INTRODUCTION
The increasing antimicrobial resistance in commensal and zoonotic bacteria of food-producing animals is a threat for human and animal health [Reference Wernli1]. The problem is exemplified by the emergence of methicillin-resistant Staphylococcus aureus (MRSA) [Reference Graveland2–Reference Weese, Avery and Reid-Smith4], extended spectrum β-lactamase (ESBL)-producing Escherichia coli [Reference Leverstein-van Hall5] and resistant Salmonella serovars [Reference Koluman and Dikici6] in food-producing animals and their products and the occurrence of these resistant strains in humans.
Commensal bacteria may serve as a reservoir of antimicrobial resistance genes that can be transferred to other (pathogenic) bacteria [Reference Hunter7, Reference van den Bogaard and Stobberingh8]. Knowledge about determinants associated with antimicrobial resistance can possibly contribute to target intervention strategies to reduce or limit selection and spread of antimicrobial resistance in commensal and zoonotic bacteria.
Several risk-factor studies in commensal E. coli of fattening pigs showed that antimicrobial usage, especially the use of in-feed medication, is associated with antimicrobial resistance [Reference Dewulf9–Reference Dunlop13]. Moreover, other management or farm-related factors, e.g. hygienic measures, feed changes, and breed and origin of animals may influence the occurrence of antimicrobial resistance in commensal E. coli of broilers and fattening pigs [Reference Dewulf9, Reference Persoons14].
Studies on risk factors associated with antimicrobial resistance in commensal E. coli of veal calves are limited. In a study of Di Labio et al., the administration of medicated feed upon arrival, non-participation in a quality assurance programme and calf purchase were seen to be risk factors associated with antimicrobial resistance. Conversely, antimicrobial injection upon arrival on the farm and having more than 20 calf suppliers were protective factors [Reference Di15]. However, these results may not reflect the situation at farm level because faecal samples were taken at slaughterhouses, which could lead to significantly increased resistance levels and changed resistance patterns of the commensal flora due to transportation [Reference Langlois and Dawson16]. Further, the farmer was asked afterwards for information on antimicrobial usage, which could lead to information bias.
The aim of this study was to identify determinants associated with the occurrence of antimicrobial resistance in commensal E. coli isolates of veal calves at the herd level.
METHODS
Study design
A cross-sectional study of white veal calf farms at the end of the fattening period was conducted. Farms were selected, stratified by farm size, from a database of 193 Dutch white and rosé veal calf farms that were monitored for their antimicrobial usage by the Agricultural Economics Research Institute (LEI), part of Wageningen University and Research Centre [17]. This database resulted from a disproportionally stratified random sampling strategy of the Dutch veal calf population to ensure that this subsample of farms was representative of the whole population, as described in the MARAN 2009 report [17]. The selection of farms for this cross-sectional study was performed in close consultation with LEI and corresponds to a disproportionally stratified random sampling of farms from the LEI database. The database was arranged into three strata based on farm size: <450, 450–750 and >750 veal calves present on the farm. The number of farms selected per stratum is proportionate to the total number of veal calves within each stratum. Besides farms of different sizes the study population also covered farms that belonged to large veal calf production and smaller veal calf production operations.
In total 69 farmers were requested to participate in the study by phone call and 49 agreed (participation rate 71%). The main reason for farmers not participating in the study was lack of time. The 49 participating farms were divided over the different strata as follows: farm size <450 calves (n = 5), farm size of 450–750 calves (n = 17), farm size >750 calves (n = 26). The final number of farms totalled 48 as one farm was excluded from the study due to overgrowth of swarming bacteria other than E. coli on the MacConkey No 3. agar plate.
Isolation and susceptibility testing of E. coli
In the period April 2009–May 2010 all 49 selected white veal calf farms were visited 2 weeks before slaughter, when animals were ∼27 weeks old. Per farm, 20 clinically healthy calves, which were not under antimicrobial medication, were selected by convenience from compartments housing calves close to slaughter, proportionate to the number of calves present in those compartments. Our previous study on sampling strategies revealed that a sampling strategy based on sampling 20 calves close to slaughter was most accurate for estimating antimicrobial resistance at herd level as this led to the smallest 2·5–97·5th percentile interval around the estimated mean proportion of E. coli isolates resistant to amoxicillin, tetracycline, ciprofloxacin and trimethoprim/sulfamethoxazole (TMP/SMX) [Reference Bosman18]. The present study was approved by the Animal Ethics Committee of Utrecht University with all calves being cared for according to the guidelines of Dutch Animal Welfare Law.
Faecal samples were collected manually per rectum and suspended 1:10 (w/v) in buffered peptone broth with 30% glycerol within 24 h. From the 20 individual samples one pooled sample was made by mixing 1 ml of every individual faecal suspension. All faecal suspensions were stored at −20°C until further processing.
