins. This same defensin was also found in the gut of P. duboscqi after ingestion of E. carotovora (Boulanger et al., Reference Boulanger, Lowenberger, Volf, Ursic, Sigutova, Sabatier, Svobodova, Beverley, Spath, Brun, Pesson and Bulet2004). Interestingly, this same study showed that this defensin was produced both in the hemolymph and in the gut of P. duboscqi following L. major infection, and the recombinant molecule was shown to be active against Gram− bacteria, yeast, fungi, and also L. major. This is initial evidence for a strong interplay between the production of immune molecules and their effect on bacteria and Leishmania, and the complexity of putative mechanisms involved in this interplay.
Further studies on the role of an AMP on sand fly bacterial infections were carried out in adult L. longipalpis by investigating the expression of a defensin in relation to infection with different bacteria and the route of bacteria acquisition on the outcome of immune responses. Insects were exposed to Gram– bacteria (E. coli, Ochrobactrum sp., S. marcescens, P. agglomerans) or the Gram+ bacteria M. luteus. When administered per-os all bacteria, with the exception of P. agglomerans, induced an increased transcription of the defensin gene with slight temporal differences depending on the microrganism the insects were fed on (Telleria et al., Reference Telleria, Sant'Anna, Alkurbi, Pitaluga, Dillon and Traub-Cseko2013). Interestingly, among the four bacteria that produced an increased defensin gene expression, an earlier and stronger response was found in insects infected with M. luteus, an interesting finding since it has been reported that insect defensins are more effective against Gram+ bacteria (Nimmo et al., Reference Nimmo, Ham, Ward and Maingon1997; Bulet et al., Reference Bulet, Hetru, Dimarcq and Hoffmann1999; Boulanger et al., Reference Boulanger, Lowenberger, Volf, Ursic, Sigutova, Sabatier, Svobodova, Beverley, Spath, Brun, Pesson and Bulet2004). The inability to detect increased defensin expression in L. longipalpis fed on P. agglomerans is interesting in light of reports that this Gram− bacteria is a commensal found in the gut of many insects, and hence is considered a symbiont bacteria. In this same report, L. longipalpis was injected intrathoracically with E. coli, which brought a strong and lasting production of defensin. This is not unexpected, since the control injection itself brought a quite strong production of the AMP, most probably due to injury and the fact that microbiota is normally restricted to the insect gut and the presence of an intestinal bacteria in the hemolymph must be considered a main aggression by the sand fly, thus explaining the strong response.
Since the discovery of the Toll pathway in Drosophila (Lemaitre et al., Reference Lemaitre, Nicolas, Michaut, Reichhart and Hoffmann1996), fruit flies have become well established as an excellent model to study immunity conferred by this pathway as well as other well-described pathways of insect immunity, IMD and JAK-STAT. Since then, the involvement of these pathways in immune responses of many insect vectors to parasites and viruses has been investigated (Cirimotich et al., Reference Cirimotich, Dong, Garver, Sim and Dimopoulos2010; Sim et al., Reference Sim, Jupatanakul and Dimopoulos2014).
The involvement of the IMD pathway in L. longipalpis infection with Leishmania and bacteria was studied (Telleria et al., Reference Telleria, Sant'Anna, Ortigao-Farias, Pitaluga, Dillon, Bates, Traub-Cseko and Dillon2012). In this report, the role of bacteria in controlling the expression of Caspar, a negative regulator of the IMD pathway, was documented in adult female flies using several different approaches. The report showed that when insects were treated with antibiotics, the expression of Caspar was increased in relation to untreated insects. This is consistent with a role of the IMD pathway in microbiota homeostasis since a decreased population of bacteria led to a higher Caspar expression resulting in a depressed IMD pathway. Also in this paper, the expression of Caspar was investigated in relation to L. longipalpis ingestion of exogenous bacteria. The expectation was that activation of the pathway would be associated with decreased expression of this negative regulator. This was seen when the insects were fed bacteria considered more pathogenic to insects: M. luteus, E. coli and S. marcescens, although, as expected, there were differences in the timing and extent of the decreased expression among the insects fed these pathogens. Interestingly, no increased expression in Caspar was observed with the symbiotic bacteria P. agglomerans and Ochrobactrum sp, indicating the insects’ immunological indifference to these harmless bacteria.
