Introduction
Dogs play an important role in human life as companion animals; however, they may be carriers of various pathogens, constituting a potential reservoir of zoonotic infections for their owners. Many of these infectious agents reside in the intestinal tract; therefore, their dispersive forms are excreted with animal stool and may easily be spread to other hosts by fecal–oral transmission through direct contact or indirectly via water or food contamination. This includes unicellular enteric organisms such as Giardia intestinalis (syn. duodenalis or lamblia), Cryptosporidium spp. or microsporidia from genera Encephalitozoon and Enterocytozoon, which can be responsible for gastrointestinal symptoms like abdominal pain, diarrhoea or flatulence (Xiao, Reference Xiao2010; Liao et al., Reference Liao, Lin, Sun, Qi, Lv, Wu, Li, Hu, Yu, Cai, Xiao, Sun and Li2020). Since these organisms belong to the group of opportunistic pathogens, infection is of particular importance for individuals with impaired immunity (e.g. HIV-infected patients, cancer-treated patients or transplant recipients), in whom it may lead to the development of hazardous, even life-threatening symptoms.
Giardia intestinalis is the most common intestinal pathogenic protozoan in humans and animals (Kváč et al., Reference Kváč, Hofmannová, Ortega, Holubová, Horčičková, Kicia, Hlásková, Květoňová, Sak and McEvoy2017), consisting of 8 distinct assemblages (A–H) differing in host specificities. Assemblages A and B display a broad host range and are most commonly reported in humans, while the remaining 6 seem to be host-specific for non-human species, with assemblages C and D predominantly found in dogs (Bouzid et al., Reference Bouzid, Halai, Jeffreys and Hunter2015; Ryan and Zahedi, Reference Ryan and Zahedi2019). Among nearly 51 valid Cryptosporidium species (Tůmová et al., Reference Tůmová, Ježková, Prediger, Holubová, Sak, Konečný, Květoňová, Hlásková, Rost, McEvoy, Xiao, Santín and Kváč2023), Cryptosporidium hominis and Cryptosporidium parvum represent the major causes of human cryptosporidiosis, whereas Cryptosporidium meleagridis, Cryptosporidium mortiferum, Cryptosporidium felis and Cryptosporidium canis are rare causative agents of zoonotic infections, with the latter being the most prevalent in dogs (Xu et al., Reference Xu, Jin, Wu, Li, Wang, Li, Feng and Xiao2016; Li et al., Reference Li, Ryan, Guo, Feng and Xiao2021; Alderisio et al., Reference Alderisio, Mergen, Moessner and Madison-Antenucci2023). Out of over 1200 microsporidian species described so far, Enterocytozoon bieneusi and Encephalitozoon genus, including Encephalitozoon intestinalis, Encephalitozoon cuniculi and Encephalitozoon hellem, represent the species causing human microsporidiosis (Didier et al., Reference Didier, Didier, Snowden and Shadduck2000), especially in persons with impaired immunity (Kicia et al., Reference Kicia, Wesolowska, Jakuszko, Kopacz, Sak, Květonova, Krajewska and Kváč2014, Reference Kicia, Wesolowska, Kopacz, Jakuszko, Sak, Květonová, Krajewska and Kváč2016). These species may also be detected in a broad range of other hosts (livestock, wildlife and domesticated animals) (Dengjel et al., Reference Dengjel, Zahler, Hermanns, Heinritzi, Spillmann, Thomschke, Löscher, Gothe and Rinder2001; Mathis et al., Reference Mathis, Weber and Deplazes2005). The phylogenetic analysis of E. bieneusi allows for distinction of various genotypes differing in a host specificity. Genotypes D, EbpC and type IV are characterized by the widest host range and are also most frequently reported in humans (Li et al., Reference Li, Feng and Santin2019). In turn, genotype PtEbIX seems to be restricted to canine host population (Li et al., Reference Li, Feng and Santin2019), although dogs can serve as a reservoir of many other zoonotic genotypes as well, including those most often reported in humans (Li et al., Reference Li, Feng and Xiao2020). Regarding the genus Encephalitozoon, the ability of these 3 species to inhabit a wide variety of organisms has been shown, with E. cuniculi having the widest host range, mainly among mammals and birds. Four E. cuniculi strains have been identified (I–IV); ‘canine’ strain III has been shown to cause high mortality in dogs, while the recently discovered ‘human’ strain IV has so far been documented in humans, cats and dogs. Although there appears to be some host preference in each strain, this specificity is not exact; humans have been found to be infected with all known strains (though rarely with strain III). In turn, E. hellem is the most common species among birds, while E. intestinalis is the most prevalent Encephalitozoon species in humans (Hinney et al., Reference Hinney, Sak, Joachim and Kváč2016).
