INTRODUCTION
Data relating to the antibiotic susceptibilities of Staphylococcus aureus isolates obtained from non-selected patients are lacking. Antibiotic susceptibilities of S. aureus are often reported from selected groups, such as cohorts of patients colonized with methicillin-resistant S. aureus (MRSA) or patients with healthcare-associated infections. National and international surveillance schemes for these infections generally include S. aureus; however, they usually concentrate on the susceptibility of isolates identified from blood cultures or sterile sites [1, Reference Hope2]. Currently, strains of S. aureus are commonly resistant to penicillin due to the production of a penicillinase and resistance to macrolides and quinolones is also widespread [Reference Moreillon, Que., Glauser, Mandell, Bennett and Dolin3]. The threat of resistance to glycopeptides, in particular to vancomycin, is a notable concern; however, high-level resistance to vancomycin (MIC ⩾16 mg/l) has so far been reported rarely [Reference Moreillon, Que., Glauser, Mandell, Bennett and Dolin3].
Bacteraemia caused by S. aureus is subject to mandatory surveillance in the UK as part of a national programme to monitor and control healthcare-associated infections [4]. System-wide interventions to reduce the number of S. aureus bacteraemia cases such as improved line and catheter care, the use of patient isolation and MRSA decolonization have led to substantial declines in the incidence of MRSA cases, although cases of both MRSA and methicillin-susceptible (MSSA) bacteraemia still persist. In the UK, mupirocin and chlorhexidine are the principal antimicrobial agents used for decolonization of patients with S. aureus, in particular MRSA, but also to reduce such infections in specific patient populations, such as low birth-weight neonates and those requiring implant surgery [Reference Popoola5–7].
Development of mupirocin resistance is an important consideration given its use for decolonization of patients with MRSA and any reduction in the effectiveness of chlorhexidine due to the development of bacterial resistance is of clear public health importance. A recent survey (2015, unpublished local surveillance data) has revealed inconsistency in testing for mupirocin susceptibility across the Yorkshire & Humber (YH) region of the UK, with half of the laboratories testing mupirocin susceptibility of MRSA isolates only, and no mechanism in place to monitor development of resistance in MSSA strains. Indeed, the prevalence and nature of mupirocin resistance and reduced susceptibility to chlorhexidine in staphylococci has not been systematically monitored in England.
Panton-Valentine leukocidin (PVL) is an extracellular, pore-forming cytotoxin [Reference Dinges, Orwin and Schlievert8] that has been strongly associated with S. aureus strains causing skin and soft tissue infections and severe, life-threatening infections, such as necrotizing pneumonia or fasciitis [Reference Lina9]. While any increased incidence of different manifestations of staphylococcal disease may trigger a public health response, staphylococcal bacteraemia and infections known to be associated with PVL-positive strains are subject to enhanced monitoring and initiate local interventions to prevent progression and/or spread of severe disease. National guidance is available for the diagnosis and management of such infections [10], but no such guidance exists for other toxin-related manifestations of staphylococcal disease, such as toxic shock syndrome or staphylococcal enterotoxin disease. Reported rates of PVL-positive strains vary considerably according to the S. aureus population tested; from 1·6% for isolates referred to a reference laboratory for epidemiological typing [Reference Holmes11] to 31% for those tested during a national sentinel surveillance scheme [Reference Kearns12]. These numbers are possibly inaccurate estimates of the true burden of PVL, as a large proportion of suspected PVL-associated S. aureus infections are treated based purely on clinical findings without confirmatory testing [Reference Ellington13].
Although there has been extensive research into staphylococcal disease and prevention of infection, there remain gaps in key knowledge areas. Currently, an up-to-date description of the antibiotic susceptibilities of clinical S. aureus from non-selected populations is lacking and there is uncertainty about the susceptibility of S. aureus to the two principal antimicrobials used in standard MRSA decolonization regimens. In addition, the true prevalence of S. aureus strains producing PVL is unknown. This study aimed to determine the antibiotic susceptibilities and characteristics of S. aureus isolates circulating within the YH region (population 5·3 million) in particular the prevalence of mupirocin resistance, chlorhexidine non-susceptibility, and PVL carriage, in order to directly inform public health control strategies and contribute to national knowledge on this topic.