Other defense mechanisms, beyond the IMD pathway, have also been investigated in the bacteria–sand fly interaction. An important group of effector molecules are ROS, oxygen-derived radical species produced during cell respiration. ROS molecules include superoxide anion (O2−), the hydroxyl radical (OH−) and hydrogen peroxide (H2O2). ROS are involved both in defense against entomopathogens and in selection and control of commensal gut microbiota in various insects. In Drosophila ROS are produced in response to gut infections (Ha et al., Reference Ha, Oh, Bae and Lee2005), in A. gambiae ROS are produced following infection with the parasite Plasmodium falciparum (Molina-Cruz et al., Reference Molina-Cruz, DeJong, Charles, Gupta, Kumar, Jaramillo-Gutierrez and Barillas-Mury2008) and also in Anopheles aquasalis infected with Plasmodium vivax (Bahia et al., Reference Bahia, Oliveira, Kubota, Araujo, Lima, Rios-Velasquez, Lacerda, Oliveira, Traub-Cseko and Pimenta2013). The involvement of ROS in bacterial control has been suggested from studies using A. gambiae. When these mosquitoes were treated with antioxidants, they were found to be more susceptible to bacterial infection than untreated insects (Molina-Cruz et al., Reference Molina-Cruz, DeJong, Charles, Gupta, Kumar, Jaramillo-Gutierrez and Barillas-Mury2008). In the sand fly L. longipalpis, the involvement of ROS in responses to the pathogenic bacteria S. marcescens has also been investigated (Diaz-Albiter et al., Reference Diaz-Albiter, Sant'Anna, Genta and Dillon2012). This study showed that the production of ROS was increased for up to 72 h post-infection in insects fed with the bacteria. Furthermore, the production of H2O2 was also found to be increased in S. marcescens fed flies. Feeding the insects with the ROS-scavenger uric acid caused premature L. longipalpis death that might be caused by a parallel increase of the native microbiota. On the other hand, under these circumstances S. marcescens numbers were decreased, which was interpreted by the authors as a consequence of the resident microbiota competition (Diaz-Albiter et al., Reference Diaz-Albiter, Sant'Anna, Genta and Dillon2012).
Taken together, these studies reveal the complex interconnection of sand fly responses to bacteria that contribute to gut homeostasis. The importance of bacteria on the establishment of Leishmania infection in sand flies will be discussed below.
Sand fly and Leishmania: a complex relationship
The Leishmania life-cycle in the insect begins when sand fly females ingest blood from an infected mammal. Within the sand fly, the acquired Leishmania develops exclusively inside the digestive tract (Sacks and Kamhawi, Reference Sacks and Kamhawi2001). During their development, the parasites undergo changes to adapt to their new environment and develop into the form infective to another mammal host. In the process named metacyclogenesis, the parasites change from the intracellular spherical aflagellated amastigotes acquired from the ingested vertebrate blood cells to elongated flagellated, extracellular infective metacyclic forms. These morphological changes are accompanied by molecular modifications. For example, alterations on the surface of the parasite enable the interaction with the insect midgut, a fundamental step for parasite survival, development and subsequent infectivity to the vertebrate host (Bates, Reference Bates2008).
To be successfully transmitted the parasite needs to overcome several obstacles, among which is infecting the correct sand fly species host. Among more than 900 species of sand flies recorded, just 98 are proven vectors of human leishmaniasis, 42 Phlebotomus species in the Old World and 56 Lutzomyia species in the New World (Volf and Myskova, Reference Volf and Myskova2007; Maroli et al., Reference Maroli, Feliciangeli, Bichaud, Charrel and Gradoni2013).
The sand fly vector competence depends on several factors such as a preference for feeding on humans, being infected with the Leishmania species occurring in humans and being able to complete their development inside the midgut after the blood meal digestion. In nature, living in sympatry is not equivalent to vector competence, since restrictive or specific vectors transmit only particular species of Leishmania (e.g. P. papatasi and L. major) (Sacks and Kamhawi, Reference Sacks and Kamhawi2001). Other sand fly species are considered permissive or nonspecific, as they are able to harbour experimental infections of several Leishmania species (e.g. L. longipalpis and Leishmania infantum chagasi or Leishmania mexicana).
