1. Introduction
Modern science’s ability to produce, store, and analyze big datasets is changing the way that scientific research is practiced (Anderson Reference Anderson2008; Boyd and Crawford Reference Boyd and Crawford2012; Mayer-Schönberger and Cukier Reference Mayer-Schönberger and Cukier2013; Harford Reference Harford2014; Kitchin Reference Kitchin2014). In light of these changes, philosophers have sought to better comprehend the nature of scientific reasoning in this age of “big data” (Floridi Reference Floridi2012; Leonelli Reference Leonelli2012; Pietsch Reference Pietsch2015; Pietsch Reference Pietsch2016). Notable among these attempts is Sabina Leonelli’s (Reference Leonelli2016) book-length treatment of data-centrism in model organism research, which addresses the “gaping hole” in our collective understanding of data’s methodological role within the practice of contemporary science (Leonelli Reference Leonelli2016, 198).
Data-centric sciences, as Leonelli defines them, are those that prioritize the production and dissemination of data to enhance data’s evidentiary value. Leonelli builds this notion of data-centric science through an analysis of model organism research that traces the paths that data “travel” from their original location of production to other applications. Central to data-centric model organism research are databases organized using sophisticated labeling systems, which are a way of packaging data for greater travel amongst different but related research contexts. However, the success of these databases has both social and epistemic consequences: As databases become standard tools for research, they begin to function as a filter, allowing certain kinds of data to travel to certain places, but also restricting data journeys based on location or kind of data. Particular kinds of data, such as genetic or behavioral data, may consequently become preferred in an area of research, or data may only flow to and from researchers of certain kinds, such as genomicists or phylogeneticists, marginalizing some research and social groups while rewarding others.
To what extent such sophisticated database structures and the consequences of their use are constitutive of data-centric science is an open question. To help address it, we analyze data management and dissemination in a different area of research: regional climate modeling. We extend our analysis to a feature that we claim is crucial for understanding data-centrism—the information architectures underlying databases—revealing that data-centric sciences can come in at least two forms, one of which avoids the use of sophisticated labeling systems and the potentially pernicious consequences that they bring. These two forms of data-centric practice advance different epistemic goals and employ different methods to do so. They are dissimilar in the extent to which the information architectures employed are “phenomena-laden.” Phenomena-laden data architectures organize data, typically in relational databases, by appealing to the stable entities and processes that are widely recognized by practitioners in an area of research. We demonstrate that data-centric science can proceed without appeal to phenomena-laden information architectures. However, this alternate approach may have trade-offs: While likely to avoid unintended—and potentially pernicious—effects on research downstream, it also may limit the value of data for certain classes of users. We refer to these approaches as “phenomena-laden data-centrism” and “phenomena-agnostic data-centrism.”Footnote 1
We develop this argument in several steps. In Section 2, we explore the origins and nature of large datasets within climate science and model organism research. We briefly explicate Leonelli’s framework for analyzing data and its value in Section 3. In Section 4, we show that there are notable differences between data journeys in the two fields resulting from the way the data is stored, managed, and disseminated. In Section 5, we argue that these differences reflect two distinct approaches to data-centrism, while in Section 6 we analyze the trade-offs apparent in each approach. We examine how the introduction of machine learning (ML) algorithms may compromise certain advantages in section seven before offering a brief conclusion.
2. Data and its origins in climate science and model organism research
“Big data” describes data that has certain characteristics, for example, large volume, velocity, or variety, typically analyzed using certain tools, for example, ML or artificial intelligence (AI) (Knüsel et al. Reference Knüsel, Marius Zumwald, Gertrude Hirsch Hadorn, Bresch and Knutti2019). We adopt Leonelli’s notion of data as the material and informational outputs of research that serve as evidence for knowledge claims. “Volume” is the size of the data, often discussed in terms of the computer storage memory required to hold it. “Velocity” is the speed at which the data must be processed, and “variety” the extent of heterogeneity of data with which a field or investigation might deal. “Large” is typically cashed out operationally, that is, when challenges for effective use of the data are encountered and new strategies must be developed. Through this lens, aspects of both regional climate modeling and model organism research qualify as “big data.” As we will see, model organism research must overcome challenges related to high velocity and especially variety, whereas regional climate modeling primarily faces challenges of massive volume.
Data-centrism is developed as a counterpoint to narratives that portray big data as revolutionizing science, perhaps even bringing about the “end of theory” (Anderson Reference Anderson2008). Rather than viewing big data approaches as constituting the rise of a new data-driven mode of research (a mode that has arguably long been present to some degree), data-centrism views big data as making salient a different innovation: the newfound attention given to data management and dissemination practices. Leonelli associates data-centrism with an approach to science that prioritizes efforts to gather, mobilize, integrate, and visualize data (Reference Leonelli2016, chap. 1). In this mode of science, data management and dissemination are considered valuable scientific practices even if they do not directly contribute to the development of theory. Both model organism research and regional climate modeling constitute data-centric science on this view.