From the pooled faecal samples 10–1, 10–2 and 10–3 dilutions were made in saline. From each dilution 50 μl was inoculated onto a MacConkey No. 3 agar plate (bioTRADING Benelux B.V., The Netherlands) and incubated overnight at 37°C. Ninety colonies with the morphological appearance of E. coli were randomly selected from one MacConkey No. 3 agar plate containing up to 300 solitary colonies. From each sample, one colony with the typical morphology of E. coli was biochemically confirmed as E. coli by testing positive for indole production and with a negative reaction on citrate conversion. The 90 selected isolates were each suspended in a separate well of a 96-well plate containing 100 μl cation-adjusted Mueller–Hinton broth (CAMHB) per well (Trek Diagnostic Systems, UK). The isolates were tested for their resistance to five different antimicrobial agents by replica plating using Mueller–Hinton (MH) agar plates, each of which contained an antimicrobial agent in breakpoint concentration, as described previously [Reference Bosman18]. These antimicrobial agents were chosen because they belong to antimicrobial classes commonly used in veal calf farming. The antimicrobial concentrations in the MH agar plates were as follows; 25 mg/l amoxicillin, 25 mg/l tetracycline, 0·5 mg/l cefotaxime, 0·125 mg/l ciprofloxacin and 8/152 mg/l TMP/SMX, as described previously [Reference Bosman18–Reference Nijsten22]. Moreover, each replica plating series started and ended with a plain MH agar plate as a negative control to ensure proper transfer and inoculation of all 90 isolates per sample on each agar plate.
Our previous study [Reference Bosman18] on quantifying resistance levels within herds showed that this test method and the antimicrobial concentrations used provided similar proportions of isolates classified as resistant compared to those determined with the reference broth microdilution test method using EUCAST epidemiological cut-off values.
Of the 48 remaining farms all 90 isolates grew on the plain MH agar plate at the end of each replica plating series. The proportion of resistant isolates per antimicrobial agent per pooled sample was determined by dividing the number of isolates grown on each antimicrobial-containing MH agar plate by the total number of isolates (n = 90) grown on the plain MH agar plate at the end of each replica plating series.
Questionnaire
On the day of sampling a questionnaire was completed by the first author while interviewing the farmer. The questionnaire (available from the corresponding author upon request) has been used before in a study on MRSA in veal calves [Reference Graveland3]. The questionnaire contained questions about general farm and herd characteristics and management factors. Factors which were hypothesized to be biologically relevant for the presence and spread of resistance determinants within the faecal flora at host level were selected and included in the analysis. These factors were farm size, number of calves/pen, feeding of roughage (straw or silage), frequency of sorting of calves, cleaning and disinfection of stables, pest control of rodents and flies, and frequency of changing work clothes by the farmer.
The questionnaire did not contain questions directly related to the use of antimicrobials on the farm as quantitative data on antimicrobial usage within the farms were provided by LEI.
Antimicrobial usage
The total amount of antimicrobials administered to veal calves during the production cycle (fattening period of ∼175 days) on the selected farms was monitored by LEI in cooperation with the veal calf sector. Detailed data were collected on the number of animals present and the amount of antibiotics used during fattening based on veterinary prescriptions, as described and presented in the MARAN 2009 report [17]. For each delivered volume of an antimicrobial drug during the production cycle on the farms the animal daily dosage (ADD) was calculated. The ADD is a measure of drug usage, which is independent of the variations in the potency of the active compound and the formulation of the pharmaceutical product. Therefore, the ADD provides a measure of the relative contribution of a certain drug to the total antimicrobial drug exposure and facilitates the comparison of antimicrobial drug use between herds [Reference Jensen23].
The ADD was calculated by dividing the treatable weight (the total number of kilograms of animal that can be treated with the active ingredient of the antimicrobial drug) by the total weight of the calves present within the herd at the time the drugs were delivered (weight of calves based on an average growth curve provided by one of the veal calf operations). For example, on a farm with 100 veal calves, 20 litres of antibiotic X (concentration 100 mg/ml) was administered when the calves had an average weight of 160 kg. The dosage of antibiotic X is 10 mg/kg body weight per day. The treatable weight is (20 000 × 100)/10 = 200 000 kg. The total average live weight of the animals on the farm was 16 000 kg, which results in 200 000/16 000 = 12·5 ADD as a quantitative measure of the usage of antibiotic X on this farm.
Delivered but still intact volumes of antimicrobial drugs which were not administered were returned to the veterinarian and corrections in the registered amounts of drugs were made by subtracting the ADD of the unopened volume. The total number of ADD administered during the production cycle (pc) is presented per antimicrobial class by ADD/pc and reflects the number of days an average veal calf on the farm was exposed to an antimicrobial drug of a certain antimicrobial class.