The success of sand fly midgut colonization by Leishmania is determined by several molecular factors. The first challenge of Leishmania within the sand fly is to resist the digestive process, which is achieved by interfering with the sand fly digestive enzymes activity (Dillon and Lane, Reference Dillon and Lane1993; Schlein and Jacobson, Reference Schlein and Jacobson1998; Sant'anna et al., Reference Sant'anna, Diaz-Albiter, Mubaraki, Dillon and Bates2009; Telleria et al., Reference Telleria, de Araujo, Secundino, d'Avila-Levy and Traub-Cseko2010) and modulating the transcription of several digestive enzymes genes (Ramalho-Ortigao et al., Reference Ramalho-Ortigao, Jochim, Anderson, Lawyer, Pham, Kamhawi and Valenzuela2007; Jochim et al., Reference Jochim, Teixeira, Laughinghouse, Mu, Oliveira, Gomes, Elnaiem and Valenzuela2008; Pitaluga et al., Reference Pitaluga, Beteille, Lobo, Ortigao-Farias, Davila, Souza, Ramalho-Ortigao and Traub-Cseko2009; Dostalova et al., Reference Dostalova, Votypka, Favreau, Barbian, Volf, Valenzuela and Jochim2011). Furthermore, according to one publication, Leishmania secretes a myoinhibitory peptide that arrests hindgut peristalsis, thus delaying fecal elimination and increasing parasite persistence within the insect (Vaidyanathan, Reference Vaidyanathan2004). Also, Leishmania damages the insect stomodeal valve interfering in the blood ingestion process. These strategies facilitate parasite colonization of the midgut and increase the likelihood of subsequent transmission (Schlein et al., Reference Schlein, Jacobson and Messer1992; Volf et al., Reference Volf, Hajmova, Sadlova and Votypka2004). From the sand fly point of view, the Leishmania is an unwanted guest that causes indigestion, gut constipation and difficulties to swallow the blood meal.
A fundamental element in the sand fly digestive process is the PM, a semi-permeable membrane composed of glycoproteins associated with chitin fibrils which isolates the digestive bolus from the midgut epithelia and might have a role Leishmania midgut colonization (Hegedus et al., Reference Hegedus, Erlandson, Gillott and Toprak2009).
Studies with P. papatasi have demonstrated that the PM biogenesis homeostasis is fundamental for the parasite colonization success. The inhibition of the PM formation, by addition of an exogenous chitinase to the blood meal, resulted in sand flies refractory to L. major infection (Pimenta et al., Reference Pimenta, Modi, Pereira, Shahabuddin and Sacks1997). More recently it was suggested that Leishmania mortality is not caused directly by sand fly proteases and might result from toxic products of the blood meal digestion (Pruzinova et al., Reference Pruzinova, Sadlova, Myskova, Lestinova, Janda and Volf2018). Nevertheless, the interruption of the PM degradation through the silencing of an insect chitinase gene (Coutinho-Abreu et al., Reference Coutinho-Abreu, Sharma, Robles-Murguia and Ramalho-Ortigao2010a), also resulted in refractoriness to Leishmania infection, indicating the need for more experiments approaching this subject.
Although in comparison with more aggressive parasites, such as Plasmodium, which transverses the mosquito gut and is exposed to the insect hemolymph, Leishmania can be considered a more mellow parasite, since it inhabits the sand fly gut in an apparently less aggressive fashion. Nevertheless, the evidence is mounting to show that this passage through the digestive system does not go unnoticed.
Sand fly immunity to Leishmania: dealing with an unwanted passenger
In contrast to the many studies on other insect immune response to parasites, not much is known about the sand fly immune response to infections with Leishmania. As described above, insect immune responses lead to the stimulation of various immune mechanisms. Among these are RNAi, JAK-STAT, IMD and Toll, that will determine the production of effectors molecules (Lemaitre et al., Reference Lemaitre, Kromer-Metzger, Michaut, Nicolas, Meister, Georgel, Reichhart and Hoffmann1995; Brennan and Anderson, Reference Brennan and Anderson2004; Blair, Reference Blair2011; Zeidler and Bausek, Reference Zeidler and Bausek2013).