Regional climate modeling is a specific way of producing data within climate science research. Climate science is the investigation of the Earth’s climate system to understand how processes generate a region’s climate and to project how that climate may change in the future. While climate science is often associated with geoscience or geology, it also draws on chemistry, biology, oceanography, ecology, physics, and meteorology, among others. This research involves analyzing data that are taken to represent a variety of aspects of the climate, including air temperature, wind speed and direction, water vapor, pressure, precipitation, cloud properties, radiation budget, and atmospheric composition (e.g., CO2, CH4, Ozone).
Climate models are mathematical equations representing how the climate evolves over time. In such models, a representation of the Earth is often divided into a three-dimensional grid, and, at each time-step, the values of the variables in each grid-cell are updated based on their present value and that of their neighbors in accordance with the model’s equations. A computer algorithm specifies how solutions to these equations should be estimated, and the process iterates, with the results of one time-step serving as the starting conditions for the next time-step. The outputs from climate models are large arrays of numbers, where each value in the array corresponds to the value of a given variable (or “field”) over some discrete chunk of space and time determined by its position in the array. Modelers run climate models to generate a record of the evolving state of the simulated Earth system. One important use of these models is to create projections of future climate that provide insight into climatic changes for which adaptation planning may be required.
Regional climate models (RCMs), the source of data we focus on in this article, are specifically designed for regional-scale analyses, which separates them from global climate models (or “general circulation models” [GCMs]), their more well-known counterparts. GCMs, as the name implies, are global in scope and produce results that span the planet. To remain computationally tractable, the spatial and temporal scales they use to represent the Earth are somewhat coarse. RCMs offer more finely resolved results for particular regions. These models “dynamically downscale” the results of GCMs by representing a region of interest with a tighter spatial grid and using shorter time-steps. They can thus better represent smaller-scale processes relevant to the variables being computed that are difficult or impossible to capture with a GCM. The results of GCMs provide boundary conditions for the region of interest examined by the RCM. One can think of an RCM as embedded in a GCM and used to zoom in on a specific region.
In an effort to examine uncertainties, climate scientists try to avoid relying on a single model run; climate models are usually deployed as parts of ensembles containing many runs of the same or different models. For example, climate models are chaotic and thus sensitive to changes in the initial conditions. Scientists explore this uncertainty by running many simulations of the same model with slightly altered initial conditions to determine how sensitive that model’s results are to these changes.Footnote 2 In the case of RCMs, one can similarly explore uncertainties arising from different boundary conditions by downscaling different GCMs. This use of ensembles significantly enlarges the volume of data produced.
The data gathered in model organism research—the icon of big data in biology—vary in kind. Sequencing projects, for example, generate large quantities of data on model organisms that specify the order of DNA nucleotides, or bases, in a genome. Presently available sequencing data covers a wide variety of model organisms, including bacteria (e.g., Echerichia coli), plants (e.g., Arabidopsis thaliana), and animals (e.g., Danio rerio, or zebrafish), as well as humans, though humans are not “model organisms.” These sequences are often produced by automated means, such as DNA sequencing machines, and often without connection to specific research questions.
Sequencing data, on its own, is not very useful (Lloyd Reference Lloyd and Cranor1994). For example, one cannot identify a gene straight from sequencing data. Thus, making sequencing data valuable requires connecting it to other high-velocity types of data regarding subcellular biology. Leonelli refers to this kind of data colloquially as “omics” data: metabolomics (metabolite behavior), transcriptomics (gene expression), and proteomics (protein functions and structures). The objects that make up the data across these areas of interest vary, and include photographs, measurements, specimens, observations from experiments or the field, and statistical surveys. The goal of gathering and amalgamating this variety of data is to enhance scientific understandings of organisms as a whole, including advancing knowledge of evolutionary processes, environmental impacts, and immune system responses.
One point of similarity between data generated by RCMs and by model organism biology is that both are used by multiple stakeholder groups. For climate model data, the primary stakeholders are often climate science researchers—some of whom are downstream and reliant on model data—but others include researchers in other related fields, government regulators, and policy makers. Regional model data is increasingly being used by local decision makers for infrastructure and emergency planning. This is not unlike the cases Leonelli examines: Biological researchers, including pharmaceutical companies, and public health organizations, are the primary stakeholders, with private companies like 23andMe becoming increasingly interested in the data.
