Introduction
Giardia duodenalis (syn. Giardia lamblia, Giardia intestinalis) is a common intestinal parasite of humans, domestic pets and livestock animals worldwide (Feng and Xiao, Reference Feng and Xiao2011). The infectious stages of the Giardia life cycle (cysts) are released in feces, are immediately infectious (Huang and White, Reference Huang and White2006) and can remain infectious for several months in the environment (Erickson and Ortega, Reference Erickson and Ortega2006). Giardia was first described as a species complex in 1989, with the identification of four discreet genetic groups in what was originally known as G. intestinalis (from humans) (Nash and Keister, Reference Nash and Keister1985; Nash et al., Reference Nash, McCutchan, Keister, Dame, Conrad and Gillin1985). These four groups were later further reclassified as assemblage A (I and II) and assemblage B (III and IV) (Monis et al., Reference Monis, Mayrhofer, Andrews, Homan, Limper and Ey1996). This classification is still used today, with the remaining six assemblages (C–H) appearing to be more host-specific and are not considered zoonotic (Ryan and Caccio, Reference Ryan and Caccio2013).
Across the UK, there are an average of 3500 human cases of giardiasis per annum, this equates to approximately 5.5 cases per 100 000 population. In Scotland, between 2006 and 2015, on average 198 diagnosed cases of human giardiasis were reported per year (http://www.hps.scot.nhs.uk/giz/giardia.aspx). However, as giardiasis in humans can present with mild symptoms or even be asymptomatic, the true number of cases is likely to be greatly under-reported (Currie et al., Reference Currie, Stephenson, Palmer, Jones and Alexander2017). The main reason why most cases of giardiasis in Scotland are not investigated is due to the fact that the laboratory testing algorithm is based on only testing feces from individuals with a recent history of travel to certain regions of the world. A recent study in Scotland genotyped Giardia isolated from 30 confirmed clinical cases of human giardiasis demonstrating the presence of both assemblages A and B. Most of the cases consisted of assemblage A (72%); assemblage B and mixed A/B infections were far less common than assemblage A infections (14 and 10%, respectively) (Alexander et al., Reference Alexander, Jones, Inverarity and Pollock2014).
Infections with Giardia are common amongst livestock animals and have been demonstrated in the UK. In a previous study conducted in England and Wales by Minetti et al. (Reference Minetti, Taweenan, Hogg, Featherstone, Randle, Latham and Wastling2014), assemblages A, C, D, E and F were all identified in cattle sheep, pigs and goats, though interestingly assemblage B parasites were not demonstrated in any of the livestock species tested (Minetti et al., Reference Minetti, Taweenan, Hogg, Featherstone, Randle, Latham and Wastling2014). Cattle, like humans may not display any overt clinical gastro-intestinal signs other than a loose fecal consistency. However, untreated infections can impair the growth and development of young cattle (Geurden et al., Reference Geurden, Vandenhoute, Pohle, Casaert, De Wilde, Vercruysse and Claerebout2010a, Reference Geurden, Vercruysse and Claerebout2010b). This reduction in the feed conversion in cattle could have a profound economic impact to farmers, particularly if infections are widespread throughout their herds.
The aim of this current study was to determine the prevalence of Giardia infections in beef and dairy cattle from across Scotland and identify the parasite assemblages (sub-assemblages) present to determine the extent of the infection in cattle and whether cattle have the potential to disseminate human infectious (zoonotic) assemblages of the parasite into the environment.
Materials and methods
Animals and sample collection
Three hundred and eighty-eight fecal samples were collected from age-appropriate cattle (available on the day of sampling) from 19 locations spread across Scotland, of which 128 and 253 were from beef and dairy cattle, respectively, while seven samples had no associated breed information. An average of 20 samples were collected from each farm (range n = 1–114). Samples were also sub-divided by host age, from 1, 2, 3, 4, 5 and 6 weeks, between 7–24 and ⩾25 weeks (see Table 1). Fecal samples were collected directly from each animal per rectum with a fresh gloved hand into a sterile tube then stored at 4°C prior to DNA extraction.
Table 1. Breeds and ages of cattle tested for the presence of G iardia duodenalis DNA
a Seven samples from cattle with no assigned breed.
DNA extraction from fecal samples
For each fecal sample, DNA was extracted using a modified Macherary–Nagel tissue kit protocol as previously described (Wells et al., Reference Wells, Thomson, Ensor, Innes and Katzer2016). In brief, samples from animals aged ⩾25 weeks of age underwent a sedimentation step to concentrate parasite cysts prior to DNA extraction (Thomson et al., Reference Thomson, Innes, Jonsson and Katzer2016). For calf samples (ages 1–24 weeks), a 250 mg loop of feces was resuspended in 1 mL of TE buffer (10 mm Tris-HCl, 0.5 mm EDTA) and mixed by vortexing. Samples were then pelleted by centrifugation at 5000 × g for 10 min. All samples were resuspended in 200 µL of lysis buffer (Macherey–Nagel Buffer T1), then underwent 10 rounds of freeze–thawing (alternating between liquid nitrogen and a 56 °C water bath) to disrupt any Giardia cysts. Samples were incubated with Proteinase K (100 µg sample−1) at 56°C overnight (~18 h) then vigorously mixed by vortexing. Samples were then centrifuged at 11 000 × g to remove any insoluble debris and the supernatant was then mixed with ethanol. DNA was then extracted using NucleoSpin Tissue DNA columns (Macherey–Nagel, NZ740952250) as per the manufacturer's instructions. For the final elution of the DNA, each sample was resuspended in 100 µL of DNase/RNase-free water. All DNA samples were stored at −20 °C prior to analysis by Giardia PCR.