The dispersive forms of the discussed pathogens are very resistant, and many species of wild and domesticated animals, as well as humans, may serve as their hosts which facilitates their spread and maintenance in the environment. Previous research on the occurrence of these species in central Europe, including Poland and the Czech Republic, shows different data depending on the population studied and the detection methods used. According to a review by Plutzer et al., the reported incidence of Cryptosporidium spp. and G. intestinalis per 100 000 inhabitants is 0.006 and 5.43 for Poland and 0.01 and 0.51 for the Czech Republic, respectively (Plutzer et al., Reference Plutzer, Lassen, Jokelainen, Djurković-Djaković, Kucsera, Dorbek-Kolin, Šoba, Sréter, Imre, Omeragić, Nikolić, Bobić, Živičnjak, Lučinger, Stefanović, Kučinar, Sroka, Deksne, Keidāne, Kváč, Hůzová and Karanis2018). In Poland, estimates of the prevalence of these 2 species in humans are available based on the results of research limited to specific population groups and regions (Plutzer et al., Reference Plutzer, Lassen, Jokelainen, Djurković-Djaković, Kucsera, Dorbek-Kolin, Šoba, Sréter, Imre, Omeragić, Nikolić, Bobić, Živičnjak, Lučinger, Stefanović, Kučinar, Sroka, Deksne, Keidāne, Kváč, Hůzová and Karanis2018). However, studies of Czech residents regarding the seroprevalence of Cryptosporidium spp. show the frequency of antibodies at the level of approximately 67–72% (Kozisek et al., Reference Kozisek, Craun, Cerovska, Pumann, Frost and Muller2008). In turn, the prevalence of Cryptosporidium spp. in dogs in Poland ranges from approximately 3.5 to 12.5% (Bajer and Bednarska, Reference Bajer and Bednarska2007; Piekara-Stępińska et al., Reference Piekara-Stępińska, Piekarska and Gorczykowski2021a), and G. intestinalis from 6 to 36%, with mainly canid-specific genotypes detected, which suggests that they do not represent an important source of Giardia infection for humans (Bajer and Bednarska, Reference Bajer and Bednarska2007; Piekarska et al., Reference Piekarska, Bajzert, Gorczykowski, Kantyka and Podkowik2016; Piekara-Stępińska et al., Reference Piekara-Stępińska, Piekarska, Gorczykowski and Bania2021b). A similar prevalence refers to dogs from the Czech Republic, especially those from animal shelters (Zemanová et al., Reference Zemanová, Husník and Svobodová2005; Dubná et al., Reference Dubná, Langrová, Nápravník, Jankovská, Vadlejch, Pekár and Fechtner2007). Microsporidia, however, occur in dogs at a low level, only a few per cent, which may nevertheless pose a risk for immunodeficient individuals, as zoonotic genotypes are often detected (Piekarska et al., Reference Piekarska, Kicia, Wesołowska, Kopacz, Gorczykowski, Szczepankiewicz, Kváč and Sak2017). Importantly, frequent exposure to microsporidia has been confirmed among immunocompetent people in the Czech Republic (Sak et al., Reference Sak, Brady, Pelikánová, Květoňová, Rost, Kostka, Tolarová, Hůzová and Kváč2011), while in studies conducted in Poland, up to 26% of tested immunocompromised individuals were found to be infected with at least 1 microsporidian species (Kicia et al., Reference Kicia, Wesolowska, Kopacz, Jakuszko, Sak, Květonová, Krajewska and Kváč2016, Reference Kicia, Szydłowicz, Cebulski, Jakuszko, Piesiak, Kowal, Sak, Krajewska, Hendrich, Kváč and Kopacz2019).
It has been previously shown that specific genotypes and assemblages of these enteric pathogens may be detected in both humans and animals (Xiao et al., Reference Xiao, Cama, Cabrera, Ortega, Pearson and Gilman2007; Soliman et al., Reference Soliman, Fuentes and Rubio2011; Karim et al., Reference Karim, Dong, Yu, Jian, Zhang, Wang, Zhang, Rume, Ning and Xiao2014a, Reference Karim, Wang, Dong, Zhang, Li, Zhang, Rume, Qi, Jian, Sun, Yang, Zou, Ning and Xiao2014b; Hinney et al., Reference Hinney, Sak, Joachim and Kváč2016), suggesting the possible zoonotic route of transmission. One of their sources may be shelter dogs, which often live in poor sanitary conditions and crowded spaces that favour the spread of such microorganisms (Raza et al., Reference Raza, Rand, Qamar, Jabbar and Kopp2018). Currently, there are 226 registered animal shelters in Poland (General Veterinary Inspectorate, 2023) and 248 in the Czech Republic (State Veterinary Administration, 2024). Therefore, the aim of the present study was to investigate the occurrence of Cryptosporidium spp., G. intestinalis, Encephalitozoon spp. and E. bieneusi in dogs living in animal shelters in central Europe (Poland and the Czech Republic) and to assess the host specificity and zoonotic potential of these organisms at the genotype level.