METHODS
Bacterial isolates
Clinical isolates of S. aureus were collected over randomly allocated, consecutive 2-day periods throughout July 2015 from the 14 NHS clinical microbiology services laboratories in YH. S. aureus was detected and identified from clinical samples according to the UK Standards for Microbiological Investigations (SMI) document [14] by participating laboratories. The dates of collection did not fall on Mondays, Fridays or weekends in order to minimize intra-site variations in practise. MRSA screening samples and duplicate isolates from the same patient were excluded from collection. Pure colonies of S. aureus were transferred onto a nutrient agar slope and transported to a central laboratory for testing. On receipt, the slopes were given a unique study number and stored at 4 °C until ready to test. Isolates were tested in batches according to procedure. Antibiotic susceptibility testing and DNA extraction for molecular testing were completed first from subcultures of the original isolates from slopes; however, isolates were frozen in glycerol broth for longer term storage and recovered from the freezer for phenotypic chlorhexidine susceptibility.
Data collection
The following data were collected: source laboratory; laboratory specimen number; date of specimen collection; date of S. aureus detection; specimen type, whether or not the isolate was from a pure or mixed bacterial culture; the patient's hospital number and National Health Service (NHS) number (or, if missing, patient name); gender; age at specimen collection (or date of birth if age was unavailable) and source of specimen (community or hospital). No clinical data were collected as part of the surveillance, instead a proxy method based on the reported specimen type at time of collection was used to categorize isolates as associated with invasive or non-invasive infection, or colonization.
Isolates were categorized as being associated with an invasive or non-invasive infection based on the reported specimen type at time of collection. Infection was defined as S. aureus isolated from an invasive sample type, regardless of whether the culture was pure, or S. aureus isolated in pure culture from a non-invasive sample type. Colonization was defined as S. aureus isolated from a urine sample collected via a catheter or isolated from a non-invasive sample type in the presence of other bacteria. Specimen source was defined as either hospital (all hospital inpatients) or community (GP surgeries, Accident & Emergency departments or hospital outpatients).
Antibiotic susceptibility testing
Susceptibility to a range of antibiotics was tested (benzyl penicillin, chloramphenicol, ciprofloxacin, clindamycin, erythromycin, fusidic acid, nitrofurantoin, gentamicin, linezolid, mupirocin, oxacillin, rifampicin, tetracycline, teicoplanin, trimethoprim, vancomycin) using AST P634 cards and the VITEK2® analyser (bioMérieux, France). A cefoxitin screen for presumptive identification of MRSA and a screen for inducible clindamycin resistance were also included on the AST P634 cards. EUCAST (European Committee on Antimicrobial Susceptibility Testing) breakpoints were used by VITEK2 for the interpretation of susceptibility results. Isolates identified as resistant to mupirocin by VITEK2 were tested further with a 200 μg mupirocin disc using the EUCAST disc susceptibility method [15].
Chlorhexidine susceptibility testing
In the absence of an agreed standardized method, chlorhexidine phenotypic susceptibility [minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC)] was tested according to methods described by Wand et al. [Reference Wand16]. Briefly, MICs were determined using an overnight tryptone soya broth culture in a broth microdilution method with a concentration range of 0–128 mg/l chlorhexidine in microtitre plates (Sigma Aldrich, USA). All broth microdilution cultures were incubated at 37 °C for ~20 h; the MIC was determined by visual inspection against a control and for the MBC, 25 μl of non-turbid wells were spotted on tryptic soy agar plates and incubated at 37 °C for 24 h. Non-susceptibility to chlorhexidine was defined as MIC ⩾4 mg/l [Reference Horner, Mawer and Wilcox17].
Molecular methods
DNA was extracted from isolates using the QIAxtractor™ automated column extraction platform (Qiagen, UK) according to the manufacturer's instructions. The presence of genes encoding methicillin resistance genes (mecA and mecC), PVL (lukS-PV), and the species-specific nuc gene were detected simultaneously using a real-time quadruplex PCR assay [Reference Pichon18]. The following strains were used as positive controls: ATCC 25923 (lukS-PV-positive), NCTC 10442 (mecA-positive), and a S. aureus strain known to be positive for the mecC gene (provided by Professor A. Kearns, Staphylococcus Reference Unit, London). The presence of the efflux-mediated chlorhexidine resistance gene (qacA) was determined by the method of Otter et al. [Reference Otter19] with ST22C64·2 as a positive control. Isolates were genotyped using spa-typing [Reference Larsen, Stegger and Sorum20] and spa-types were assigned with Ridom StaphType software [Reference Ridom21]. S. aureus ATCC 25923 was used as a spa-typing control (t021). MLST-CC were assigned by reference to spa server (http://spa.ridom.de/mlst.shtml) and MLST (http://saureus.mlst.net) databases and assignment was completed by Professor A. Kearns, Staphylococcus Reference Unit, Public Health England, London.