PCR amplification of Giardia DNA
The PCR reaction mixture used in this study has been previously described (Burrells et al., Reference Burrells, Bartley, Zimmer, Roy, Kitchener, Meredith, Wright, Innes and Katzer2013). Briefly each reaction (20 µL) contained final concentrations of: bovine serum albumin 0.113 mg mL−1, EDTA 4.4 µ m, MgCl2 4.5 mm, (NH4)2SO4 11 mm, Tris-HCl 45 mm, each of deoxyribonucleotide triphosphates (dATP, dCTP, dGTP and dTTP) at 1.0 mm, forward and reverse primers (Table 2) (Eurofins MWG Operon) 0.25 pM each, Taq polymerase 0.75U (Bioline Ltd., London, UK) and 2 µL of template DNA. To increase the sensitivity of the assays, each sample was analysed in duplicate. PCR results were only accepted when all appropriate positive and negative controls passed; a single positive result was still accepted as positive.
Table 2. Primers used for the detection of G iardia doudenalis DNA in fecal samples from beef and dairy cattle in Scotland

All PCR annealing temperatures were optimized by gradient PCR using a positive control sample, the highest temperature where clear amplification was observed was used to analyse the samples. Initial screening of all 388 samples was performed using a nested PCR to detect the G. duodenalis (β-giardin) (bg) gene DNA (Table 2). The G7 and G759 primers (Liu et al., Reference Liu, Zhang, Zhang, Wang, Li, Shu, Zhang, Shen, Zhang and Ling2012) were used in the primary amplification using the PCR reaction mixture described above. Following amplification, the primary amplicons were each diluted with 100 µL of DNase/RNase-free water, and 2 µL of this diluted product was used as the template for the second round of amplification using the GiarF and GiarR primers (Table 2). The basic reaction conditions for all the PCR amplifications were as follows: 95 °C for 5 min followed by 35 cycles at 95 °C for 1 min, annealing (see Table 2 for temperatures) for 1 min and 72 °C for 1 min with a final extension period at 72 °C for 5 min. Following secondary amplification, 6 µL of each PCR product was analysed by agarose gel electrophoresis (2% agarose in 1× Tris Acetate EDTA buffer), stained with gel red (used at 1:10 000) (Biotonium, Hayward, CA, USA) and visualized using UV light (alpha imager system).
Samples that tested positive using the Giardia bg PCR were then tested using the G. duodenalis triose-phosphate isomerase (tpi) (Caccio et al., Reference Caccio, Beck, Lalle, Marinculic and Pozio2008) and G. duodenalis glutamate dehydrogenase (gdh) (Caccio et al., Reference Caccio, De Giacomo and Pozio2002) PCRs. The reaction mixture and general reaction conditions are identical to those listed above, with the primers and optimal annealing temperatures listed in Table 2.
PCR purification and sequence analysis
A selection of positive bg gene PCR amplicons was prepared for sequence analysis. Amplicons were first purified using a commercially available kit (Wizard® SV Gel and PCR Clean-up System) (Promega, Madison, WI, USA), as per manufacturers’ instructions. The final elution of the PCR product was in 30 µL of DNase/RNase-free water. The nucleic acid concentration of each sample was determined using a Nanodrop ND1000 spectrophotometer. For sequencing (MWG Operon), 100 ng of each PCR product was analysed with each appropriate set of second round primers, GiarF and GiarR (bg), to create overlapping forward and reverse consensus sequences for each gene of interest. To augment the bg gene data, the tpi and gdh PCR amplicons from selected samples (see Table 4) were sent for sequence analysis using the AL3544 and AL3545 (tpi) and Gdh3 and Gdh4 (gdh) primers (Table 2).
Phylogenetic analysis
Consensus sequences were generated for each G. duodenalis gene (bg, gdh and tpi) sample using SeqMan Pro™ software (v 12.3.1). These consensus sequences were compared against published sequences using the NCBI BLAST tool (nucleotide collection database), to produce a percentage identity. The phylogenetic analysis was carried out where possible on all three gene sequences (bg, gdh and tpi) from individual samples, using the Maximum Likelihood method based on the Kimura two-parameter model that included bootstrap values (1000 replicates), using the MEGA7 software.
The different bg, gdh and tpi sequences identified during this study have been submitted to GenBank (Accession numbers MG988428–MG988451)
Statistical analysis
Significant differences between non-infected and infected animals, depending on breed and age, were compared using χ 2 tests using RStudio software (RStudio Team, 2016); a value of P = <0.05 was considered statistically significant. The results from the beef and dairy cattle and animal ages were analysed separately by logistic regression; this produced an odds ratio of the likelihood of Giardia infection occurring in each cattle breed and at each time point in a logistic regression model with confidence intervals (95% CI). The proportion of positive samples (i.e. prevalence), with 95% CI was calculated for Giardia DNA using Minitab 15 (v15.1.0.0).