These descriptions help demonstrate why both regional climate modeling and model organism research might embrace data-centrism’s focus on data as a scientific goal, rather than as just a means to develop theory. However, there is further evidence that these disciplines prioritize the gathering, mobilization, integration, and visualization of data. Model organism researchers have automated means of producing data that are disconnected from concerns regarding theory testing. They also, as we will see, place a high value on, and reward, data integration. Along the same lines, the data produced by regional climate modeling studies are distributed broadly and used for purposes beyond those imagined by their original producers. Often, they are used for more than theoretical insight, for example, by creating climate projections to support future planning. Regional climate modelers also have developed suites of visualization tools (Rendfrey, Bukovsky, and McGinnis Reference Rendfrey, Bukovsky and McGinnis2018) and—demonstrating a focus on data management—have implemented digital object identifiers so that those creating datasets or visualization tools can receive professional credit for their work. Having established that, like model organism research, regional climate modeling constitutes a data-centric area of research, we now turn to analyzing the character of the data-centrism that regional climate modeling displays.
3. A framework for data-centrism: Packaging for decontextualization and recontextualization
To investigate the rise of data-centrism, Leonelli introduced, and she and others have explored (Leonelli Reference Leonelli2016; Leonelli and Tempini Reference Leonelli and Tempini2020), a conceptual framework featuring data journeys as well as data decontextualization and recontextualization. Each component of this framework helps analyze how the evidentiary value of data can be expanded. “Data journeys” are the movement of data from their sites of production to other similar or dissimilar sites of investigation. The ability for data to travel on a journey is affected by several factors, including transmission speed, legal regulation, community and disciplinary norms, and demand for the data. These factors encourage scientists to package data in particular ways. Data packaging is essentially putting the data into a form that would foreseeably decrease the “friction” data faces when it travels, making it easier to produce, transfer, read, and utilize (Edwards Reference Edwards2010).
A packaging process might include storing data in a widely used file format, but also may extend to ensure that data-handling practices abide by the relevant regulations and that data are stored in a way that makes them accessible to relevant researchers. Leonelli highlights the role of databases, and their ontologies, in this packaging process. We join Leonelli in using the term “ontology” in the sense often employed in computer science (Breitman, Casanova, and Trszkowski Reference Breitman, Antonio Casanova and Trszkowski2007), denoting a set of categories with specified properties and relations linking the various categories together. An ontology in this sense serves as a classificatory or labeling system that is part of the structure, or information architecture, that organizes a database.
Successful data packaging allows for efficient and accurate decontextualization and recontextualization. Decontextualization is the process of disconnecting data, temporarily, from its origin. The whole point of decontextualization, Leonelli claims, is to strip data of as many qualifications as possible. By stripping data bare—taking away the significance the data had in its original context by relegating, for example, its method of production, precision, or uncertainty, to metadata—the data can be more easily categorized and searched within a database, helping users to find potentially useful data.
Recontextualization is the process of discovering potentially relevant data and assessing whether they could serve as evidence in a new context. Recontextualization involves researchers querying or exploring the ontological relations in a database to find potentially relevant decontextualized data and then investigating the provenance of that data through the associated metadata. Researchers thereby assess whether the data can be situated within a new context to serve as evidence; if it can, this data is considered recontextualized. Importantly, it is the successful packaging of data and its ability to be recontextualized in different contexts that permits data to gain epistemic value: The value of data as an epistemic object is enhanced when it can be situated as evidence within multiple investigative contexts.
4. Information architecture and decontextualization
To compare data-centric practices between model organism biology and regional climate modeling, we extend the level of analysis from Leonelli’s examination of the role of databases in data journeys down to the details of the information architectures underlying the databases. We use “database,” as is typical in information science, as a general term encompassing any organized collection of data. The information architecture, more properly the “data model,”Footnote 3 organizes the elements of data and specifies how they relate to one another and to the properties of real-world objects or concepts. The choice of architecture thus significantly influences how data travels, how it can be searched and accessed, and, ultimately, the objects and methods of investigation. Our contention is that this level of analysis is necessary for effective disambiguation of species of data-centrism in science, as will become apparent in the remainder of this article.
Model organism databases, like most business and government databases, are relational. Relational databases are effectively collections of tables of information related to each other (see our author-created tables, Figures 1 and 2). The columns of each table are referred to as “fields” (or “attributes”); rows are referred to as “records” (or “tuples”).Footnote 4
In normal practice, each table represents one type of entity or relation. We use the term “entity” as it would be in computer science. In this sense, an entity can represent an object type (e.g., chromosome or nucleotide) or a concept type (e.g., host gene response or taxonomical class). Each record represents an instance of that entity, each field (heading) represents an attribute or property of the entity type, and the values in the cells represent the information about those properties for that instance. A unique key identifies each row—each row must have a different value for that field,Footnote 5 typically termed a “primary key.” For example, we may have a table for genes (entity type) like the following, with “Gene ID” serving as primary key.