Statistical analysis
Logistic regression analysis for grouped data (number of resistant isolates of total tested per antimicrobial agent within sample) was performed to examine the association between the odds of an E. coli isolate testing resistant to each antimicrobial agent, the questionnaire-derived variables and the amount of ADD/pc per antimicrobial class.
Usage of the antimicrobial classes aminoglycosides, cephalosporins and trimethoprim-sulphonamide combinations was dichotomized in two categories: administration of these drugs ‘yes’ vs. ‘no’, as these drugs were administered on 27, 31 and 30 farms, respectively. Within the antimicrobial classes quinolones, macrolides, penicillins, tetracyclines and also colistin, the median ADD/pc value was used as the cut-off value in order to dichotomize the data because almost all farms (⩾40 farms) administered these drugs. For tetracyclines three levels of ADD/pc were realized to compare farms with low (<20 ADD/pc), medium (20–40 ADD/pc) and high (>40 ADD/pc) usage of tetracyclines.
One multivariable random-effects model was used to fit the data with the number of resistant isolates of the 90 isolates tested per sample as outcome of interest. Antimicrobial agent was included in the model as an explanatory variable in order to differentiate the outcome of the model for each antimicrobial agent separately. From each faecal pooled sample E. coli isolates were tested for their resistance against five different antimicrobial agents. To take into account that these observations (resistance to each antimicrobial agent) were clustered within the faecal sample a random sample effect was included in the model.
All selected questionnaire-derived factors and the antimicrobial usage data (ADD/pc per antimicrobial class) were included in the multivariable random-effects model. Moreover, a two-way interaction term between the antimicrobial agent and each risk factor was included to study the difference in odds of a resistant isolate for each antimicrobial agent for a specific level of the risk factor compared to the reference level of the risk factor. A backward stepwise selection based on the value of Akaike's Information Criterion (AIC) starting with the full model was performed to find the best-fitting model to describe the dataset and to examine the association between each risk factor and the odds of resistance to each antimicrobial agent. Because the dataset was stratified by farm size the explanatory variable farm size was kept in the model.
Cefotaxime resistance data were excluded from the analysis because the estimates of the log odds for each variable on cefotaxime resistance in the final model showed inestimable standard errors due to very low proportions of cefotaxime-resistant isolates (0–0·16) present on seven farms only.
The analyses were performed using the open-source programme R, version 2.14.0 [24] and package lme4, version 0.999 375-42 [25] for generalized linear mixed-effects models.
RESULTS
Proportion of resistance
The estimated proportions of resistant isolates per antimicrobial agent, including 95% confidence intervals (CI), were as follows: amoxicillin 0·59 (95% CI 0·52–0·66), cefotaxime 0·006 (95% CI 0·004–0·009), ciprofloxacin 0·11 (95% CI 0·08–0·14), tetracycline 0·92 (95% CI 0·89–0·94) and TMP/SMX 0·52 (95% CI 0·45–0·59).
The most common resistance pattern in the isolates was: amoxicillin-tetracycline-TMP/SMX (31·7%), followed by tetracycline only resistance (23·2%), amoxicillin-tetracycline (12·7%), amoxicillin-tetracycline-ciprofloxacin-TMP/SMX (10·5%) and tetracycline-TMP/SMX resistance (6·1%) (Table 1).
Table 1. Resistance patterns found in 4320 E. coli isolates on 48 veal calf farms
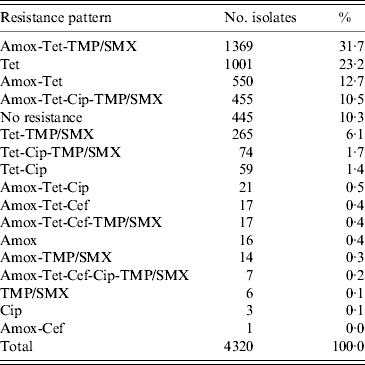
Amox, Amoxicillin; Cef, cefotaxime; Cip, ciprofloxacin; Tet, tetracycline; TMP/SMX, trimethoprim/sulfamethoxazole.
Seven (0·2%) isolates showed resistance to all five antimicrobial agents, these isolates originated from two farms. From the 4320 isolates in total, 445 (10·3%) isolates were susceptible to each of the five antimicrobial agents.