Results
Prevalence of G. duodenalis DNA in fecal samples from cattle in Scotland
Fecal samples were collected from 388 cattle from 19 farms from across Scotland, from both beef and dairy herds and from animals ranging from new born calves (<1 week) through to cattle ⩾25 weeks of age.
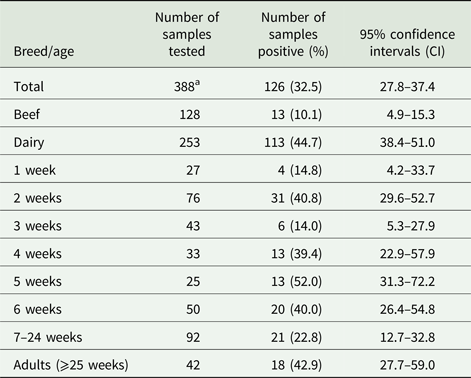
The results in Table 1 show that G. duodenalis β-giardin (bg) DNA was detected in 126/388 samples tested, giving a prevalence of 32.5% (95% CI 27.8–37.4) when combining all of the beef and dairy samples from all of the locations tested. However, when the results from the beef and dairy cattle are analysed separately, the prevalence in beef cattle is only 10.1% (95% CI 4.9–15.3) (13/128), compared with a prevalence of 44.7% (95% CI 38.4–51.0) (113/253) in the dairy animals. When the prevalence of G. duodenalis (i.e. levels of parasite DNA detection) in beef and dairy cattle were compared, it was seen that dairy cattle have statistically significantly higher numbers (χ 2 45.73, P = 0.0001) of positive samples compared with the beef cattle. A logistic regression model of the breed data demonstrated that the dairy cattle were 8.35 (95% CI 4.2–17.88) times more likely to be infected with Giardia than the beef cattle.
Positive Giardia samples were detected on 12/19 (63.15%) farms tested. The prevalence on the individual farms that tested positive for the presence of Giardia ranged from 5 to 100%. When the ages of the animals were compared, positive samples were detected for all of the age ranges tested (see Table 1). However, the highest prevalence was seen in animals 5 weeks of age, which had a prevalence of 52.0% (95% CI 31.3–72.2) (13/25), while the animals aged 3 weeks old had the lowest prevalence of parasite DNA with 14.0% (95% CI 5.3–27.9) (6/43). High prevalence of parasite DNA was also detected in 2-week-old calves and in adult cattle with a prevalence of 40.8% (95% CI 29.6–52.7) (31/76) and 42.9% (95% CI 27.7–59.0) (18/42), respectively. Positive samples were even detected in 14.8% (95% CI 4.2–33.7) (4/27) of calves at 1 week of age.
Sequence analysis of G. duodenalis DNA found in cattle fecal samples β-giardin (bg) gene
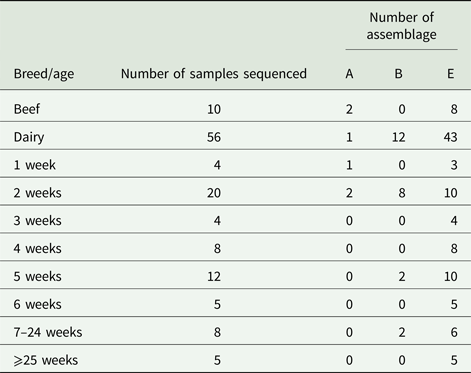
Due to cost implications, sequence analysis of the β-giardin gene was only able to be carried out on 66/126 (52.4%) of the PCR-positive amplicons. From these sequence data, we found that G. duodenalis assemblage E accounted for most of the positive samples, 77.2% (51/66), while assemblage B 18.2% (12/66), and assemblage A 4.6% (3/66) were also found, but these were much less common than assemblage E.
The results illustrated in Table 3 show that for beef cattle, 10/13 positive samples were sent for sequence analysis; of these, 8/10 (80%) were assemblage E while the remaining 2/10 (20%) were assemblage A. No assemblage B-positive samples were found in the beef cattle. In the dairy animals, we found that 43/56 (76.8%) of the sequences returned were assemblage E, 12/56 (21.4%) were assemblage B and the remaining 1/56 (1.8%) was an assemblage A (Table 3). All of the assemblage A (3/3) and 8/12 assemblage B sequences identified during this study were found in calves aged 1–2 weeks (Table 3), while the older calves and cattle were still found to be shedding some assemblage B parasites, but predominantly shed assemblage E parasites.
Table 3. Assemblages of G iardia duodenalis DNA found in fecal samples from beef and dairy cattle in Scotland
When the G. duodenalis β-giardin (bg) sequences from this study were compared with previously published data (BLAST), it was seen that the assemblage E sequences could be separated into five distinct sequence types (E-S1–E-S5) (see Table 4). Sequence type E-S1 demonstrated 100% sequence identity to KT922250. Sequence type E-S2 demonstrated 100% sequence identity to KT369772. Sequence type E-S3 demonstrated 100% sequence identity to KT922249 and sequence type E-S4 demonstrated 100% sequence identity to KT922247. All of the sequence types E-S1–E-S5 were found on multiple farms across the country. All of the assemblage E reference parasites KT922250, KT369772, KT922249 and KT922247 had all been previously identified in ruminants.