Tables are related to each other by including the primary key from one table as a field in the other, where it functions as a “foreign key.” These relationships can be one-to-one, one-to-many, or many-to-many.Footnote 6
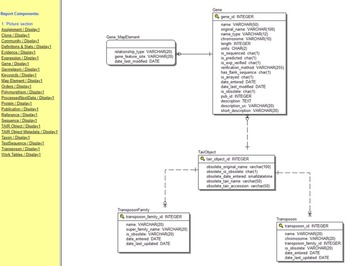
In Figure 3, “One” and “Many” describe the respective sides of the relation. Each chromosome may appear in the genes table many times because for example chromosome 1 contains around 3,000 genes. Likewise, we may have many sequences of a particular gene in our collected samples. We can more succinctly provide a blueprint for a relational database by listing the fields in each table and using lines and symbols for relations in a “database diagram” (The Arabidopsis Information Resource [TAIR] database [Phoenix Bionformatics Corporation 2021]; Figure 4).
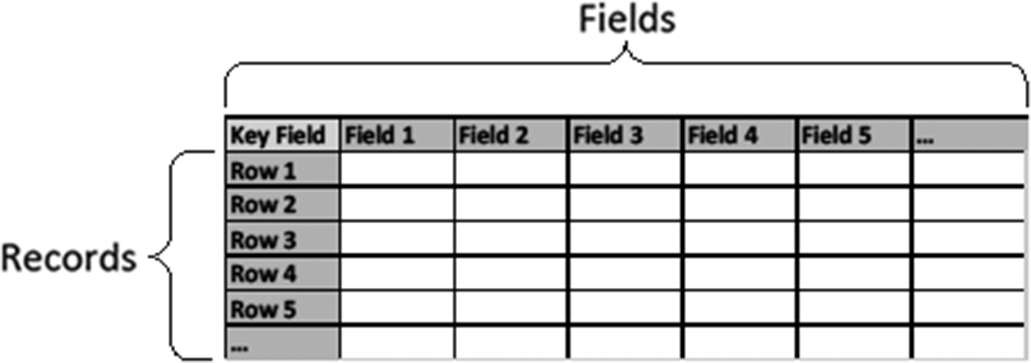
Figure 1. A database table.
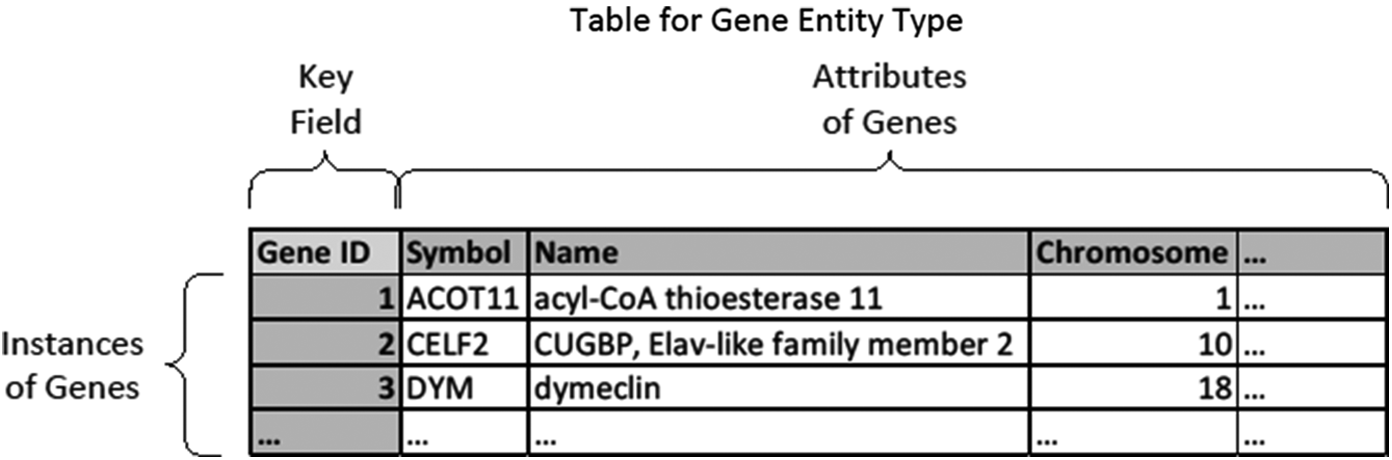
Figure 2. A table for the entity type of genes.