Antimicrobial usage
All farms administered antimicrobial drugs during the production cycle with a total mean of 51·7 ADD/pc, of which 46·8 ADD/pc were orally administered drugs (91%) and 4·9 ADD/pc were parenterally administered drugs (9%) (Table 2). Antimicrobial drugs belonging to the class tetracyclines contributed 46% to total mean medication, followed by colistin (18%), macrolides (9%), trimethoprim-sulfonamide combinations (8%), aminoglycosides (7%), penicillins (5%), combination drugs (2·5%), quinolones (2·3%), florfenicol (1·9%) and cephalosporins (0·6%).
Table 2. Overview of antimicrobial drug use on 48 veal calf farms
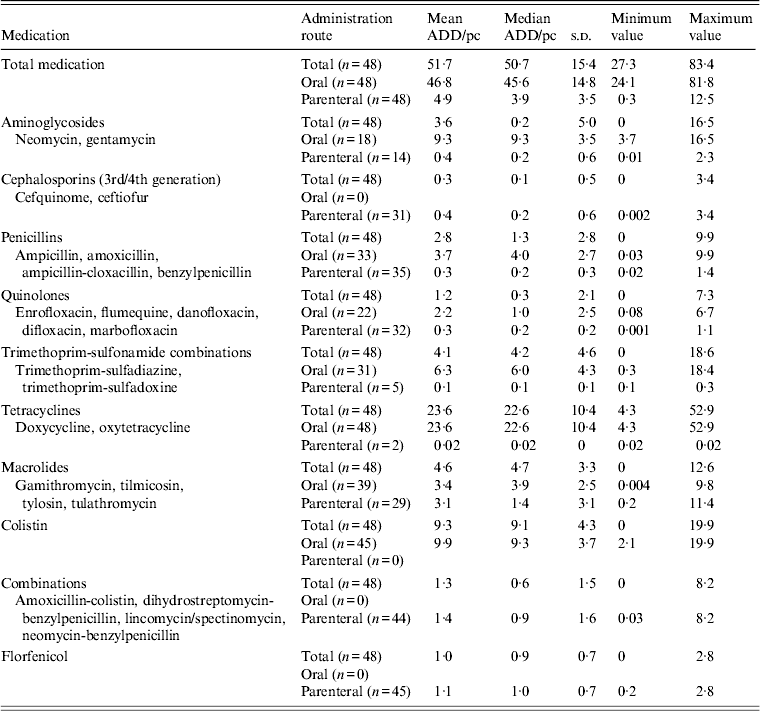
ADD/pc, Animal daily dosage per production cycle; s.d., standard deviation.
The largest amount of ADD was recorded in the first week of fattening (Figs 1 and 2). After this peak in week 1, the amount of ADD fell to a lower level for several weeks (weeks 2–10) and diminished during the following weeks until the end of the fattening period. Exceptions to this general trend were seen within the classes tetracyclines and penicillins which were delivered for oral treatments throughout the fattening period (Fig. 1), and for the class cephalosporins with a peak in the amount of ADD for parenteral treatments in week 25 (Fig. 2).
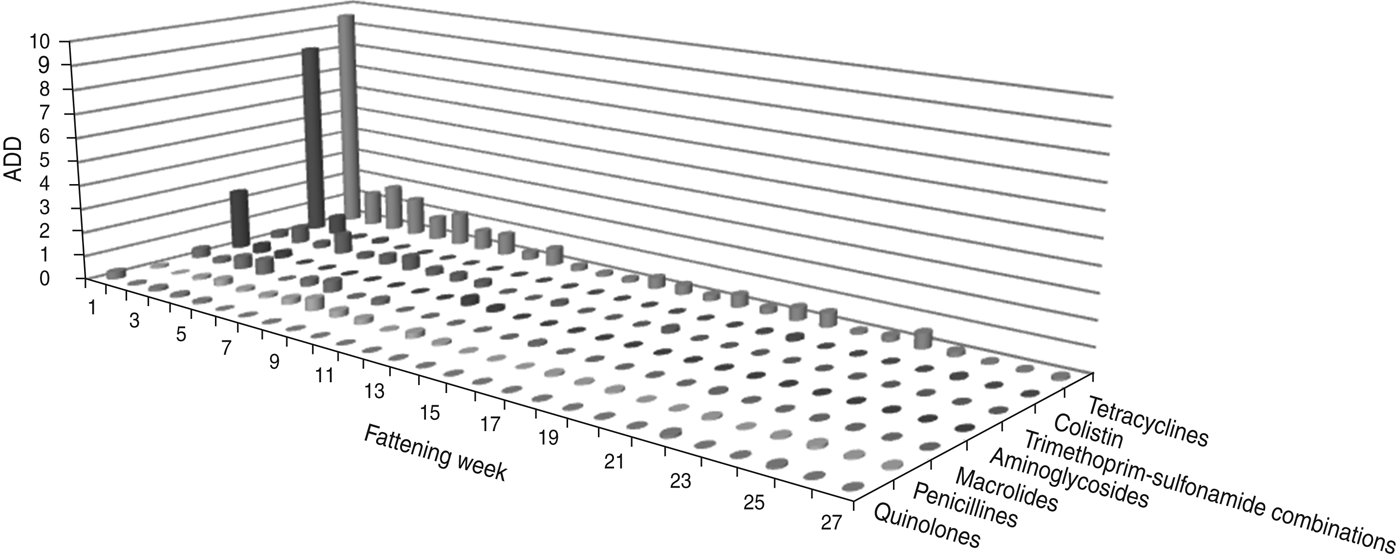
Fig. 1. The average animal daily dosage (ADD) of oral antimicrobial drugs delivered during the fattening period in 48 veal calf herds.