Materials and methods
Samples
Individual fresh fecal samples were collected from dogs in animal shelters in Poland and the Czech Republic. Samples were collected directly from the floor by study research staff immediately after defecation, with care taken to avoid sampling fecal material that came into contact with the ground (concrete surface, without contact with the soil). They were collected in the morning, before daily cleaning routinely performed in each shelter. Each sample was individually placed in a sterile tube with animal ID, refrigerated at 4°C without preservatives and transported to laboratory. None of the collected stool had an apparent diarrhoeal symptom at the time of sampling. Where possible, information about the animal, such as sex and age, was also recorded during material collection (see Supplementary Table 1). Control of intestinal protozoa in dogs and cats in both Polish and Czech shelters is carried out according to current ESCCAP guidelines (ESCCAP, 2018) – in all facilities, pyrantel and fenbendazole were routinely used once every 3 months as part of antiparasitic prophylaxis.
Molecular analysis
Stool samples were stored up to 2 months in 4°C without preservatives until DNA extraction. Initial homogenization of 200 mg of each stool sample was performed by bead disruption for 60 s at 5.5 m s−1 with 0.5 mm glass beads using a Precellys 24 Instrument (Bertin Technologies, Montigny le Bretonneux, France), followed by genomic DNA (gDNA) extraction using a GeneMATRIX Stool DNA Purification Kit (EurX, Gdańsk, Poland). Molecular detection was based on the nested polymerase chain reaction (PCR) protocols for the amplification of the chosen genes of E. bieneusi (ITS), Encephalitozoon spp. (ITS), Cryptosporidium spp. (18S rRNA) and G. intestinalis (TPI) (Didier et al., Reference Didier, Vossbrinck, Baker, Rogers, Bertucci and Shadduck1995; Katzwinkel-Wladarsch et al., Reference Katzwinkel-Wladarsch, Lieb, Helse, Löscher and Rinder1996; Xiao et al., Reference Xiao, Morgan, Limor, Escalante, Arrowood, Shulaw, Thompson, Fayer and Lal1999; Buckholt et al., Reference Buckholt, Lee and Tzipori2002; Sulaiman et al., Reference Sulaiman, Fayer, Bern, Gilman, Trout, Schantz, Das, Lal and Xiao2003). Additional PCRs amplifying selected loci were performed for subtyping in order to assess intra-species genetic diversity in the case of samples positive for Cryptosporidium spp. (partial 60-kDa glycoprotein gene – gp60) and G. intestinalis (β-giardin – BG and glutamate dehydrogenase – GDH) (see Supplementary Table 2) (Cacciò et al., Reference Cacciò, De Giacomo and Pozio2002, Reference Cacciò, Beck, Lalle, Marinculic and Pozio2008; Lalle et al., Reference Lalle, Jimenez-Cardosa, Cacciò and Pozio2005; Jiang et al., Reference Jiang, Roellig, Guo, Li, Feng and Xiao2021). Each PCR contained 0.25–2.0 μL of DNA, 200 μm each of deoxynucleoside triphosphate (dNTP), 1× PCR buffer (DreamTaq™ Green Buffer, ThermoFisher Scientific, Waltham, MA, USA), 3.0 mm MgCl2, 0.125 U of Taq polymerase (ThermoFisher Scientific), 10 μg of bovine serum albumin (BSA) and 200 nm of each primer in a total of 20–25 μL reaction. The reactions were performed in a C1000 Bio-Rad thermocycler, with an initial hot start (94°C for 5 min) and a final extension (72°C for 10 min), according to the conditions described in Supplementary Table 2. An aliquot of primary PCR was used as a template for the secondary PCR. Its conditions were identical to the primary PCR, except that BSA was not added to the secondary reaction. Negative (molecular grade water) and positive controls (DNA extracted from E. bieneusi genotype CZ3, E. cuniculi genotype III spores, Cryptosporidium serpentis oocysts or G. intestinalis assemblage F cysts) were included in each PCR amplification. Secondary PCR products were electrophoresed on a 1% agarose gel containing 0.2 mg mL−1 Midori Green DNA stain in TAE buffer at 75 V for approximately 1 h. Bands of the predicted sizes were visualized using a UV light source, cut from the gel, extracted using a Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA) and sequenced bi-directionally by a company offering this service commercially (Genomed S.A., Warsaw, Poland). The nucleotide sequences obtained were processed using Chromas Pro 2.4.1 software (Technelysium, Pty, Ltd., South Brisbane, Australia). Subsequently, BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was performed to verify the identity of the sequences. The edited and aligned sequences were further processed using BioEdit v.7.0.5 (Hall, Reference Hall1999). To align the obtained sequences with reference sequences from GenBank, the online server MAFFT version 7 was used (http://mafft.cbrc.jp/alignment/software). The best model for DNA/protein phylogeny for each alignment was selected based on the Bayesian information criterion in MEGA 7 (Kumar et al., Reference Kumar, Stecher and Tamura2016). Tamura's 3-parameter model + G + I was used for the alignments. The maximum likelihood (ML) approach was carried out in MEGA7 software. Bootstrap support was calculated based on 1000 replications to evaluate the robustness of tree branching. Finally, the resulting trees were visualized using Corel Draw X7 software (https://www.corel.draw.com). Representative nucleotide sequences of all loci used as markers for subtyping of isolates obtained in the current study were deposited in GenBank with the accession numbers OR791083, OR791084, OR791770, OR791659, OR791771–OR791785, OR807726 and OR807727.