Statistical analysis and power calculations
The optimal sample size required was calculated for a 0·8% prevalence of high-level mupirocin resistance (2008, unpublished local surveillance data) with a 95% confidence interval (CI) of ±0·7 precision as 622 isolates. A 2-day sample collection period was expected to yield ~700 S. aureus isolates in YH which would allow for precise estimates of the prevalence of PVL toxin-positivity and mupirocin and chlorhexidine resistance. The prevalence of the foregoing characteristics in addition to the qacA gene were calculated as proportions with associated 95% Wilson binomial CIs. Prevalence estimates for each primary outcome were presented separately for the following categories: invasive/non-invasive infection, methicillin sensitivity, community/hospital case, and MLST CC. Differences between subgroups were estimated by calculating prevalence ratios (PRs) with corresponding 95% CIs and tested using Pearson's χ 2. PRs for categorical variables (MLST CC) were calculated by pairwise comparisons of each subgroup against the other subgroups combined. All statistical analyses were performed in Stata v. 13.1 (StataCorp, USA).
RESULTS
Bacterial isolates
During July 2015, 547 S. aureus isolates were collected from the participating laboratories; 27 isolates were excluded from analysis owing to lack of accompanying datasheet (n = 3), S. aureus not recovered from original slope due to contamination by Gram-negative organism (n = 5); Gram-positive organism other than S. aureus submitted (n = 6); and duplicate isolates received from the same patient (n = 13). Thus, 520 clinical isolates were suitable for analysis.
The number of isolates collected by each laboratory during their allocated 2-day period ranged from 9 to 95, depending on the size and nature of the laboratory. Isolates from hospital inpatients accounted for 23·5% and community isolates comprised 74·2% (57·3%, 12·5% and 4·4% from GP surgeries, hospital outpatients and A&E locations, respectively). The location of 2·3% isolates was not recorded. Isolates from invasive infections accounted for only 14 (2·7%) of 520 isolates in the study sample, compared with 320 (61·5%) from non-invasive infections and 186 (35·8%) isolates associated with colonization. Accordingly, the most common specimen type was a wound or skin swab (413, 79·4%). Further characteristics of the study population and S. aureus isolates are presented in Table 1.
Table 1. Characteristics of the study population and S. aureus isolates
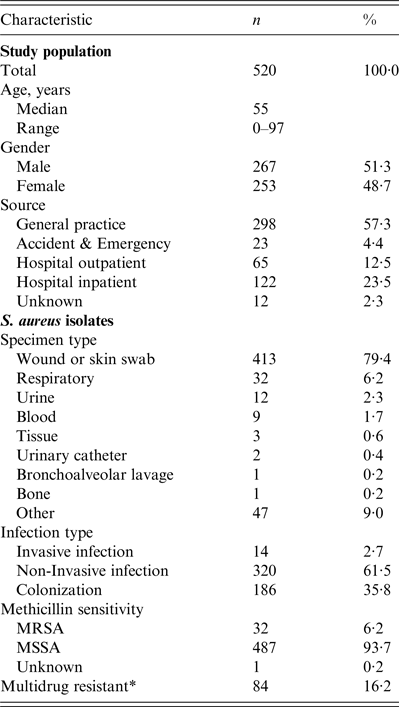
* Multidrug-resistant = non-susceptible to ⩾3 classes of antibiotic agent.
Antibiotic susceptibility
The majority of S. aureus isolates were non-susceptible to benzyl penicillin (75·8%); 18·7%, 18·3% and 14·2% isolates were non-susceptible to fusidic acid, erythromycin and clindamycin, respectively. Other uncommon non-susceptibilities were ciprofloxacin (8·5%), trimethoprim (5·8%) and tetracycline (5·8%), and <1% of isolates were non-susceptible to gentamicin (0·8%), teicoplanin (0·2%), and rifampicin (0·2%). All isolates were susceptible to vancomycin, linezolid, daptomycin, and chloramphenicol. Only four mupirocin-resistant isolates were identified (0·8%, 95% CI 0·3–2·0) (Fig. 1), these were multidrug-resistant (MDR) MRSA isolates with high-level (MIC >256 mg/l) resistance to mupirocin.
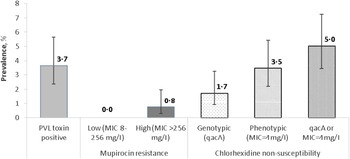
Fig. 1. Prevalence of Panton-Valentine leukocidin (PVL) toxin, mupirocin and chlorhexidine resistance in clinical S. aureus isolates (n = 520). Error bars represent 95% Wilson binomial confidence intervals around the prevalence estimate. MIC, Minimum inhibitory concentration.