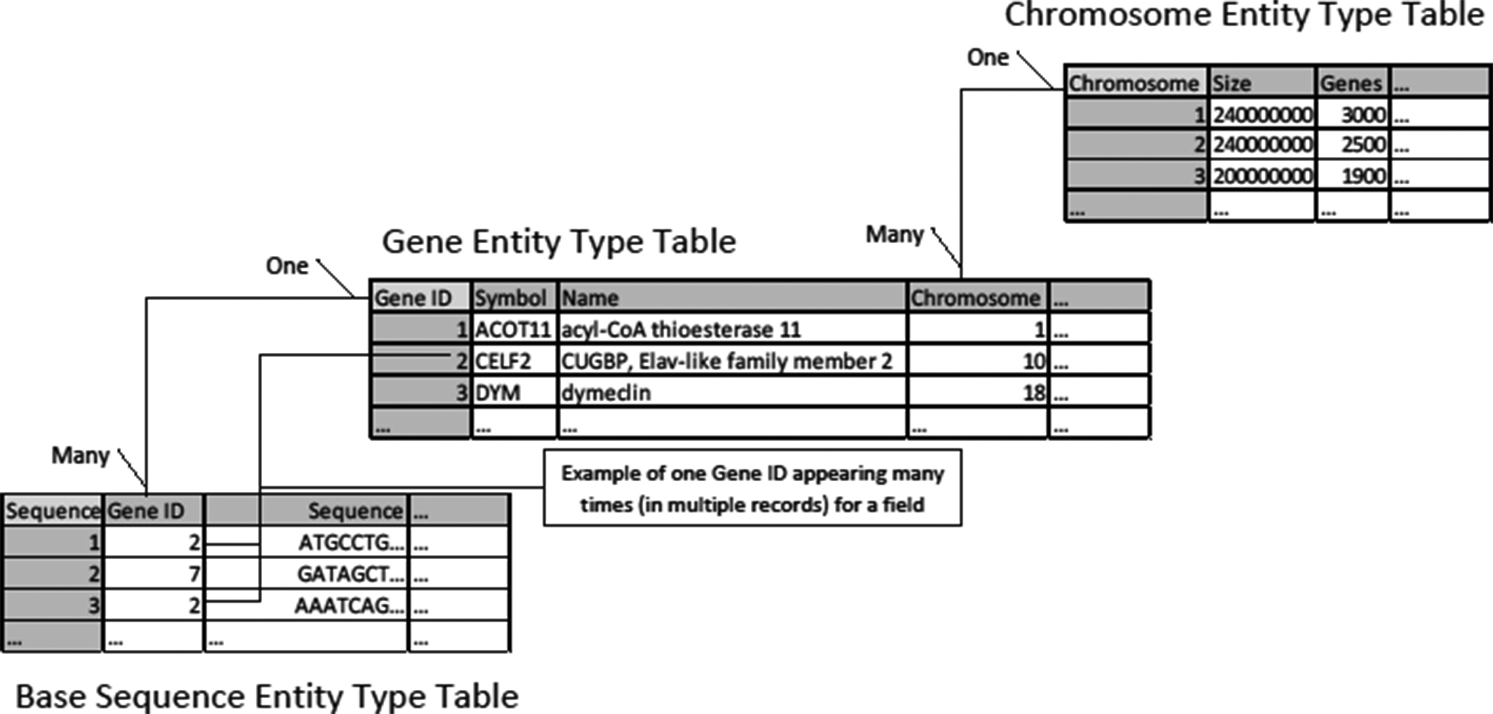
Figure 3. Relations between database tables.
Large relational databases with such sophisticated organization are not easy to create or manage. Designing a relational database requires a great deal of thought about which entity types need representation by tables and about the proper relationships between tables. Furthermore, many fields—particularly those that are not quantitative—should accept only “valid codes,” which are predetermined categories that collectively fully partition the relevant field. (These valid codes are examples of a concept Leonelli calls “labels.”) Often users experience these valid codes as the options in a drop-down box when searching for or inputting data; they are the only acceptable choices of value for attributes that can be associated with that entity type. To take a simplified example, if an entity type were “garden rose,” then each record would be an instance of a particular type of garden rose, an attribute (i.e., field) might be “color,” and the valid codes for it “red,” “white,” and “yellow.” Furthermore, a protocol or rubric must be established for selecting appropriate entity types and valid codes for the inputting of records into the database.
Choices made about the information architecture of the database and the procedure(s) for inputting records into the database inform data decontextualization and recontextualization. In model organism research, the organizing principles of the information architecture makes heavy use of phenomena terms to aid in describing the data independently from its source of production (Leonelli Reference Leonelli2016). These phenomena terms are typically represented in the data structures as not only entity types or primary keys but also as the permissible valid codes for attributes. Organizing around phenomena helps with decontextualization precisely because phenomena are stable and repeatable features that do not depend on the idiosyncrasies of the data and its production (Bogen and Woodward Reference Bogen and Woodward1988; Parker and Lusk Reference Parker and Lusk2019).Footnote 7 Because phenomena are abstracted away from underlying data in this way, they can be used to organize datasets to make them searchable without needing to examine the context of data production—the primary goal of decontextualization.
We call databases organized in this way “phenomena-laden.” In using this term, we are intentionally alluding to the phrase “theory-laden” that is so familiar to philosophers of science. When one says that observation is theory-laden, one implies that content of the theory is informing and perhaps classifying the way in which we interpret observations. Observation, even if theory-laden, is not completely determined by theory in all its aspects. Likewise, when we say that a database is phenomena-laden, we imply that the information architecture reflects claims about phenomena that are widely accepted. Nevertheless, the architecture is not completely determined by a community’s knowledge of phenomena.