Table 4. Comparison of assemblages of G iardia duodenalis found in fecal samples from beef and dairy cattle in Scotland
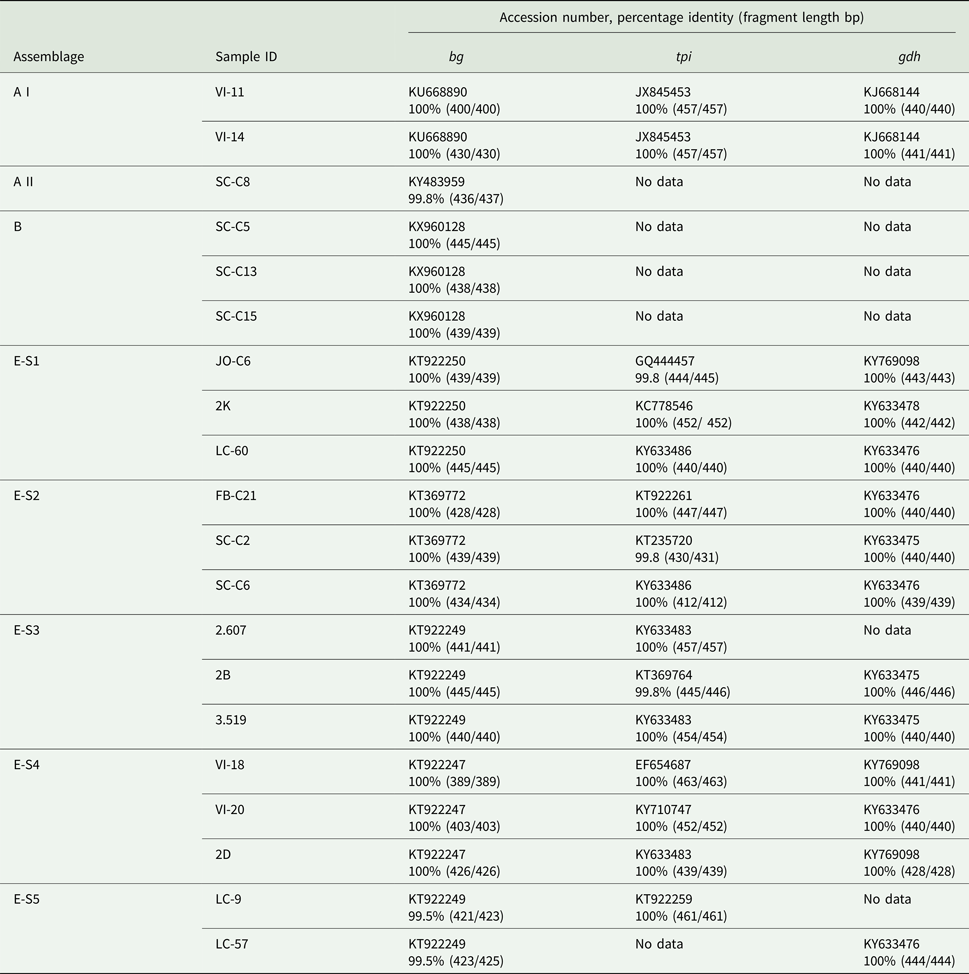
The assemblage B sequences (n = 12) were all identical to each other in relation to the G. duodenalis bg gene and were also found in multiple locations across Scotland. Following BLAST analysis, the assemblage B DNA was found to have 100% sequence identity to a G. duodenalis parasite previously isolated from human feces in Spain (KX960128) (de Lucio et al., Reference de Lucio, Bailo, Aguilera, Cardona, Fernandez-Crespo and Carmena2017). The assemblage A sequences were divided into two separate sub-assemblages: an A I (n = 2) which demonstrated 100% sequence identity to KU668890 (Wild boar, China), and a sub-assemblage A II (n = 1) which demonstrated 100% identity to KY483959 (Water, Spain).
Triose-phosphate isomerase (tpi) and glutamate dehydrogenase (gdh) genes
The primers used to amplify the tpi and gdh genes were less sensitive than those used for the bg gene amplification. Following repeated attempts, amplification of the tpi and gdh genes was not achieved for all G. duodenalis bg-positive samples. The 12 samples identified as sequence type E-S1–E-S5 using the bg gene (Table 4) were shown to have 99.8–100% identity to 10 previously identified sequences for the tpi gene. While for the gdh gene sequence types E-S1–E-S5 showed 100% identity to four previously identified sequences. The two isolates identified as assemblage A I both demonstrated 100% sequence identity to JX845453 (Alpaca, Peru) for tpi and KJ668144 (pig, China) for gdh. The samples that tested bg-positive for assemblages A II and B failed to amplify with the tpi and gdh primers, even following repeated attempts.
Phylogenetic analysis of the G. duodenalis DNA isolated from cattle fecal samples
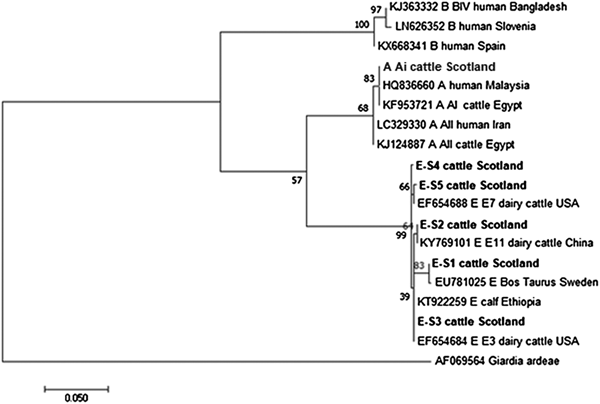
The phylogenetic analyses of the G. duodenalis β-giardin sequences identified during this current study are illustrated in Fig. 1. The assemblage E sequences (sequence type E-S1–E-S5) are all clearly distinct from each other, but all group together in a clade with other assemblage E sequences previously identified in cattle from around the world. Likewise the assemblage A (I and II) and assemblage B sequences cluster in comparable clades of samples found in cattle and humans. It is also interesting to note that the assemblage A and E sequences appear more closely related (less genetic distance) to each other than to the assemblage B parasites.