Eighty-four (16·2%) isolates were resistant to ⩾3 classes of antibiotic and classified as MDR. MRSA isolates accounted for 25% (n = 21) of MDR isolates and were commonly resistant to ciprofloxacin, erythromycin (n = 20), plus fusidic acid (n = 11). Of the 63 MSSA isolates that were also MDR, four were susceptible to benzyl penicillin but non-susceptible to erythromycin, tetracycline, trimethoprim (n = 1); clindamycin, erythromycin and tetracycline (n = 1), or clindamycin, erythromycin and fusidic acid (n = 2). The remaining 59 isolates were non-susceptible to benzyl penicillin, and another two antibiotic classes (n = 40) [most commonly plus erythromycin and clindamycin (n = 22)], three classes of antibiotics (n = 18), and in one MSSA isolate non-susceptibility to five classes of antibiotic agents was recorded. The teicoplanin-resistant isolate (MIC 4 mg/l) was confirmed by the national reference laboratory and resistance was not due to a single identifiable or transferrable genetic element.
Thirty-two isolates (6·2%) were positive for the mecA gene and classified as MRSA; no isolate harboured the mecC gene. One isolate was positive for the mecA gene but susceptible to oxacillin and was excluded from further analysis (labelled as unknown in Table 1). The majority of MRSA isolates were non-susceptible to ciprofloxacin (81·3%), erythromycin (59·4%), fusidic acid, (37·5%) and clindamycin (34·4%). Hospital inpatient isolates were more than twice as likely to be methicillin-resistant than community isolates (10·7% vs. 4·9%, PR 2·2, 95% CI 1·1–4·2, P = 0·032); however, there was no significant association between MRSA and invasive or non-invasive sample types.
Chlorhexidine susceptibility
Of the 519 isolates tested for phenotypic susceptibility to chlorhexidine (one isolate was not recovered from frozen storage), 18 (3·5%, 95% CI 2·2–5·4) had a MIC equal to 4 mg/l; no isolates exhibited a MIC >4 mg/l. Nine isolates were positive for the qacA gene (1·7%, 95% CI 0·9–3·3), one of which was also positive for PVL. None of the qacA-positive isolates carried the mecA gene. There was very little correlation between carriage of the qacA gene and phenotypic non-susceptibility to chlorhexidine as only one isolate which possessed the qacA gene also exhibited a chlorhexidine MIC of 4 mg/l whereas 27 isolates with or without qacA had a similar MIC (5%, 95% CI 3·4–7·2) (Fig. 1).
There were 181 isolates that gave a chlorhexidine MBC of >4 mg/l; however, reading the MBC results was not straightforward. For instance, 79 isolates gave variable MBC values at different dilutions (i.e. no growth at one dilution, growth at the next, no growth at the next, and growth at the final dilution). Such variable growth was attributed to the presence of variant or ‘hetero-resistant’ populations. Figure 2 shows the frequency distribution of MIC and MBC to chlorhexidine in all clinical S. aureus isolates. There were no significant differences in the prevalence of isolates with MIC 4 mg/l chlorhexidine or qacA-positive isolates between MRSA and MSSA, community and hospital settings, or non-invasive and invasive sample types (Table 2).