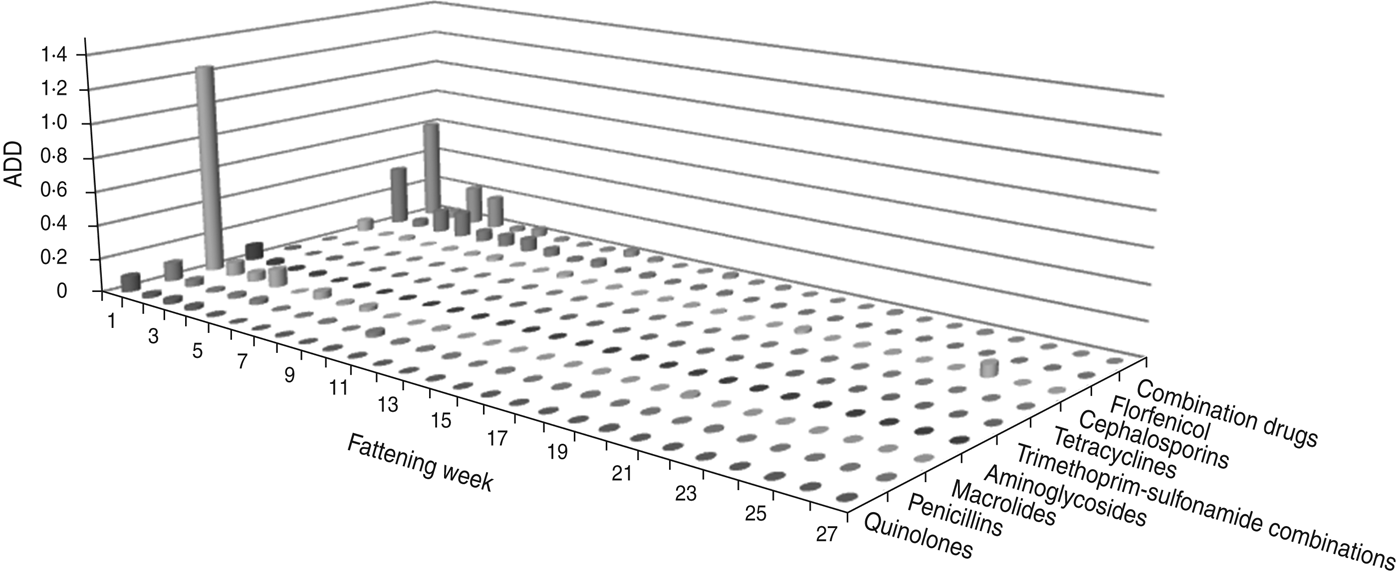
Fig. 2. The average animal daily dosage (ADD) of parenteral antimicrobial drugs delivered during the fattening period in 48 veal calf herds.
Risk-factor analysis
The best-fitting model, based on AIC value, included the variables: farm size, frequency of changing work clothes, number of calves/pen, feeding roughage, pest control of flies, the administration of aminoglycosides, cephalosporins, quinolones, penicillins, trimethoprim-sulfonamide combinations, tetracyclines and colistin, and the two-way interaction between each variable and the antimicrobial agents tested.
Variables significantly associated (P < 0·05) with increased resistance for one or more antimicrobial agents were the farmer wearing the same work clothes for several days vs. daily change of work clothes (OR 2·6 and 2·4 for ciprofloxacin and tetracycline resistance, respectively), administration of trimethoprim-sulfonamide combinations vs. no administration (OR 3·1, 3·0 and 2·6 for amoxicillin, TMP/SMX and tetracycline resistance, respectively), administration of ⩾0·3 ADD/pc quinolones vs. <0·3 ADD/pc (OR 2·8 for ciprofloxacin resistance), administration of ⩾1·3 ADD/pc penicillins vs. <1·3 ADD/pc (OR 3·4 and 3·3 for tetracycline and ciprofloxacin resistance, respectively) and administration of 20–40 ADD/pc tetracyclines vs. <20 ADD/pc (OR 3·2 for tetracycline resistance) and >40 ADD/pc tetracyclines vs. <20 ADD/pc (OR 13·1 and 6·5 for tetracycline and amoxicillin resistance, respectively) (Table 3).