Statistical analysis
Statistical analysis was performed using χ 2 or Fisher's tests to compare the frequency of occurrence of the tested pathogens between Polish and Czech shelters (Statistica software, TIBCO Software Inc., USA). A P < 0.05 was considered significant.
Results
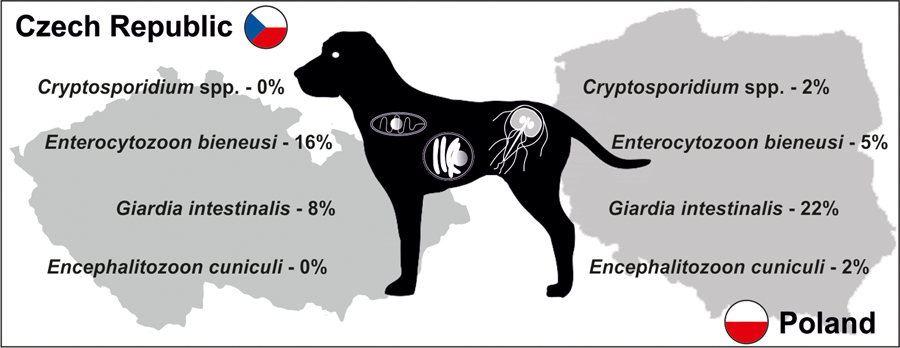
A total of 187 apparently healthy dogs from 10 shelters (Fig. 1), 5 in Poland (101 dogs) and 5 in the Czech Republic (86 dogs), were studied (Table 1). Specific DNA of targeted parasites was detected in 50 of 187 animals (26.7%), with higher occurrence observed in Polish than in Czech animals (32.6 vs 24.4%, χ 2 = 1.0254; P = 0.3112; Table 1). Most of the detected infections were monoinfections; only 2 dogs (ID 2966_CZ.1 and 3031_CZ.5) had a coinfection, E. bieneusi and G. intestinalis. Giardia intestinalis (29 dogs, 15.5%) followed by E. bieneusi (19 dogs, 10.2%) were the most frequently detected parasites, whereas each Encephalitozoon spp. and Cryptosporidium spp. were found in 2 dogs (Table 1). Overall, there was a significant trend towards more frequent occurrence of G. intestinalis in Polish (21.8%) vs Czech animals (8.1%, χ 2 = 6.5979; P = 0.0102), while E. bieneusi was seen more often in dogs from Czech shelters (16.3%) than in Polish ones (4.9%, χ 2 = 6.5305; P = 0.0106). Since detailed demographic data were obtained only from a small number of individuals, they were not subjected to statistical analysis.

Figure 1. Schematic arrangement of shelters in Poland (P.1–P.5) and the Czech Republic (CZ.1–CZ.5) from which samples were collected.
Table 1. Occurrence of Cryptosporidium spp., Giardia intestinalis, Encephalitozoon cuniculi and Enterocytozoon bieneusi in individual dogs kept in animal shelters in Poland and the Czech Republic

The results of genotyping of all pathogens performed in this study are presented in Table 2. Phylogeny analysis of partial sequences of 18S rRNA of Cryptosporidium showed the presence of C. canis identical to C. canis dog genotype (GenBank Acc. No. AB120909) in isolate 2806_P.1 and in a Cryptosporidium sp. isolate 2735_P.2, phylogenetically clustered near the gastric Cryptosporidium spp. (Fig. 2). Cryptosporidium sp. isolate 2735_P.2 differed from Cryptosporidium proliferans in 3 single-nucleotide polymorphisms (SNPs) with 99.6% sequence identity. Based on C. canis gp60 locus subtyping, isolate 2806_P.1 was assigned to the XXe family (Fig. 3). Genotype II, determined at ITS sequences, was detected in both dogs positive for E. cuniculi (Table 2; phylogeny is not shown). Phylogenetic analysis of Giardia showed different results in 12 isolates, depending on the marker used (Table 2). Subtyping based on TPI gene revealed the presence of only assemblage C in all samples examined (Fig. 4), while for BG (Fig. 5) and GDH (Fig. 6) loci the presence of both C and D assemblages has been shown. Subtyping of BG and GDH failed in 2 and 3 isolates, respectively. With the exception of genotype D, which was detected in dog 2699_P.5, all other E. bieneusi sequences were identical to the PtEbIX genotype (Fig. 7).