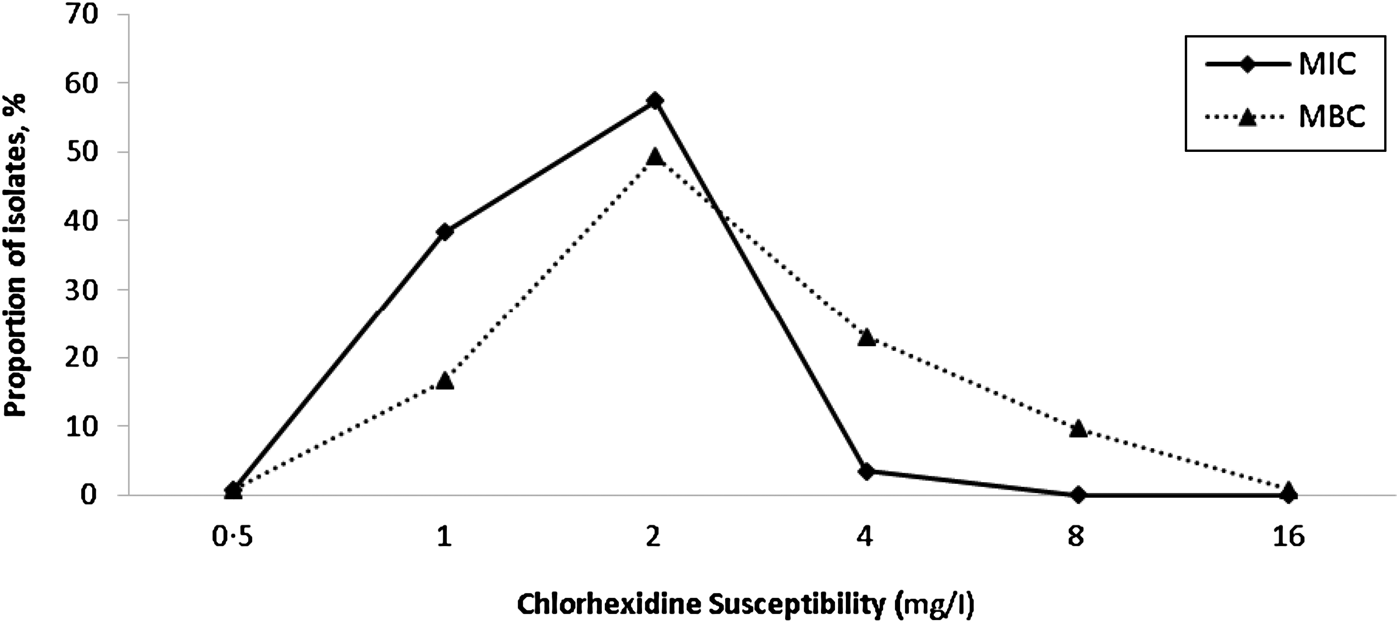
Fig. 2. Distribution of the minimum inhibitory concentrations (MIC) and minimum bactericidal concentrations (MBC) of chlorhexidine for clinical S. aureus isolates. Non-susceptibility to chlorhexidine was defined as an MIC ⩾4 mg/l [Reference Horner, Mawer and Wilcox17].
Table 2. Differences in the prevalence of mupirocin resistance, chlorhexidine non-susceptibility, and Panton–Valentine leukocidin (PVL) in clinical isolates of MSSA and MRSA*, from community and hospital settings, and from non-invasive and invasive infections*
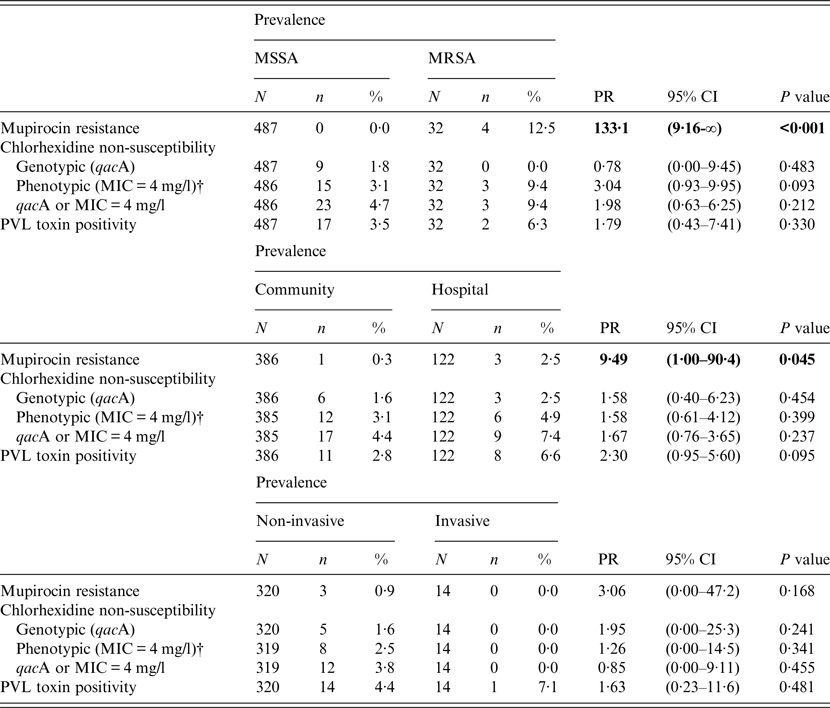
MIC, Minimum inhibitory concentration; PR, prevalence ratio; CI, confidence interval.
P value = Fisher's exact test p value; statistically significant prevalence ratios are presented in bold type.