The sand fly's ability to counteract microbial infections has been demonstrated in many studies. In the case of parasite infections, the isolation and subsequent characterization of an active antimicrobial peptide defensin of P. duboscqi induced by L. major challenge was the first study demonstrating, at the molecular level, a sand fly humoral immune response elicited by Leishmania infection (Boulanger et al., Reference Boulanger, Lowenberger, Volf, Ursic, Sigutova, Sabatier, Svobodova, Beverley, Spath, Brun, Pesson and Bulet2004). More recently the involvement of a L. longipalpis defensin in the sand fly immune response was investigated (Telleria et al., Reference Telleria, Sant'Anna, Alkurbi, Pitaluga, Dillon and Traub-Cseko2013). In this report, L. longipalpis females were challenged with different bacteria species (discussed above) or with L. mexicana either orally or through microinjection. Interestingly, contrary to what was seen with P. duboscqi, the L. longipalpis oral Leishmania challenge did not stimulate the defensin transcription. It is possible that other L. longipalpis AMPs (not examined in this report), rather than this defensin, are induced by L. mexicana challenge. An interesting aspect of these different responses is the fact that P. duboscqi is an Old World restrictive vector, whereas the L. longipalpis is a New World permissive vector (Dostalova and Volf, Reference Dostalova and Volf2012). While attachment to the gut in specific vectors (P. papatasi, P. duboscqi and P. sergenti) involves parasite lipophosphoglycan (LPG), this molecule is not required for parasite attachment in other sand fly species permissive for various New World Leishmania species, including L. longipalpis (Myskova et al., Reference Myskova, Svobodova, Beverley and Volf2007).
The lower induction of defensin in P. duboscqi challenge with L. major LPG defective mutants showed the involvement of the LPG molecule in the P. duboscqi immune response (Boulanger et al., Reference Boulanger, Lowenberger, Volf, Ursic, Sigutova, Sabatier, Svobodova, Beverley, Spath, Brun, Pesson and Bulet2004).
Sand fly transcriptomic studies have reported changes in the expression of several immune-related genes in insects challenged with Leishmania, including components of the Toll, IMD and JNK pathways as well as oxidative stress-related molecules such as the antioxidants glutathione s-transferase, catalase, copper-zinc superoxide dismutase and peroxiredoxin, responsible to control ROS levels (Thannickal and Fanburg, Reference Thannickal and Fanburg2000; Dillon et al., Reference Dillon, Ivens, Churcher, Holroyd, Quail, Rogers, Soares, Bonaldo, Casavant, Lehane and Bates2006; Ramalho-Ortigao et al., Reference Ramalho-Ortigao, Jochim, Anderson, Lawyer, Pham, Kamhawi and Valenzuela2007; Pitaluga et al., Reference Pitaluga, Beteille, Lobo, Ortigao-Farias, Davila, Souza, Ramalho-Ortigao and Traub-Cseko2009; Abrudan et al., Reference Abrudan, Ramalho-Ortigao, O'Neil, Stayback, Wadsworth, Bernard, Shoue, Emrich, Lawyer, Kamhawi, Rowton, Lehane, Bates, Valenzeula, Tomlinson, Appelbaum, Moeller, Thiesing, Dillon, Clifton, Lobo, Wilson, Collins and McDowell2013). Among genes related to the immune signaling pathways, an L. longipalpis Caspar gene was identified. The Caspar gene is a homologue of the human Fas-associating factor 1 protein, which negatively controls the IMD pathway (Kim et al., Reference Kim, Lee, Lee, Kim and Chung2006). The L. longipalpis Caspar gene expression profile has been recently investigated following challenge with bacteria (discussed above) and Leishmania. The effects of experimental Caspar RNAi silencing on the Leishmania infection success was also established. Importantly, L. mexicana challenge downregulated Caspar expression at the third and sixth days after infection while the silencing of Caspar led to a decreased Leishmania population size and infection prevalence (Telleria et al., Reference Telleria, Sant'Anna, Ortigao-Farias, Pitaluga, Dillon, Bates, Traub-Cseko and Dillon2012). A role for the IMD pathway on parasite control was also seen in Anopheles sp. infected with P. falciparum, where the RNAi-mediated knockdown of Caspar reduced the protozoa survival in the insect (Garver et al., Reference Garver, Dong and Dimopoulos2009).