Table 2. Results of genotyping of Cryptosporidium spp., G. intestinalis, E. cuniculi and E. bieneusi in all tested samples

Accession numbers in square brackets indicate the isolates deposited in GenBank as representative nucleotide sequences derived from the present study.
TPI, triosephosphate isomerase; BG, β-giardin; GDH, glutamate dehydrogenase; 18S rRNA, small ribosomal subunit rRNA; gp60, 60-kDa glycoprotein; ITS, internal transcribed spacer region of rRNA.
a Cryptosporidium sp. isolate 2735_P.2, phylogenetically clustered near the gastric Cryptosporidium spp., differed from C. proliferans in 3 SNPs with 99.6% sequence identity of the 18S rRNA region.
b N/A, assemblage not available (subtyping failed).

Figure 2. Phylogenetic relationships between Cryptosporidium spp. detected in dogs in this study (highlighted in green) and other Cryptosporidium available in GenBank using an ML analysis of partial sequences of 18S rRNA (sequence alignment length: 820 bp). Percentage supports (>50%) from 1000 pseudoreplicates are indicated next to the supported node. The branch length scale bar indicates the number of substitutions per site. Sequences from this study are identified by an isolate number (e.g. 2806) followed by region and location (P.1, Poland location 1).

Figure 3. Phylogenetic relationship between Cryptosporidium canis detected in 1 dog in this study (highlighted in green) and other C. canis available in GenBank using an ML analysis of a region of gp60 gene (sequence alignment length: 540 bp). Percentage supports (>50%) from 1000 pseudoreplicates are indicated next to the supported node. The branch length scale bar indicates the number of substitutions per site. Sequences from this study are identified by an isolate number (2806) followed by region and location (P.1, Poland location 1).

Figure 4. Phylogenetic relationships between Giardia intestinalis assemblages detected in dogs in this study (highlighted in green – Poland or in grey – Czech Republic) and other G. intestinalis assemblages available in GenBank using an ML analysis of a region of TPI gene (sequence alignment length: 467 bp). Percentage supports (>50%) from 1000 pseudoreplicates are indicated next to the supported node. The branch length scale bar indicates the number of substitutions per site. Sequences from this study are identified by an isolate number (e.g. 2966) followed by region and location (P.1, Poland location 1, CZ.1, Czech Republic location 1).

Figure 5. Phylogenetic relationships between G. intestinalis assemblages detected in dogs in this study (highlighted in green – Poland or in grey – Czech Republic) and other G. intestinalis assemblages available in GenBank using an ML analysis of a region of BG gene (sequence alignment length: 820 bp). Percentage supports (>50%) from 1000 pseudoreplicates are indicated next to the supported node. The branch length scale bar indicates the number of substitutions per site. Sequences from this study are identified by an isolate number (e.g. 2966) followed by region and location (P.1, Poland location 1, CZ.1, Czech Republic location 1).

Figure 6. Phylogenetic relationships between G. intestinalis assemblages detected in dogs in this study (highlighted in green – Poland or in grey – Czech Republic) and other G. intestinalis assemblages available in GenBank using an ML analysis of a region of GDH gene (sequence alignment length: 439 bp). Percentage supports (>50%) from 1000 pseudoreplicates are indicated next to the supported node. The branch length scale bar indicates the number of substitutions per site. Sequences from this study are identified by an isolate number (e.g. 2966) followed by region and location (P.1, Poland location 1, CZ.1, Czech Republic location 1).

Figure 7. Phylogenetic relationships between Enterocytozoon bieneusi genotypes detected in dogs in this study (highlighted in green – Poland or in grey – Czech Republic) and other E. bieneusi genotypes available in GenBank using an ML analysis of ITS region of rRNA gene (sequence alignment length: 309 bp). Percentage supports (>50%) from 1000 pseudoreplicates are indicated next to the supported node. The branch length scale bar indicates the number of substitutions per site. Sequences from this study are identified by an isolate number (e.g. 2973) followed by region and location (P.1, Poland location 1, CZ.1, Czech Republic location 1).