When examining the structuring of regional climate model data, a very different picture emerges, one that lacks the phenomena-ladenness displayed in the architecture of model organism databases. For ease of reference, we refer to this architecture as phenomena-agnostic. The databases used in regional climate modeling are not relational, but rather collections of “flat files,” or plain tables. In most cases, the output is stored in a standard community-developed format (netCDF)Footnote 8 that automatically saves metadata about provenance (e.g., the model used, grid size, and other important technical information).
One way to visualize this kind of data architecture (see Figure 5) is to consider a workbook of tables, as is commonly used in spreadsheet programs, as a heuristic. In practice, spreadsheet programs are not well-suited for analyzing climate model outputs, in part because the data volumes are much too large, but the data structures are conceptually similar. When representing the surface layer (of the Earth), for example, the rows and columns of the workbook table would represent latitude and longitude, so that each “cell” (location in the array) locates a unique spatial cube in the simulation. Initially, the RCM generates one file per time-step, and each file contains numerous sheets, each containing the values for one variable. The research teams rearrange the initial outputs of simulations to create files representing single variables, such as temperature, and each “sheet” in the file would then represent a single time-step.
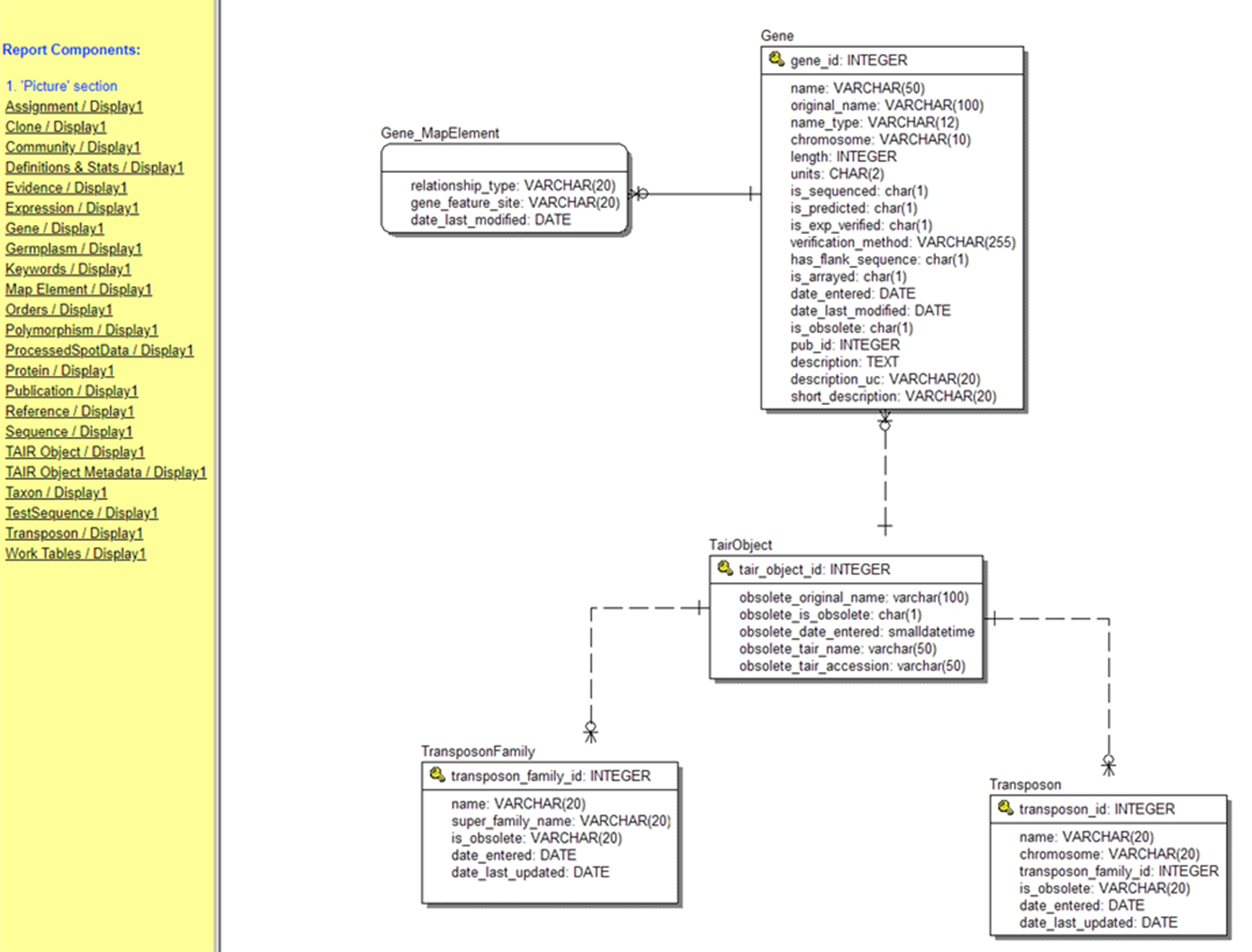
Figure 4. The database (entity relationship) diagram for just the transposon portion of the TAIR (The Arabidopsis Information Resource) Database (Huala et al. Reference Huala, Dickerman, Garcia-Hernandez, Weems, Reiser, LaFond and Hanley2001). The list on the left displays other portions of the database, most of which are far larger.