Table 3. Risk factors associated with antimicrobial resistance in commensal E. coli isolates of veal calves in statistical end model
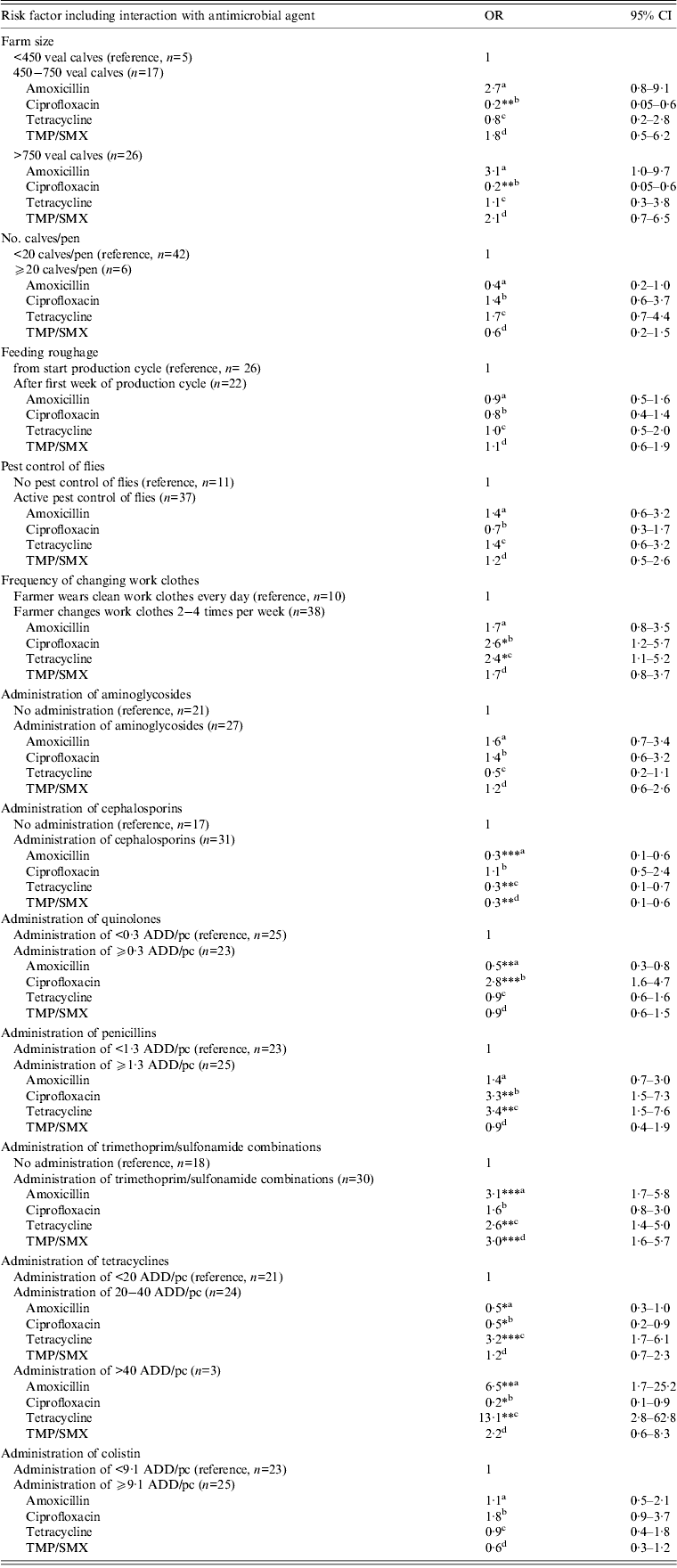
OR, Odds ratio; CI, confidence interval; ADD/pc, animal daily dosage per production cycle.
a Denotes the odds ratio for amoxicillin resistance for the risk factor level of interest compared to the reference level.
b Denotes the odds ratio for ciprofloxacin resistance for the risk factor level of interest compared to the reference level.
c Denotes the odds ratio for tetracycline resistance for the risk factor level of interest compared to the reference level.
d Denotes the odds ratio for TMP/SMX resistance for the risk factor level of interest compared to the reference level.
* P < 0·05,** P < 0·01, *** P < 0·001
Variance random sample effect = 0·7964, ICC rho = 0·195.