Discussion
In the present study, the screening of fecal samples collected from shelter dogs in central Europe was performed in order to analyse the prevalence of zoonotic unicellular pathogens (Cryptosporidium spp., G. intestinalis, E. bieneusi and Encephalitozoon spp.) at the molecular level. Over a quarter of the tested dogs were carriers of at least one of the studied pathogens, among which the most often observed was G. intestinalis – one of the most common intestinal parasites infecting humans and animals (Kváč et al., Reference Kváč, Hofmannová, Ortega, Holubová, Horčičková, Kicia, Hlásková, Květoňová, Sak and McEvoy2017). Nevertheless, its prevalence in other European canine populations, including shelter dogs, has shown to be even higher [27–36.5% in various regions of Spain (Gil et al., Reference Gil, Cano, de Lucio, Bailo, de Mingo, Cardona, Fernández-Basterra, Aramburu-Aguirre, López-Molina and Carmena2017; Adell-Aledón et al., Reference Adell-Aledón, Köster, de Lucio, Puente, Hernández-de-Mingo, Sánchez-Thevenet, Dea-Ayuela and Carmena2018; Remesar et al., Reference Remesar, García-Dios, Calabuig, Prieto, Díaz-Cao, López-Lorenzo, López, Fernández, Morrondo, Panadero and Díaz2022), 33.8% in Portugal (Pereira et al., Reference Pereira, Teixeira, Sousa, Parreira, Campino, Meireles and Maia2021), over 45% in Serbia (Sommer et al., Reference Sommer, Zdravković, Vasić, Grimm and Silaghi2017), while in central Italy this value ranged from about 7% (Scaramozzino et al., Reference Scaramozzino, Carvelli, Iacoponi and De Liberato2018) to 41% (Agresti et al., Reference Agresti, Berrilli, Maestrini, Procesi, Loretti, Vonci and Perrucci2022)]. Generally, such a high prevalence is most likely due to the simple and direct life cycle of Giardia, with easily spread dispersive forms excreted in feces, which facilitates transmission in highly dense populations, such as those found in animal shelters. Enterocytozoon bieneusi was also found to be a common pathogen in dogs in the present study, with an overall prevalence of 10.2%, which agrees with previous reports considering European dogs, with infection rates ranging between 4.9 and 11.7% (Mathis et al., Reference Mathis, Weber and Deplazes2005; Santín and Fayer, Reference Santín and Fayer2011; Piekarska et al., Reference Piekarska, Kicia, Wesołowska, Kopacz, Gorczykowski, Szczepankiewicz, Kváč and Sak2017). In turn, considerably low prevalences were observed for Encephalitozoon ssp. and Cryptosporidium spp. (~1% for both), comparably to previous reports regarding canine populations: 0–2.4% for Encephalitozoon spp. (Piekarska et al., Reference Piekarska, Kicia, Wesołowska, Kopacz, Gorczykowski, Szczepankiewicz, Kváč and Sak2017; Delrobaei et al., Reference Delrobaei, Jamshidi, Shayan, Ebrahimzade, Ashrafi Tamai, Rezaeian and Mirjalali2019) and 0.6–4.9% for Cryptosporidium spp. (Giangaspero et al., Reference Giangaspero, Iorio, Paoletti, Traversa and Capelli2006; Simonato et al., Reference Simonato, Frangipane di Regalbono, Cassini, Traversa, Tessarin, Di Cesare and Pietrobelli2017; Yu et al., Reference Yu, Ruan, Zhou, Chen, Zhang, Wang, Zhu and Yu2018; Piekara-Stępińska et al., Reference Piekara-Stępińska, Piekarska and Gorczykowski2021a), although in 1 German study the Cryptosporidium prevalence was as high as 10% (Murnik et al., Reference Murnik, Daugschies and Delling2022). Notwithstanding, the true prevalence of these pathogens among healthy hosts may, in fact, be higher as their forms are excreted periodically and irregularly, which may be overlooked with a single sampling (Sak et al., Reference Sak, Kašičková, Kváč, Květoňová and Ditrich2010). It would therefore be recommended to collect samples several times from the same animals, which may prove difficult due to the conditions specific to the shelters, such as the rotation of animals or irregular hours of cleaning the excrement. Nevertheless, the fact that the demonstrated prevalence of pathogens such as G. intestinalis was high even with only 1 sampling underscores the importance of their likely distribution in the population and the wide reservoir of the pathogen. Differences in the frequencies of the studied species between Polish and Czech dogs, as well as the higher prevalence of specific pathogens in individual facilities, may be related to some specific conditions typical of a particular location.