* Prevalence ratios and confidence intervals have been estimated for categories with zero cell counts using a Woolf–Haldane correction.
† One isolate was not recovered from frozen storage.
Prevalence of PVL-positive S. aureus
Nineteen (3·7%, 95% CI 2·4–5·6) isolates were identified as PVL-positive (Fig. 1). There was no significant difference in prevalence of PVL-positivity between MRSA and MSSA, community and hospital settings, or non-invasive and invasive sample types (Table 2).
Genetic characterization
Two hundred and thirty-four different spa-types were identified from 514 isolates, mapping to 22 different MLST CCs. Six isolates were spa non-typable, and 22 spa-types were unable to be mapped to MLST CC without further investigation. Sixteen new spa-types were identified and submitted to RIDOM.
As expected, MSSA isolates belonged to a range of CC (n = 16 lineages), compared with MRSA isolates. The majority of isolates (n = 421, 81%) were grouped in one of nine CCs; CC30 (13·5%), CC15 (12·5%), and CC1 (11%) being the most predominant (Fig. 3). MRSA isolates fell in six CCs with CC59 and CC22 (EMRSA-15) being the most prevalent (n = 13 isolates per CC). All four mupirocin-resistant MRSA isolates belonged to CC59. PVL-positive isolates were distributed in 11 CCs with the highest prevalence in CC8 (10·4% of isolates; PR 3·45, 95% CI 1·29–9·27) and CC121 (22·2% of isolates; PR 6·53, 95% CI 1·75–24·3) compared to isolates in all other identified CCs. Isolates that carried the qacA gene were distributed among four lineages (CC30, CC5, CC8 and CC88), and the prevalence of qacA-positive isolates was significantly higher in CC88 [33·3% of isolates (n = 2); PR 26·3, 95% CI 6·59–105].

Fig. 3. Most commonly occurring multilocus sequence type clonal complexes (CCs) of clinical S. aureus isolates (n = 520). Other comprise: CC7 (n = 12), CC25 (n = 12), CC121 (n = 9), CC97 (n = 8), CC80 (n = 6), CC88 (n = 6), CC12 (n = 5), CC9 (n = 3), CC152 (n = 2), CC182 (n = 2), CC101 (n = 1), CC1290 (n = 1), CC20 (n = 1); unknown comprise: no CC match (n = 22), no spa-type data (n = 6).
DISCUSSION
These data from a prospective cross-sectional prevalence survey provide a reassuring picture of S. aureus within a single healthcare region of the UK at a time when antibiotic resistance is a threat to the treatment of infectious disease.
The prevalence of non-susceptibility in S. aureus isolates to various antibiotic agents identified in the present study bears comparison with rates reported by the British Society for Antimicrobial Chemotherapy (BSAC), which summarize the non-susceptibility trends in staphylococci from cases of bacteraemia in the UK and Ireland (2001–2006) [Reference Hope2], and with contemporaneous but non-published data of the antibiotic susceptibilities of S. aureus (n = 2640) from clinical samples of a non-selected community population in two large UK cities (Leeds and Bradford) during 2010–2012. Of note, the prevalence of non-susceptibility to ciprofloxacin in the present study was similar to local data but lower than that of BSAC; prevalence of non-susceptibility to erythromycin and fusidic acid were higher in the present study compared to previous local data or with BSAC data; whereas prevalence of non-susceptibility to trimethoprim has decreased (Table 3). Nevertheless, isolates defined as resistant to ⩾3 antibiotic classes remained susceptible to at least four classes of antibiotics tested, leaving options for treatment.
Table 3. Comparison of the prevalence of non-susceptibility of S. aureus recorded by British Society for Antimicrobial Chemotherapy (BSAC) surveillance data (2001–2006), non-published local surveillance data (2010–2012) and the present study (July 2016)
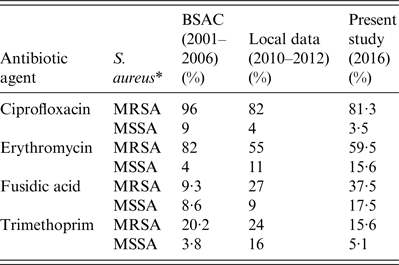
* MRSA, Methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus.
