Article contents
Rapid and label-free detection of COVID-19 using coherent anti-Stokes Raman scattering microscopy
Published online by Cambridge University Press: 13 November 2020
Abstract
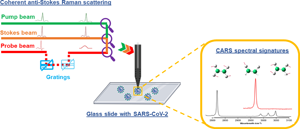
From the 1918 influenza pandemic (H1N1) until the recent 2019 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, no efficient diagnostic tools have been developed for sensitive identification of viral pathogens. Rigorous, early, and accurate detection of viral pathogens is not only linked to preventing transmission but also to timely treatment and monitoring of drug resistance. Reverse transcription-polymerase chain reaction (RT-PCR), the gold standard method for microbiology and virology testing, suffers from both false-negative and false-positive results arising from the detection limit, contamination of samples/templates, exponential DNA amplification, and variation of viral ribonucleic acid sequences within a single individual during the course of the infection. Rapid, sensitive, and label-free detection of SARS-CoV-2 can provide a first line of defense against the current pandemic. A promising technique is non-linear coherent anti-Stokes Raman scattering (CARS) microscopy, which has the ability to capture rich spatiotemporal structural and functional information at a high acquisition speed in a label-free manner from a biological system. Raman scattering is a process in which the distinctive spectral signatures associated with light-sample interaction provide information on the chemical composition of the sample. In this prospective, we briefly discuss the development and future prospects of CARS for real-time multiplexed label-free detection of SARS-CoV-2 pathogens.
- Type
- Prospective Articles
- Information
- Copyright
- Copyright © The Author(s), 2020, published on behalf of Materials Research Society by Cambridge University Press
References
- 14
- Cited by