Sequence analyses of detected pathogens showed that most infections involved dog-specific genotypes or species, the transmission of which may be favoured by intensive contact among large numbers of dogs living together. In the case of 12 isolates, assignment to the appropriate G. intestinalis assemblage was difficult because subtyping results varied depending on the locus used. Similar discrepancies have been reported in previous studies (Read et al., Reference Read, Monis and Andrew Thompson2004; Traub et al., Reference Traub, Monis, Robertson, Irwin, Mencke and Thompson2004), emphasizing the importance of using multilocus genotyping in the molecular analysis of Giardia diversity. Nevertheless, all G. intestinalis-positive samples harboured assemblage C or D, which have a strong host specificity for dogs and other canines (Feng and Xiao, Reference Feng and Xiao2011). These assemblages were also found to be highly prevalent in different dog populations in Europe (Simonato et al., Reference Simonato, Frangipane di Regalbono, Cassini, Traversa, Beraldo, Tessarin and Pietrobelli2015; Adell-Aledón et al., Reference Adell-Aledón, Köster, de Lucio, Puente, Hernández-de-Mingo, Sánchez-Thevenet, Dea-Ayuela and Carmena2018; Pereira et al., Reference Pereira, Teixeira, Sousa, Parreira, Campino, Meireles and Maia2021) and although sporadically they have been reported in humans as well (Broglia et al., Reference Broglia, Weitzel, Harms, Cacció and Nöckler2013; Liu et al., Reference Liu, Shen, Yin, Yuan, Jiang, Xu, Pan, Hu and Cao2014; Villamizar et al., Reference Villamizar, Higuera, Herrera, Vasquez-A, Buitron, Muñoz, Gonzalez-C, Lopez, Giraldo and Ramírez2019), their zoonotic relevance seems to be low and limited to individuals at risk, for instance, children or immunocompromised persons. Likewise, cases of C. canis colonization in humans were described in individuals at increased risk (children, HIV-infected adults) and immunocompetent people as well (Learmonth et al., Reference Learmonth, Ionas, Ebbett and Kwan2004; Gatei et al., Reference Gatei, Das, Dutta, Sen, Cama, Lal and Xiao2007; Feng et al., Reference Feng, Wang, Duan, Gomez-Puerta, Zhang, Zhao, Hu, Zhang and Xiao2012; Liao et al., Reference Liao, Lin, Sun, Qi, Lv, Wu, Li, Hu, Yu, Cai, Xiao, Sun and Li2020). However, due to the relatively transient nature of these infections in humans, dogs do not seem to represent an important source of cryptosporidiosis for people (Villamizar et al., Reference Villamizar, Higuera, Herrera, Vasquez-A, Buitron, Muñoz, Gonzalez-C, Lopez, Giraldo and Ramírez2019; Liao et al., Reference Liao, Lin, Sun, Qi, Lv, Wu, Li, Hu, Yu, Cai, Xiao, Sun and Li2020). To date, 9 families of C. canis subtypes (XXa–XXi) have been identified based on gp60 locus subtyping, occurring not only in canids, but also in minks, foxes and humans (Jiang et al., Reference Jiang, Roellig, Guo, Li, Feng and Xiao2021; Murnik et al., Reference Murnik, Daugschies and Delling2022; Wang et al., Reference Wang, Wei, Cao, Wu, Zhao, Guo, Xiao, Feng and Li2022). According to the study of Jiang et al., the zoonotic potential may concern XXa family, detected in both dog and human samples (Jiang et al., Reference Jiang, Roellig, Guo, Li, Feng and Xiao2021). In our study, analysis of the C. canis gp60 locus in the 2806_P.1 isolate revealed that it belongs to the XXe family (Fig. 3), which was also the most prevalent among dogs in the report from Germany (Murnik et al., Reference Murnik, Daugschies and Delling2022). In turn, the newly detected Cryptosporidium sp. isolate 2735_P.2 also does not pose a significant risk to humans and probably not to dogs as well. An incidental infection/contamination was likely caused by rodent feces. Cryptosporidium sp. isolate 2735_P.2 is closely related to Cryptosporidium muris and C. proliferans, whose hosts are rodents. However, the phylogenetic position is not related to host specificity, and therefore another host cannot be excluded (Kváč et al., Reference Kváč, Vlnatá, Ježková, Horčičková, Konečný, Hlásková, McEvoy and Sak2018). To summarize the subtyping results, as in previous studies (de Lucio et al., Reference de Lucio, Bailo, Aguilera, Cardona, Fernández-Crespo and Carmena2017; Rehbein et al., Reference Rehbein, Klotz, Ignatius, Müller, Aebischer and Kohn2019), zoonotic transmission of giardiasis or cryptosporidiosis between dogs and humans is most likely a rare event.