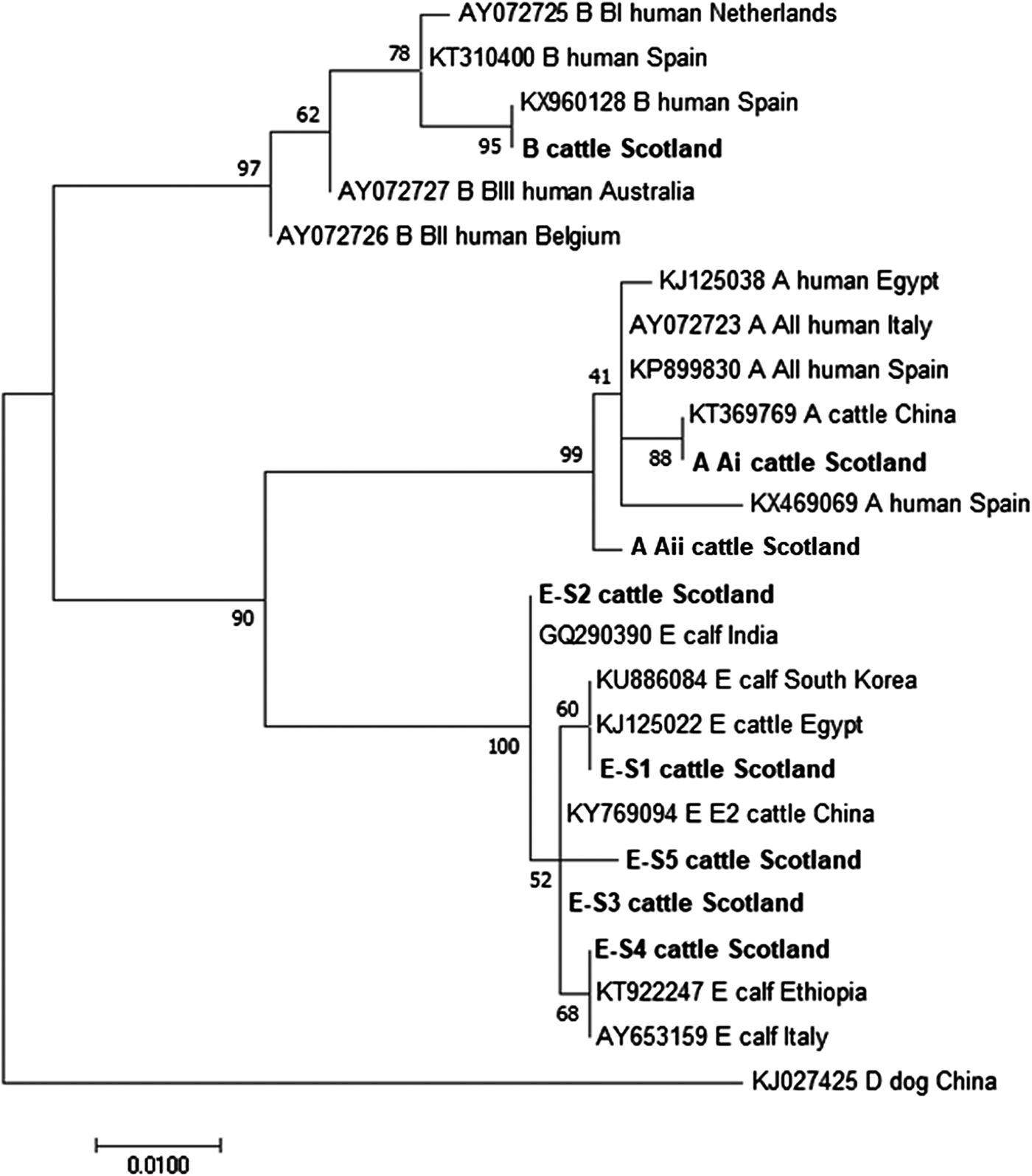
Fig. 1. Phylogenetic analysis of G iardia duodenalis β-giardin gene found in fecal samples from beef and dairy cattle in Scotland. Note: Phylogenetic analysis of G. duodenalis β-giardin gene found in fecal samples from beef and dairy cattle in Scotland using the Maximum Likelihood method based on the Kimura two-parameter model including bootstrap values (1000 replicates), shown next to the nodes and branch lengths scaled to the same units as those of the evolutionary distances. The tree was rooted against G. duodenalis assemblage D.
Figures 2 and 3 illustrate the phylogenetic analyses of the G. duodenalis tpi and gdh gene data, respectively. A similar pattern can be seen emerging for both the tpi and gdh genes, as was seen for bg, with individual isolates within an assemblage clustering together but still being clearly distinct from each other.