Insect immunity to Leishmania has also been investigated using insect cell lines. As mentioned earlier in this review, multiple reports have confirmed the immune competence of diverse insect cell lineages, including the LL5 embryonic cell line from L. longipalpis. Tinoco-Nunes et al. (Reference Tinoco-Nunes, Telleria, da Silva-Neves, Marques, Azevedo-Brito, Pitaluga and Traub-Cseko2016) challenged LL5 cells with different microorganisms, including L. i. chagasi. The presence of Leishmania led to the upregulation of the Cactus gene, the negative regulator of the Toll pathway, whereas the expression of Caspar, the IMD pathway negative regulator, did not change significantly. The Dorsal and Relish genes, positive modulators of the Toll and IMD pathways, were upregulated by the Leishmania challenge and the expression of the AMPs attacin, cecropin and defensin 2 increased at different time points (Tinoco-Nunes et al., Reference Tinoco-Nunes, Telleria, da Silva-Neves, Marques, Azevedo-Brito, Pitaluga and Traub-Cseko2016). These results revealed that the Toll and IMD pathways are involved in the sand fly LL5 cell line immune response against Leishmania.
Sand flies and mosquitoes present divergent oxidative responses to protozoan parasite infection. Whereas the presence of Plasmodium in the Anopheles gut produces an intense oxidative response with increased ROS production, Leishmania infected L. longipalpis do not show significant changes in ROS gut level when compared with control insects (Molina-Cruz et al., Reference Molina-Cruz, DeJong, Charles, Gupta, Kumar, Jaramillo-Gutierrez and Barillas-Mury2008; Diaz-Albiter et al., Reference Diaz-Albiter, Mitford, Genta, Sant'Anna and Dillon2011). These opposite responses between sand flies and mosquitoes might be related to differences between the parasites more than differences between the insects hosts. Studies on macrophage infection by Leishmania revealed a direct relation between parasite virulence and antioxidant enzymes expression and cell ROS levels control, with the parasites lacking antioxidant enzymes being less virulent than normal parasites. The decrease of sand fly gut Leishmania population after silencing of the sand fly antioxidant enzyme catalase suggests that Leishmania oxidative escape also depends on manipulation of vector antioxidative elements (Pal et al., Reference Pal, Dolai, Yadav and Adak2010). The hypothesis is that Leishmania modulates the ROS levels in sand fly midgut during digestion, diminishing the activity of endogenous and exogenous antioxidant enzymes, to produce a friendlier developing environment.
Since Leishmania development occurs exclusively in the vector gut in the presence of a dynamic microbiota, our understanding of the parasite infection process inside the vector gut is challenged by multipartite factors. In the subsequent section, we discuss this aspect under the light of some recent and important publications.
Sand fly, microbiota and Leishmania: an even more complex interrelation
Leishmania is not alone in the sand fly midgut. When the parasites reach the insect digestive tract they encounter the gut microbiota, a rich commensal microorganism community, with bacterial predominance that naturally colonizes the sand fly gut (discussed above).
As mentioned above, the insect microbiota plays important roles in vector physiology, such as nutrition and digestion (Dillon and Dillon, Reference Dillon and Dillon2004) and can also act on the maturation of the innate immune system (Weiss et al., Reference Weiss, Wang and Aksoy2011). The relationship between vector-borne pathogens and insect gut microbiota has been highlighted in several reports. The data produced suggest that microbiota can influence the parasite infection through the activation of vector innate immune pathways leading to induction of effectors molecules that will help in the control of infection by insect-vectored disease agents. As an example, the L. longipalpis midgut ROS suppression revealed the significant role of microbiota in facilitating Leishmania infection (Diaz-Albiter et al., Reference Diaz-Albiter, Mitford, Genta, Sant'Anna and Dillon2011).
Immune responses mounted against midgut bacteria in the mosquito and symbiotic bacteria in the tsetse fly have a protective effect against infections by insect-vectored viruses and parasites. Studies have shown that the mosquito employs some of the same immune factors to combat bacteria and Plasmodium parasite infection (Beier et al., Reference Beier, Pumpuni, Beier and Davis1994; Dong et al., Reference Dong, Aguilar, Xi, Warr, Mongin and Dimopoulos2006; Weiss et al., Reference Weiss, Wang, Maltz, Wu and Aksoy2013). Furhermore, A. gambiae previously treated with antibiotics to eliminate the midgut commensal bacteria were more susceptible to Plasmodium infection than untreated mosquitoes, and the reintroduction of various midgut bacteria restored the resistance phenotype (Dong et al., Reference Dong, Manfredini and Dimopoulos2009; Meister et al., Reference Meister, Agianian, Turlure, Relogio, Morlais, Kafatos and Christophides2009).