Source: https://www.arabidopsis.org/search/ERwin/Tair.htm.
Note: The precise relationships between the tables are indicated by the symbols at the end of the lines joining the tables. The line with a short perpendicular line denotes “one,” and the circle with multiple lines denotes “zero or many.”

Figure 5. A heuristic to represent NetCDF architecture.
The overall information architecture is therefore quite straightforward, and quite different from the phenomena-laden relational databases used in model organism research. In particular, the data are not classified into entity types with associated attributes. This is because, whereas relational databases are organized around records as instances of the entity type defined by the primary key, here there are no primary keys and no records. Rather, each file constitutes a single variable, which is akin to a field in the relational database. The (typically quantitative) values are distributed throughout the “cells,” which are the unique intersections of latitude and longitude for that time step. In short, each “cell” contains a single datum. This structure is optimized for storing and making computations with data, rather than for instantiating and being searchable in terms of phenomena.
RCM databases organized in this way obviously are not phenomena-laden. The absence of phenomena as structural organizing principles reflects the relative lack of decontextualization. Certain highly idiosyncratic features of the data are resolved—replacing the spatial gridding that was most efficient for computation of the model on a specific computer by one that is more geographically intuitive, for example. Overall, though, the data made available to users in netCDF format preserves the context of production of the data, and the storage structure more or less preserves its form (organized by variable and spatio-temporal location). In the following sections, we will discuss reasons why these ontologies and associated information architectures are best situated for their respective scientific practices as well as associated trade-offs. For now, we note that whereas model organism relational databases contain decontextualized information, RCM output archives contain data effectively still in context, so to speak.
However, the analysis of the two different approaches to data architecture discussed in the preceding text are sufficient to demonstrate that the centrality of phenomena-laden relational databases and their associated ontologies is not a universal aspect of data-centric science. The data-packaging process—which influences the way data is decontextualized and recontextualized—differs significantly between the model organism context and the regional climate model context.
5. Varieties of data-centrism: Phenomena-laden and phenomena-agnostic
These significant dissimilarities in the information architectures employed in model organism research and RCMs suggest the existence of different varieties of data-centrism, that is, distinct sets of practices that reflect the prioritization of data which differ in epistemically significant ways. The information architectures, and the associated practices linked with them, reflect the different challenges to effectively producing, managing, disseminating, and making use of data in each case.
Big data in model organism biology features a high velocity and especially extensive variety. Many research groups from many specializations are producing many datasets that can be quite different from each other. The model organism research community is “highly fragmented, encompassing a wide variety of epistemic cultures, practices, and interests and multiple intersections with other fields” (Leonelli Reference Leonelli2016, 4). In the past, the flow of data was restricted so that it remained within siloed communities of interest. Information that would be highly valuable to scientists beyond the area of production was difficult for them to discover and thus underutilized. The prioritization of data in model organism biology has been driven by technological advances that have enabled various modes of useful integration (Leonelli Reference Leonelli2016, chap. 6).
To promote data travel, data-centrism in model organism biology embraces data decontextualization using the phenomena-laden relational databases discussed in the preceding section. In such databases, data is “ontologized” through reference to phenomena, noting relationships between phenomena, and providing a relatively user-friendly database interface for identifying and downloading appropriate information. Phenomena-terms (as primary keys) function as crucial organizing principles for revealing to users large-scale and stable structures in the data, enhancing a user’s ability to get an overview of potential connections between types of data. Because the phenomena terms associated with primary keys typically reflect fundamental concepts that would be familiar to many different kinds of researchers working in a particular area, they are widely recognized, and even those unfamiliar with the motivations or techniques that produced the data categorized by that key can easily browse through them. Such methods support data integration by allowing data to travel further and among more researchers, thus enhancing its value.