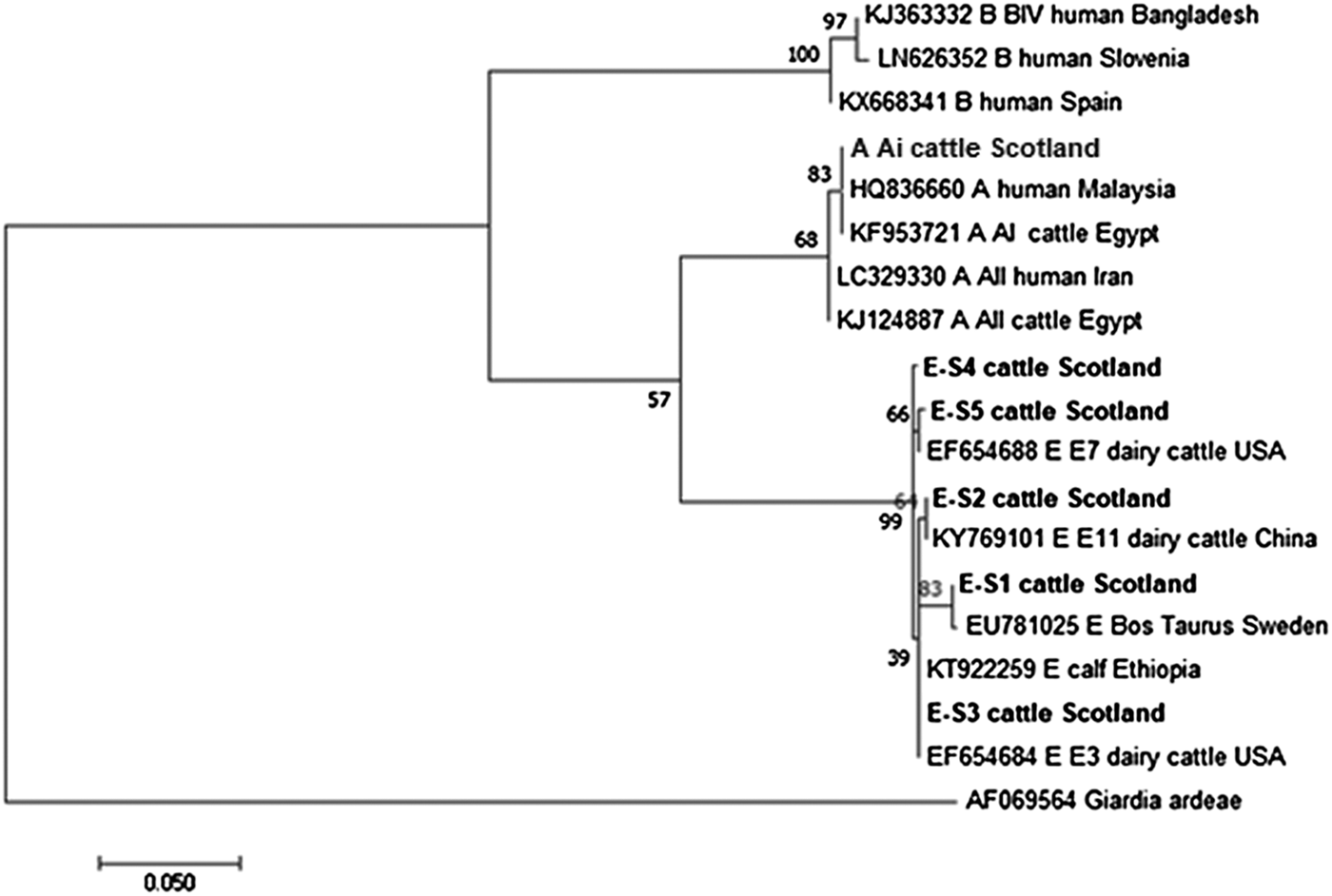
Fig. 2. Phylogenetic analysis of the G iardia. duodenalis triose-phosphate isomerase (tpi) gene found in fecal samples from beef and dairy cattle in Scotland. Note: Phylogenetic analysis of the G. duodenalis triose-phosphate isomerase (tpi) genes found in fecal samples from beef and dairy cattle in Scotland using the Maximum Likelihood method based on the Kimura two-parameter model including bootstrap values (1000 replicates), shown next to the nodes and branch lengths scaled to the same units as those of the evolutionary distances. The trees were rooted against Giardia ardeae.