All E. bieneusi isolates detected in the studied dogs, except for 1 clustering to genotype D, were identical to E. bieneusi genotype PtEbIX. This genotype appears to be specific to dogs; to date, it has been detected almost exclusively in dogs and sporadically in wolves, cats and swans (Santín et al., Reference Santín, Vecino and Fayer2008; Abe et al., Reference Abe, Kimata and Iseki2009; Santín and Fayer, Reference Santín and Fayer2011; Mori et al., Reference Mori, Mahittikorn, Thammasonthijarern, Chaisiri, Rojekittikhun and Sukthana2013; Karim et al., Reference Karim, Dong, Yu, Jian, Zhang, Wang, Zhang, Rume, Ning and Xiao2014a; Piekarska et al., Reference Piekarska, Kicia, Wesołowska, Kopacz, Gorczykowski, Szczepankiewicz, Kváč and Sak2017; Kváč et al., Reference Kváč, Myšková, Holubová, Kellnerová, Kicia, Rajský, McEvoy, Feng, Hanzal and Sak2021). In turn, both E. bieneusi genotype D and E. cuniculi genotype II detected in the present study have been reported in a broad range of hosts so far, including humans (Li et al., Reference Li, Xiao, Wang, Zhao, Zhao, Duan, Guo, Liu and Feng2012; Kváč et al., Reference Kváč, Hofmannová, Ortega, Holubová, Horčičková, Kicia, Hlásková, Květoňová, Sak and McEvoy2017; Piekarska et al., Reference Piekarska, Kicia, Wesołowska, Kopacz, Gorczykowski, Szczepankiewicz, Kváč and Sak2017; Delrobaei et al., Reference Delrobaei, Jamshidi, Shayan, Ebrahimzade, Ashrafi Tamai, Rezaeian and Mirjalali2019). Observation of microsporidian genotypes with a human-infection capacity in companion animals suggests that pets may be of importance as one of the potential sources of infection. However, the presented results do not indicate that dogs in shelters in Poland and the Czech Republic represent a significant source of zoonotic species and genotypes of the studied parasites for humans.
Our study had some limitations. Firstly, due to its screening nature, a detailed analysis in the context of the demographic data of the tested dogs or the drugs used was not possible to conduct. Moreover, the study groups differed in size – in Poland it was possible to collect material from a larger number of animals than in the Czech Republic, which may have an impact on the differences in prevalence.
This study included clinically healthy animals without signs of intestinal infection, yet dispersive forms of potentially pathogenic and infectious organisms were observed. It should also be borne in mind that asymptomatic hosts could shed cysts, oocysts or spores occasionally and irregularly, and thus their screening with multiple sampling could increase the real observed prevalence (Sak et al., Reference Sak, Kašičková, Kváč, Květoňová and Ditrich2010). Nevertheless, since infections of the studied pathogens in dogs can often be asymptomatic, they may not be detected by routine veterinary examinations, and their occurrence in companion animals may be underestimated. On the contrary, the majority of species and genotypes observed in canine samples herein are not commonly associated with human infections, and aforesaid transmission routes seem to be rare. The exceptions are genotypes D (E. bieneusi) and II (E. cuniculi) observed in the present study, whose zoonotic potential should be emphasized due to their occurrence in a wide range of different hosts (Hinney et al., Reference Hinney, Sak, Joachim and Kváč2016; Li et al., Reference Li, Feng and Santin2019, Reference Li, Feng and Xiao2020). Despite the low likelihood of transmission of the studied pathogens and due to the fact that they mainly affect immunosuppressed individuals, in whom the consequences of opportunistic infections may be life-threatening, the awareness among new dog owners is recommended, especially those with various levels of immunosuppression, on the relevance of diagnosing and treating zoonotic diseases.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S003118202400009X
Data availability statement
The data that support the findings of this study are available from the corresponding author, MS, upon reasonable request.
Acknowledgements
The authors acknowledge the staff of dog shelters for their assistance in sample collection and the technician staff of the Laboratory of Veterinary and Medical Protistology, Biology Centre CAS, for sample processing during this study.
Author's contribution
M. S., M. Ki. and Ż. Z. conceived and designed the study. M. S., M. Ki., Ż. Z., M. Ka., M. Kv., B. S. and N. H. conducted data gathering. M. S., M. Ki., Ż. Z., M. Ka., B. Ł. and A. L. prepared the samples and conducted molecular analysis. M. S. conducted statistical analyses. M. Kv. prepared phylogenetic analyses. M. S., M. Ki. and M. Kv. wrote the article.
Financial support
This work was supported with a subvention by the Polish Ministry of Health (M. S., grant number SUBK.A060.23.027 from the IT Simple system of Wroclaw Medical University) and the Grant Agency of the Czech Republic (M. Kv., GACR 21-23773S and B. S. 23-06571S).
Competing interests
None.
Ethical standards
The collection of samples carried out in a non-invasive way, without interfering with the organism of the animals included in the study.