Variables significantly associated with reduced resistance for one or more antimicrobial agents were farm size (OR 0·2 for ciprofloxacin resistance for farms with 450–750 and >750 calves vs. farm size <450 calves), administration of cephalosporins vs. no administration (OR 0·3 for amoxicillin, tetracycline and TMP/SMX resistance), administration of ⩾0·3 ADD/pc quinolones vs. <0·3 ADD/pc quinolones (OR 0·5 for amoxicillin resistance) and administration of 20–40 ADD/pc tetracyclines (OR 0·5 for amoxicillin and ciprofloxacin resistance) or >40 ADD/pc tetracyclines (OR 0·2 for ciprofloxacin resistance) both vs. <20 ADD/pc tetracyclines.
DISCUSSION
The aim of this study was to identify determinants associated with the occurrence of antimicrobial resistance within veal calf herds. For this purpose the resistance levels to five antimicrobial agents, belonging to antimicrobial classes commonly used within veal calf farming, were determined at the end of the production cycle. In this study, isolates were most frequently resistant to tetracycline, followed by amoxicillin, TMP/SMX, ciprofloxacin and cefotaxime. Although the same ranking in prevalence of resistance is seen at national level [17] the levels of tetracycline, amoxicillin, trimethoprim, and sulfamethoxazole resistance in our study were higher. This might be explained by the fact that only white veal calf herds were included in this study whereas the monitoring at national level comprises both white and rosé veal herds. The facts that on white veal farms more antimicrobials (daily dosages per animal year) are used during the production cycle [26] and calves are slaughtered at a younger age (6 months) could have led to the higher proportions of resistant isolates found in this study.
Antimicrobial drugs were mainly administered orally with tetracyclines the most abundantly used class. These results are in accordance with antimicrobial usage data of white veal calves in Belgium [Reference Pardon27]. The large contribution of tetracyclines in the total ADD/pc in this study can be explained by the fact that these antimicrobials were orally administered to all herds in the first week of fattening as a prophylactic treatment, mostly in combination with colistin.
The data were analysed using a generalized linear mixed-effects model. Unfortunately, the data on cefotaxime resistance could not be analysed as the estimates of the log odds showed inestimable standard errors due to the very low proportions of resistant isolates present in the dataset. As resistance levels to the other antimicrobial agents did not give any problems in analysing the data it could be stated that generalized linear mixed-effects models are only suited for long-standing and well established high levels of resistance as they will not converge in situations with low prevalences of resistance.
Logistic regression analysis of the data showed that the administration of antimicrobial drugs was especially associated with higher odds of resistance to one or more antimicrobial agents. The administration of quinolones, trimethoprim-sulfonamide combinations and tetracyclines was associated with homologue resistance. The data show that the amount of ADD/pc influences the prevalence of resistance as the odds of tetracycline resistance was further increased when more ADD/pc of tetracyclines were administered. The association between antimicrobial drug usage and homologue resistance has been demonstrated before in studies in pigs [Reference Dewulf9, Reference Akwar12, Reference Dunlop13, Reference Alali28]. Surprisingly, the association between administration of ⩾1·3 ADD/pc of penicillins and the odds of amoxicillin-resistant isolates (also homologue resistance) was positive, but not significant. The lack of a significant association is difficult to explain especially because the largest part of the ADD/pc of penicillins was due to the oral administration of ampicillin, which was used on most farms (n = 33, mean ADD/pc = 3·7) and throughout the production cycle. Further analysis of penicillin usage data using several cut-off values compared to no antimicrobial usage as a reference revealed there was no significant association between any amount of penicillins administered and the odds of amoxicillin-resistant isolates (data not shown). These results suggest that the association between penicillin usage and amoxicillin resistance is not that strong or it might be that the association has been interfered with by other unknown factors.
Besides homologue resistance the administration of antimicrobial drugs was also associated with co-resistance to other antimicrobial agents. An explanation for this phenomenon is the frequent occurrence of multi-resistant isolates due to linkage of resistance genes [Reference Aalbaek29–Reference Hinton31]. Our study revealed that 56·4% of the tested isolates had a combined amoxicillin-tetracycline resistance phenotype and 42·8% had a combination of amoxicillin-tetracycline-TMP/SMX as resistance phenotype. Due to the existence of multi-resistant isolates administration of one certain antimicrobial drug could lead to increased resistance levels to several antimicrobial agents.