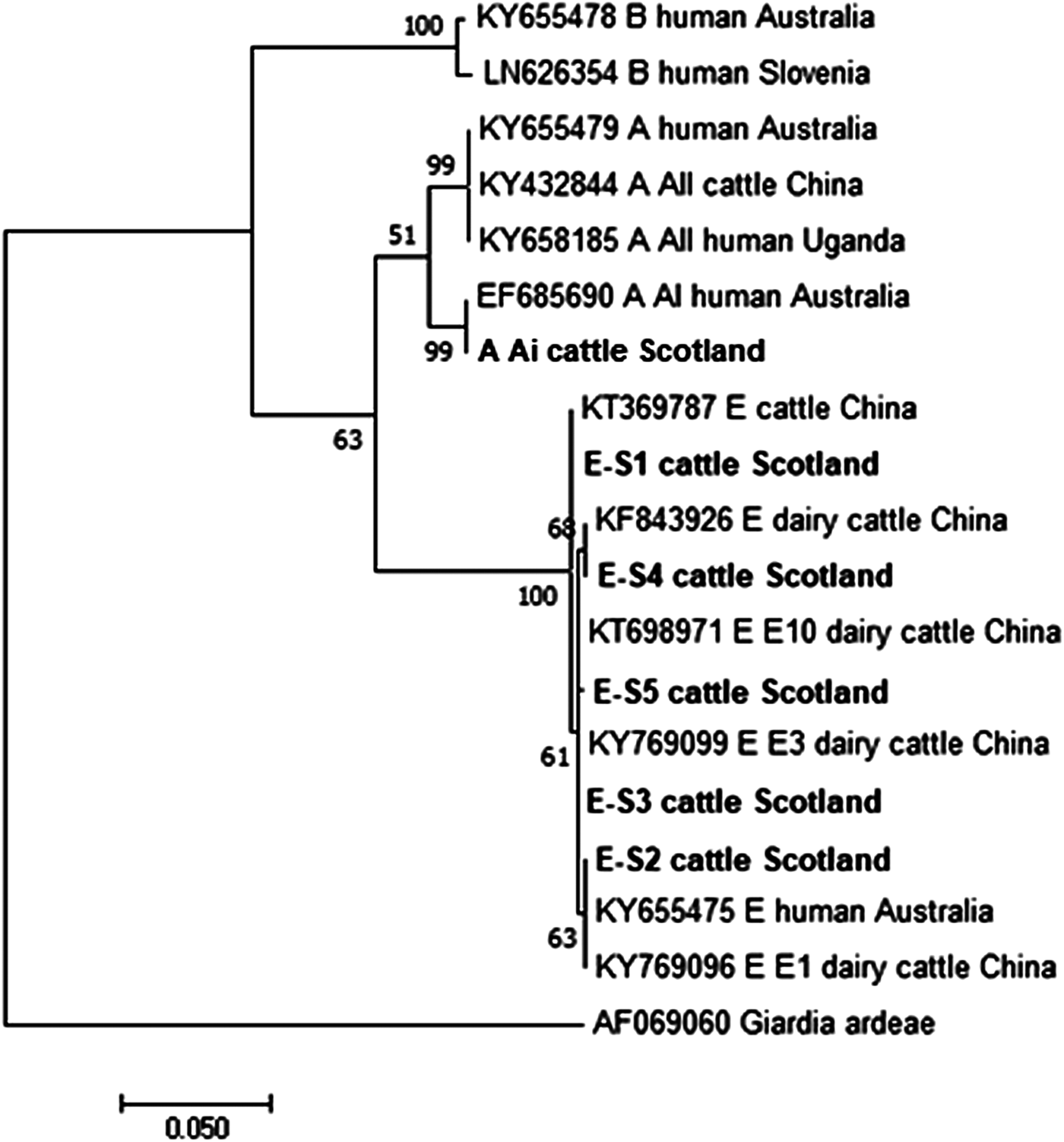
Fig. 3. Phylogenetic analysis of the G iardia duodenalis glutamate dehydrogenase (gdh) gene found in fecal samples from beef and dairy cattle in Scotland. Note: Phylogenetic analysis of the G. duodenalis glutamate dehydrogenase (gdh) genes found in fecal samples from beef and dairy cattle in Scotland using the Maximum Likelihood method based on the Kimura two-parameter model including bootstrap values (1000 replicates), shown next to the nodes and branch lengths scaled to the same units as those of the evolutionary distances. The trees were rooted against Giardia ardeae.
Discussion
The data presented in this paper demonstrate that fecal samples from both beef and dairy cattle in Scotland have the potential to be involved in the zoonotic spread of giardiasis, as evidenced by the presence of potentially human infectious G. duodenalis assemblage A (sub-assemblages A I and A II) and assemblage B. This study also demonstrates that cattle of all ages, from as early as 1 week of age through to full-grown adult cattle are contributing to the spread of Giardia cysts into the environment and that G. duodenalis was the only species of Giardia identified using multi locus PCR screening. Only assemblages A, B and E were identified and there was no evidence of assemblages C, D or F, which have been found previously on rare occasions in one study testing cattle in England and Wales (Minetti et al., Reference Minetti, Taweenan, Hogg, Featherstone, Randle, Latham and Wastling2014).
The pre-patent period of G. duodenalis in cattle is 3–10 days (Mark-Carew et al., Reference Mark-Carew, Wade, Chang, Schaaf and Mohammed2012), so some animals must become infected almost immediately after birth, as animals aged 1 week were found to be positive for G. duodenalis. We know that even the very young animals are infected with G. duodenalis as samples were collected per rectum (direct from each animal), so that we avoided sampling error or environmental contamination.
Prevalence rates of G. duodenalis infection differ greatly between the beef and dairy cattle, with dairy cattle showing a significantly greater prevalence than that seen in the beef cattle. However, we must bear-in-mind that many factors such as management systems, stocking density, water supplies and the overall hygiene conditions of the farm will greatly influence the likelihood of infection. Though the overall G. duodenalis prevalence differs significantly between the beef and dairy cattle, the proportions of animals infected with assemblages E and B are comparable. In beef cattle, we had 80% assemblage E and 20% assemblage B, while in the dairy cattle 76.8 and 21.4% of samples were positive for assemblages E and B, respectively. The results from this current study indicate that both beef and dairy cattle are involved in the spread of potentially zoonotic assemblages of G. duodenalis.