The stimulation of tsetse flies antibacterial immune responses by the endosymbiont bacteria Wigglesworthia has been implicated in trypanosome control. The tsetse peptidoglycan recognition protein LB (PGRP-LB) induced by the presence of Wigglesworthia and effector molecules of the activated fly IMD pathway presented direct anti-trypanosome activity (Wang et al., Reference Wang, Smith and Kappe2009). On the other hand, bacteria may also directly inhibit pathogen development, either by disrupting necessary interactions between the pathogen and vector epithelium or through the production of anti-parasite molecules (Azambuja et al., Reference Azambuja, Garcia and Ratcliffe2005). An Enterobacter bacterium isolated from wild Anopheles populations in Zambia was found to decrease Plasmodium development through the production of ROS, suggesting that a mosquito-mounted response was not required for the observed infection inhibition (Cirimotich et al., Reference Cirimotich, Ramirez and Dimopoulos2011).
As seen in other vector–parasite interaction models, the microbiota influences the sand fly vector competence, although existing work presents some divergent results. In experiments with colony-raised L. longipalpis, pre-feeding the insects with the bacteria Asaia sp., Ochrobactrum intermedium and a yeast-like fungus Pseudozyma sp. previously isolated from wild-caught and laboratory-reared female L. longipalpis, prevented Leishmania establishment in the sand fly midgut (Sant'Anna et al., Reference Sant'Anna, Diaz-Albiter, Aguiar-Martins, Al Salem, Cavalcante, Dillon, Bates, Genta and Dillon2014). In the same work, the authors verified that L. longipalpis previously infected with L. mexicana were resistant to Serratia infection, thus identifying a protective effect of Leishmania infection against Serratia challenge. These results pointed to a competitive relationship between Leishmania and microbiota.
Recently this interpretation was revised in two papers, both demonstrating that Leishmania development in the vector is dependent on microbiota. Firstly, the presence of bacterial phylogenies in midguts of sugar-fed, blood-fed and L. infantum-infected colony-reared L. longipalpis was studied. Also, the effect of antibiotics treatment on Leishmania infection was investigated. The results of these studies showed that while Leishmania infection led to an escalating loss of bacterial diversity throughout the course of infection, the depletion of L. longipalpis midgut microbiota after antibiotics treatment impaired L. infantum replication and development to infective metacyclic forms (Kelly et al., Reference Kelly, Bahr, Serafim, Ajami, Petrosino, Meneses, Kirby, Valenzuela, Kamhawi and Wilson2017). The second work, conduced by Louradour and collaborators (Reference Louradour, Monteiro, Inbar, Ghosh, Merkhofer, Lawyer, Paun, Smelkinson, Secundino, Lewis, Erram, Zurek and Sacks2017), carried out experiments of L. major infection in P. duboscqi under various conditions. As observed in Kelly and collaborators, treatment with antibiotics prevented the development of Leishmania in the midgut of P. duboscqi, and feeding with bacterial culture supernatants was not able to reverse the antibiotic effect. The introduction of engineered antibiotic-resistant bacteria isolated from natural P. duboscqi microbiota to the midgut did not impair the Leishmania infection as reported in Sant'Anna et al. (Reference Sant'Anna, Diaz-Albiter, Aguiar-Martins, Al Salem, Cavalcante, Dillon, Bates, Genta and Dillon2014). Differences in the number of time points assessed and differences in the number of bacteria used in these studies might explain these different outcomes. In the work by Kelly et al. (Reference Kelly, Bahr, Serafim, Ajami, Petrosino, Meneses, Kirby, Valenzuela, Kamhawi and Wilson2017) the authors used a high concentrations of bacteria (>107 CFU mL−1) and investigated only one early time point, while Lauradour and collaborators (2017) employed low bacterial doses (104 CFU mL−1) and investigated diverse time points throughout the Leishmania development. Experiments with sand flies treated with antibiotics and fed with different sugar meal concentrations showed that sugar concentrations usually used to maintain the insects (15–30%) impaired L. major growth. These latter results suggest that the role of microbiota in the sand fly and Leishmania interaction might be to ‘buffer’ the ‘milleu’ osmolarity.