We have identified a low prevalence of MRSA (6·2%) compared to 11% noted previously from the earlier local data (2010–2012). As such, flucloxacillin remains a reasonable first-line empirical choice for treatment of non-complicated S. aureus infections in this geographical area. It is important, however, that GPs remain aware of the prevalence of MRSA circulating in the local community so that patients with previous history or risk factors for MRSA colonization/infection can be treated accordingly. It is reassuring that isolates carrying the mecC gene were not identified during this surveillance. mecC, previously known as mecAL251, was first reported in 2011, initially from dairy cattle but subsequently identified in clinical samples from humans [Reference Garcia-Alvarez22]. The prevalence of mecC-positive MRSA remains low in England and throughout most of Europe [Reference Hetem23, Reference Paterson24]; however, with an increased prevalence in Denmark [Reference Petersen25] vigilance is recommended for widespread emergence of S. aureus carrying this gene.
Despite the widespread use of mupirocin and chlorhexidine in MRSA decolonization, non-susceptibility to these agents remains low. Although the emergence of resistance to mupirocin with increasing use has been documented in centres in Brazil [Reference Vivoni26] and Canada [Reference Simor27], other studies have not demonstrated such an association. Low levels of mupirocin resistance over a 10-year period have been identified in Belgium; of 1971 isolates tested, 39 MRSA (3·1%) and four MSSA (0·6%) isolates showed high-level mupirocin resistance (>256 mg/l) [Reference Nagant28]. A 4-year repeated point-prevalence study between 1999 and 2003 in the surgical units of one of the major teaching hospitals in the YH region found no evidence for the sustained emergence of mupirocin resistance after the introduction of a mupirocin-based peri-operative prophylaxis regimen for the prevention of MRSA [Reference Fawley29]. Over a decade later, our study has shown that the prevalence of mupirocin resistance in S. aureus remains low, both in the same teaching hospital and across other locations within the region. Interestingly, the only mupirocin-resistant isolates (n = 4) identified here belonged to CC59, a lineage that was associated with mupirocin resistance and whose prevalence increased during a 4-year period of surveillance of MRSA in care homes for the elderly in the YH region [Reference Horner30]. The four mupirocin-resistant isolates were submitted by four different laboratories and were not associated with PVL carriage.
The low prevalence of PVL-positive S. aureus reported here does not appear to indicate a large, undetected pool of such strains circulating in our region. A similarly low prevalence of PVL-positive isolates (1·6%, 95% CI 0·7–3·0) was found in 515 S. aureus isolates referred to the national reference laboratory for epidemiological typing from laboratories in England and Wales in 2002 [Reference Holmes11]; the same study reported a PVL prevalence of 4·9% in isolates associated with skin and soft tissue infections. Other sources have since recorded a higher prevalence of PVL-positive S. aureus, notably a 2007 cross-sectional survey of 390 clinical S. aureus isolates at a major London hospital with 9·7% of isolates harbouring the gene [Reference Shallcross31] and with PVL-positivity being four times more frequent in isolates from skin and soft tissue infections than other isolates. A national sentinel surveillance study of community staphylococcal pyogenic skin infections completed by PHE in 2013 found that 36% (n = 279/771) cases were PVL-positive [Reference Kearns12].
Chlorhexidine is the most frequently used topical antiseptic for skin decolonization in UK healthcare and is an integral part of the control and management of MRSA-positive individuals [7]. Such widespread use may engender resistance and enable certain bacteria to persist in the clinical setting. For a number of reasons, accurate detection of chlorhexidine resistance is technically challenging [Reference Horner, Mawer and Wilcox17]. These include lack of a consensus on the definition of ‘resistance’, commonly defined as a raised MIC or MBC ⩾4 mg/l or the presence of genes known to encode efflux-mediated biocide resistance, and lack of an agreed standardized method to assay susceptibility. As a result, the ability of bacteria to withstand chlorhexidine exposure is not routinely tested and possible resistance is largely unmonitored. In addition, the relationship between the carriage of chlorhexidine resistance genes and phenotypic reduced susceptibility remains unclear [Reference Horner, Mawer and Wilcox17].
Our work has shown that there is little evidence of chlorhexidine non-susceptibility in S. aureus circulating in the YH region and there was poor correlation of carriage of qacA with chlorhexidine susceptibility as only eight isolates were positive for this gene. Despite a published method being available to detect qacB and smr chlorhexidine efflux genes [Reference Otter19], only a qacA-positive control organism could be sourced. In a survey of MRSA bloodstream infection isolates, Otter et al. tested 602 MRSA isolates and identified the qacA gene in 26·2% isolates; qacB and smr positive isolates were less frequently identified (0·3% and 8·1%, respectively) [Reference Otter19]. Although the setting is different (the present study did not just include MRSA isolates) we may infer that screening for qacB and smr genes may only have identified a small number of additional isolates carrying these genes. It is possible that investigation of the levels of qacA expression in isolates that are phenotypically chlorhexidine susceptible compared with non-susceptible strains may shed further light on this issue. Expression of other genes, such as the regulatory gene, qacR, or examination of the sequence of the qacA gene itself may help to elucidate if key mutations are necessary for biocide resistance.