The prevalence of G. duodenalis observed in this study (32.5%) was almost identical to that seen in a previous study conducted in cattle in the North of England and Wales (32.86%) (Minetti et al., Reference Minetti, Taweenan, Hogg, Featherstone, Randle, Latham and Wastling2014). However, the prevalence in the UK appears much higher than that seen in other countries, for example, the prevalence in Henan Province, China was 7.2% (Wang et al., Reference Wang, Zhao, Chen, Jian, Zhang, Feng, Wang, Zhu, Dong, Hua, Wang and Zhang2014), while in Ethiopia it was 9.6% (Wegayehu et al., Reference Wegayehu, Karim, Erko, Zhang and Tilahun2016), but as previously mentioned, differences in farming practices, general hygiene and water supply can make comparisons between countries difficult. The differences in the prevalence of Giardia observed between the beef and dairy cattle might be caused by the housing systems used. Dairy cattle in Britain are more commonly either continuously housed or housed for prolonged periods of time, which may allow the rapid spread of pathogens such as Giardia cysts over a comparatively short period of time, compared with beef cattle which are more often pasture grazed and only housed during the period of inclement weather and around calving.
The proportion of assemblage E found in this study (80%) is comparable to another study in the USA, where 77.9% of Giardia-positive dairy cattle were shedding assemblage E (Mark-Carew et al., Reference Mark-Carew, Wade, Chang, Schaaf and Mohammed2012). It appears that assemblage B parasites are not often found in cattle compared with assemblage A. In a recent review, it is suggested that cattle in countries where assemblages A and B are found should be considered a greater potential reservoir of zoonotic Giardia infection (Ryan and Caccio, Reference Ryan and Caccio2013). In another study of Giardia assemblages found in cattle, they found that 75% of sequences were assemblage E, 23% were assemblage A (predominantly A II) while only 2% of samples were assemblage B (Sprong et al., Reference Sprong, Caccio and van der Giessen2009). However, the findings from our study have assemblage B parasites as the second most common parasite after assemblage E, with comparatively few incidents of assemblage A being recorded. However, as all the assemblage B sequences described in this study showed 100% sequence identity to a Giardia sequence previously isolated from humans in Spain, it would strongly suggest that cattle in Scotland have a potential role in the spread of zoonotic assemblages of Giardia.
In our study, we found the highest prevalence of Giardia (52%) in calves of 5 weeks of age; similar observations were made by Santin et al. (Reference Santin, Trout and Fayer2009) where 5-week-old dairy cattle had the highest prevalence (83.3%). Interestingly, in previous studies, pre-weaned calves have either not been found to be shedding assemblage A (Santin et al., Reference Santin, Trout and Fayer2009) or have not been found to be important in the spread of zoonotic Giardia cysts (Ng et al., Reference Ng, Yang, McCarthy, Gordon, Hijjawi and Ryan2011). In our study, all the animals (3/3) that shed assemblage A parasites and a majority of the animals (8/12) shedding assemblage B were pre-weaned; they were amongst the youngest animals at only 1–2 weeks of age. Our data support the observation made by Mark-Carew et al. (Reference Mark-Carew, Wade, Chang, Schaaf and Mohammed2012) that younger cattle are an important reservoir of potentially zoonotic assemblages (both A and B from our study) of G. duodenalis.
One thing that became evident when analysing the data from this current study was that there is limited availability of gene sequences for tpi and gdh, compared with the bg gene. Both tpi and gdh are routinely used (Minetti et al., Reference Minetti, Lamden, Durband, Cheesbrough, Fox and Wastling2015), but there are considerably fewer sample sequences available in GenBank for comparison. This paucity of data could be due to the primers being less sensitive than the β-giardin primers. Previous studies have also shown lower prevalence of Giardia amplification using the gdh (Wang et al., Reference Wang, Zhao, Chen, Jian, Zhang, Feng, Wang, Zhu, Dong, Hua, Wang and Zhang2014) and tpi (Geurden et al., Reference Geurden, Vanderstichel, Pohle, Ehsan, von Samson-Himmelstjerna, Morgan, Camuset, Capelli, Vercruysse and Claerebout2012) genes compared with bg. The β-giardin gene appears to be a good indicator for the presence of the parasite DNA; however, for a more detailed analysis of genetic diversity within the Giardia population, we need to consider analysing more than a single locus. The tpi and gdh primers show that there is a considerable genetic diversity in the Giardia samples found in Scotland, which would have been overlooked had only a single locus (i.e. β-giardin) been investigated; however, there is also a need to develop novel markers to help identify and distinguish assemblages of Giardia. When all of the sequence data from each isolate are combined, there are clearly considerable genetic differences between the assemblage E parasites found in Scottish cattle.
The results from this study demonstrate that G. duodenalis is prevalent across Scotland in both beef and dairy herds and that cattle (in particular pre-weaned calves) are potentially playing a role in the dissemination of human infectious assemblages (A and B) of Giardia.
Acknowledgements
The authors would like to thank Scottish Government's Rural and Environment Science and Analytical Services Division (RESAS) for supporting this work.
Financial support
Scottish Government's Rural and Environment Science and Analytical Services Division (RESAS).
Conflict of interest
None.
Ethical standards
Not applicable.