The complexity of insect vector, protozoan pathogen and microbiota relationship can be observed in another recent work where evidence was presented for the involvement of microbiota in the Anopheles PM formation. Mosquitoes treated with antibiotics showed a reduction in the expression of several genes related to the PM production. In addition, microscopy revealed disruptions in the PM integrity in antibiotic-treated insects (Rodgers et al., Reference Rodgers, Gendrin, Wyer and Christophides2017). The potential participation of sand fly PM in the establishment of Leishmania in the sand fly midgut was already addressed above. The possibility that the phenomenon is seen in Anopheles also occurs in other insect models raises doubts about the interpretation of the results associating pathogen infection and microbiota depletion after antibiotic administration. Disruptions in the sand fly PM integrity could permit the access of sand fly molecules damaging to Leishmania, before the parasite has undergone changes that enable resistance against the sand fly midgut hostile environment, leading to parasite losses.
The relevance of sand fly microbiota studies was reinforced in a recent work where the authors identified a crucial role for sand fly microbiota in Leishmania donovani infection in mammals. Dey et al. (Reference Dey, Joshi, Oliveira, Pereira, Guimaraes-Costa, Serafim, de Castro, Coutinho-Abreu, Bhattacharya, Townsend, Aslan, Perkins, Karmakar, Ismail, Karetnick, Meneses, Duncan, Nakhasi, Valenzuela and Kamhawi2018) demonstrated that during the L. longipalpis blood meal microorganisms from vector microbiota are co-egested with Leishmania (Dey et al., Reference Dey, Joshi, Oliveira, Pereira, Guimaraes-Costa, Serafim, de Castro, Coutinho-Abreu, Bhattacharya, Townsend, Aslan, Perkins, Karmakar, Ismail, Karetnick, Meneses, Duncan, Nakhasi, Valenzuela and Kamhawi2018). These microbes activate the mouse neutrophil inflammasome leading to a rapid production of interleukin-1b (IL-1b), which sustains neutrophil infiltration. These neutrophils help shield L. donovani parasites and promote infection of macrophages post transmission. The sand fly dysbiosis using antibiotic treatment impaired the L. donovani infection. This outcome suggests that the relationship between the Leishmania and the sand fly microbiota may go beyond the vector midgut, with the microorganisms presence modulating vertebrate host immune response, by producing a friendly ‘milleu’ fundamental for the success of the vertebrate infection by the transmitted parasite.
The information compiled here reveals that there is a complex relationship among the sand fly, Leishmania and commensal microbiota. Greater understanding of the relationship and the mechanism that underpin it could identify targets for the development of strategies to control the ability of the sand fly to transmit leishmaniasis to man.
Concluding remarks
In this review, we compiled and discussed the hitherto published data about the interactions among sand flies, its microbiota and sand fly-borne pathogens. Exploring available information we can say that although sand flies are important vectors of viruses in the Old World, in the New World there are still few studies on the putative sand fly vectorial competence for viral transmission. Strikingly, in sharp contrast to how much is known in mosquito–virus interactions, there is almost total absence of data on sand fly-virus interplay, pointing to the need of more studies to help understand pathways that could be involved in sand fly antiviral immune responses.
Another important point is the increasing awareness of the importance of microorganisms for the survival and development of sand flies, and for a successful parasite–vector interaction. The sand flies microbiota prospective studies disclosed the existence of a rich and variable intestinal flora. The recent growing data amassed by all these studies is a first step for the potential establishment of paratransgenesis strategies.
Since sand flies are mostly known as the vectors for leishmaniasis, a few studies have approached the question of how the insect responds to the parasite infection. Some data on the involvement of the IMD pathway, or the production of effector molecules are discussed herein, but much remains to be uncovered.
Very recent work has focused on how resident microbiota can affect the Leishmania infection of the vector with surprising results. Basically, it was found that the sand fly microbiota is fundamental for Leishmania development and transmission. One paper suggests that the removal of the microbiota alters the osmolarity of the intestinal environment and is thus deleterious for the Leishmania development. Since we do not know exactly which molecular mechanisms are responsible for this dependence of Leishmania on microbiota, this may be considered an open field of research. Interestingly, the Leishmania-microbiota relationship does not seem restricted to the sand fly midgut. Microorganisms egested during the bite together with Leishmania elicit an immune response that increases the Leishmania infectivity in mice.
The reviewed studies demonstrate the complex and intricate network among sand flies, microbiota, virus and Leishmania. More studies are needed to increase the knowledge about these very interesting interplays. This information will be crucial for the development of control and eradication measures of sand fly-borne diseases.
Acknowledgement
We thank Peter W. Mason, University of Texas Medical Branch, Galveston, TX for critical review of this work.
Financial support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Conflict of interest
None.
Ethical standards
Not applicable.