Of the selected management-related variables included in the analysis, surprisingly the frequency of changing work clothes by the farmer was the only hygiene-related variable significantly associated with higher odds of resistance. Although the association was positive (OR>1) for all four antimicrobial agents tested, it was only significant for two antimicrobial agents. It seems that the association between farm hygiene and the prevalence of resistant bacteria is not always clear. In a study of veal calves, farm hygiene (cleaning of stables before arrival of new herd on the farm) was a protective factor for the occurrence of MRSA [Reference Graveland3]. In contrast, a clean pen or a clean treatment reservoir was significantly associated with higher resistance in commensal E. coli isolates of fattening pigs [Reference Dewulf9] and broilers [Reference Persoons14]. A possible explanation why other hygiene-related and management-related variables in this study were not associated with antimicrobial resistance could be that these factors have limited impact compared to the effect of antimicrobial usage on the occurrence of antimicrobial resistance. Moreover, the fact that veal calves within a farm come from multiple suppliers makes it plausible that the resulting high infection pressure within herds cannot solely be overcome by hygiene-related interventions.
Besides positive associations between variables and the proportions of resistant isolates, there were also several significantly negative associations (OR < 1) observed in our study. A negative association implies that the presence of the explanatory variable decreased the risk of resistance to certain antimicrobial agents. In the case of antimicrobial usage this seems implausible, but has been demonstrated before in a study on pigs [Reference Akwar12] and veal calves [Reference Di15]. It could be hypothesized that the administration of certain antimicrobials (especially cephalosporins which were administered only parenterally) led to lesser use of other antimicrobial drugs, especially the classes which select for resistance to amoxicillin, ciprofloxacin, tetracycline and TMP/SMX. The only negative correlation between antimicrobial classes was present between the administration of cephalosporins and tetracycline usage (Spearman's rank correlation ρ = −0·20, P = 0·005). This significant correlation combined with the existence of multi-resistant isolates might explain the significant lower odds of amoxicillin, tetracycline and TMP/SMX resistance associated with cephalosporin administration.
For the other variables with a significantly lower odds of resistance to one or more antimicrobial agents there was no negative correlation between these variables and the administration of certain antimicrobial classes as Spearman's rank correlations were not significant (data not shown). Therefore, it seems that other factors may have influenced or interfered with these results. One of these factors could be the presence and absence of certain phenotypical resistance patterns in this dataset. For example, the administration of ⩾0·3 ADD/pc quinolones was significantly associated with higher odds of ciprofloxacin resistance, but with a negative odds ratio for amoxicillin resistance. The absence of isolates with a phenotypical resistance pattern containing solely the combination amoxicillin-ciprofloxacin in this dataset could be the reason for this negative association. Further, the low prevalence of isolates with a phenotypical resistance pattern containing the combination tetracycline-ciprofloxacin in combination with the abundance of other resistance patterns containing tetracycline resistance may have led to the significantly negative odds ratios for ciprofloxacin resistance associated with the administration of 20–40 ADD/pc or >40 ADD/pc tetracyclines, due to a selection advantage.
On the other hand, the existence of a selection pressure exerted by antimicrobial drug concentrations present within the environment of the calves, e.g. in feeding systems (remnants of oral treatments) or in manure (excreted by calves treated with antimicrobial drugs) cannot be excluded as it has recently been demonstrated that antimicrobial drug residues were present in drinking water of broiler farms [Reference Lamers, van Baak and Jehoel32]. Moreover, the presence of different age groups on some farms in this study (no all-in/all-out management) may have led to a continuous presence of drug concentrations on these farms which may have influenced the prevalence of resistant isolates within the commensal faecal flora. These factors might explain some of the associations in this study which could not be explained by the registered amounts of administered antimicrobial drugs or the presence or absence of certain phenotypical resistance patterns.
In this study the registration of antimicrobial drug usage within the sampled herds was based on veterinary prescriptions in order to obtain unbiased data. As a consequence it was assumed that the delivered volumes of antimicrobial drugs were administered in total, starting from date of prescription. For parenterally administered drugs some bias might have occurred, especially if the drug was delivered in large volumes, for example in a bottle, while only a part of this volume was needed at the time of prescription to treat a few animals. This leads to an overestimation of the ADD at the time of prescription (less kilograms per animal treated) and an underestimation of the ADD when the volume left is used at a later time point within the production cycle (more kilograms per animal to treat). Because the contribution of parenterally administered drugs on the total amount of drugs administered throughout the production cycle is small, the influence of this bias is assumed to be minimal.
In conclusion, the results of this study provide evidence that antimicrobial drug use in veal calves is associated with homologues and co-resistance in commensal E. coli isolates. Management-related variables appear to be of minor importance in the occurrence of antimicrobial resistance.
ACKNOWLEDGEMENTS
We thank all the farmers who cooperated in this study, the Task Force ABRES for constructive support and coordination and the Agricultural Economics Research Institute (LEI), part of Wageningen University and Research Centre for monitoring and providing the antimicrobial usage data of the sampled herds. This study was funded by the Ministry of Agriculture, Nature and Food Quality (project no.3 201 949); and the Product Board for Livestock and Meat (project no. 08·30·002)
DECLARATION OF INTEREST
None.