We did not collect data about mupirocin/chlorhexidine use/exposure and therefore are not able to draw conclusions about the effects of mupirocin/chlorhexidine on the prevalence of non-susceptibility. As hospital admission records were not recorded, this has restricted any inference regarding co-morbidities and speciality and we cannot estimate the proportion of patients that may have been exposed to chlorhexidine.
S. aureus strains circulating within the YH region are a diverse group and while three lineages predominate (CC30, CC15, CC1), six other lineages are circulating throughout the region. Many of the commonly occurring spa-types identified in the present study are also represented in the 20 most frequent spa-types in MSSA and MRSA isolated in 26 European countries [Reference Grundmann32]. Notable exceptions include t032 and t021, which map to CC22; t316 and t216, which map to CC59, and t579, which maps to CC398. Such differences in prevalent spa-type may be explained in part by the fact that the European study only included isolates from invasive infections and almost 10 years has passed since the data were collected (September 2006–February 2007).
As genotyping is usually requested following an increase in the number of cases of S. aureus (MSSA or MRSA) in a clinical area, the results of the present surveillance provide an important baseline of S. aureus circulating in primary care and outside an outbreak situation. In recent years, EMRSA-15 (CC22) and EMRSA-16 (CC30) were the most common MRSA strains in England and Wales; however, while CC22 remains in circulation, EMRSA-16 is no longer prevalent in the YH region. Instead MRSA isolates belonging to CC59 have become more predominant.
We acknowledge that there are limitations associated with the present study. The survey sample was designed to minimize systematic bias by capturing all S. aureus isolates from all NHS laboratories in the YH region over two consecutive days. The dates of study participation were staggered to ensure that isolates were collected in a timely manner, but each hospital trust was randomized to the dates in order to minimize the potential for over-representation of particular patient groups due to scheduling of specialist clinics or laboratory activity. Prior to isolate collection, it was estimated that the 14 participating laboratories would detect approximately 350 S. aureus isolates per day for patients in both community and hospital settings, hence a 2-day collection period would provide sufficient isolates for precise estimates of mupirocin resistance and chlorhexidine non-susceptibility prevalence, and PVL toxin-positivity, according to the sample size calculations completed. The target number of samples (n = 700) was not reached, resulting in lower levels of precision than expected around the prevalence estimates reported; however, we have tested >500 isolates from the general population presenting to a GP or hospital in a large single healthcare region of England. Data have been included from a range of hospital trusts of different size and patient mix. Isolates were drawn from all NHS diagnostic laboratories providing services to the population of YH (5·3 million), covering both rural and urban areas. All observations occurred in the same season but seasonality was not considered likely to impact upon the variables of interest and the risk of seasonality was outweighed by the value of collecting data from all trusts in the same period to permit valid comparison between them.
In summary, the findings of this surveillance suggest that the widespread use of staphylococcal decolonization regimens over the past decade has not had an adverse impact on antimicrobial and biocide resistance rates and that mupirocin and chlorhexidine continue to be suitable agents for MRSA decolonization.
ACKNOWLEDGEMENTS
The authors thank staff in the participating laboratories for collecting the S. aureus isolates and completing the datasheets, and Olivia Sherburn, Public Health England, Regional Laboratory, Leeds who completed data entry. We thank Dr Jonathan Edgeworth, Guy's and St Thomas’ NHS Trust, London the provision of a control isolate for detection of the qacA gene, and Professor Angela Kearns and colleagues at the Staphylococcus Reference Unit, Public Health England, London for the provision of a control isolate for detection of the mecC gene, for help and advice with spa-typing and assignment of spa-types to relevant MLST CC and for general discussion about the surveillance project. Thanks to Dr Matthew Wand and Dr Mark Sutton, Public Health England, Porton Down, Salisbury for input and training of chlorhexidine susceptibility testing. Finally, we thank Dr Obaghe Edeghere, Consultant Epidemiologist, Field Epidemiology Service, Public Health England, and Dr Andrew Kirby, Consultant Microbiologist, Leeds Teaching Hospitals NHS Trust, for reviewing the protocol before implementation.
This work was supported by Public Health England.
DECLARATION OF INTEREST
None.