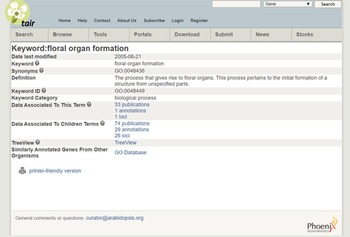
As an example, consider the cross-species integration of research on Miscanthus giganteus and Arabidopsis. The former is a plant of great interest because of its applications for biofuels but is too large to grow in a laboratory. The latter is convenient to study and similar in important (genetic) respects to Miscanthus giganteus and other plants of interest, and so has become a model organism. The Arabidopsis Information Resource (TAIR) database (Phoenix Bionformatics Corporation 2021; Huala et al. Reference Huala, Dickerman, Garcia-Hernandez, Weems, Reiser, LaFond and Hanley2001) serves to disseminate information about it, which can be recontextualized for Miscanthus. A Miscanthus researcher can enter “floral organ formation” into the TAIR Keyword Search. The result provides a definition, synonyms, a “treeview” showing how the phenomenon fits into a hierarchy with other phenomena, and citations for data on it and “child” concepts (phenomena lower in the hierarchy). (See Figure 6 displaying the search results and Figure 7 for the “treeview” [TAIR 2021]). It is then straightforward to click on a publication in “data associated to this term” and collect relevant datasets.
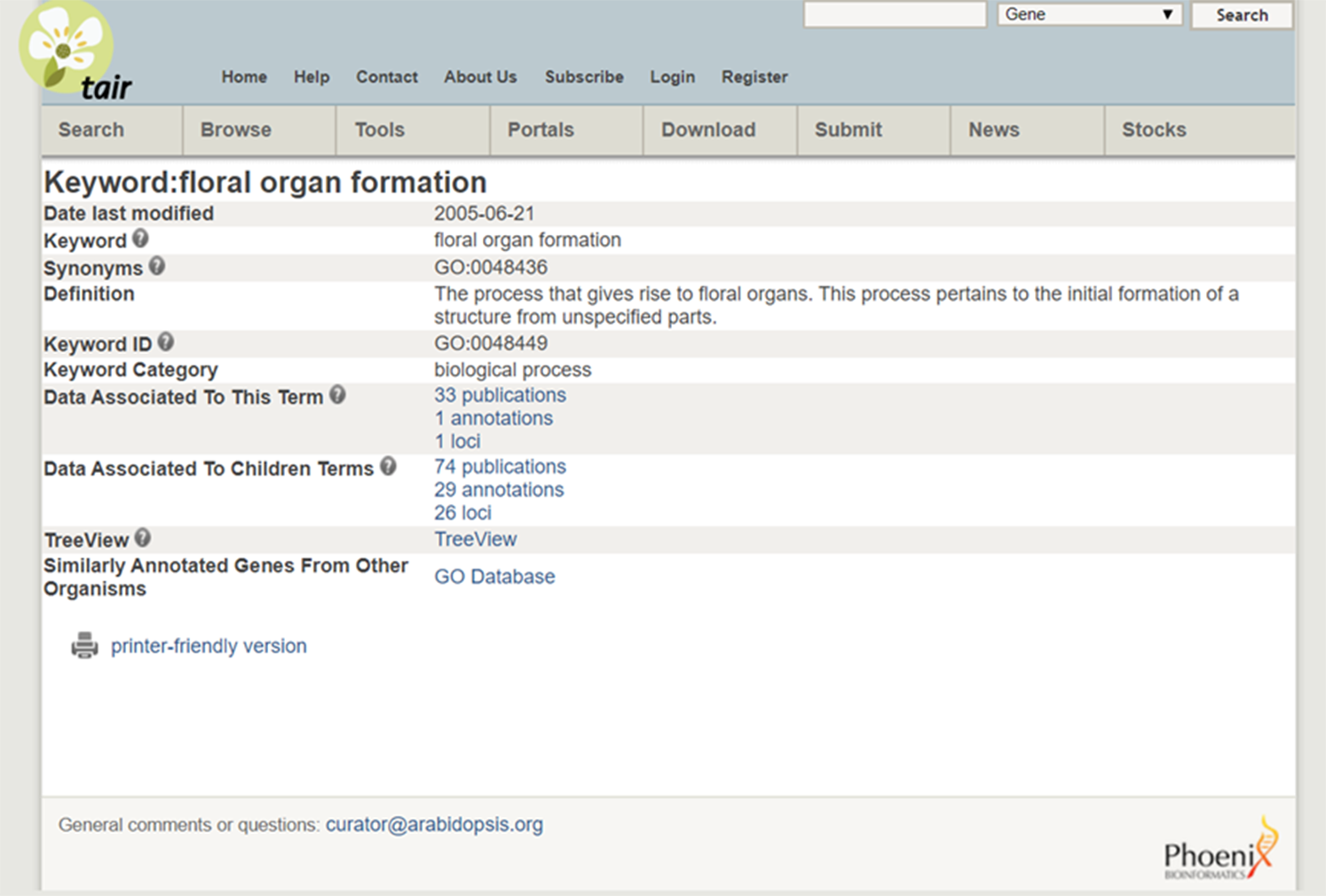
Figure 6. Search results from the TAIR database.
Source: https://www.arabidopsis.org/servlets/TairObject?type=keyword&id=18869.
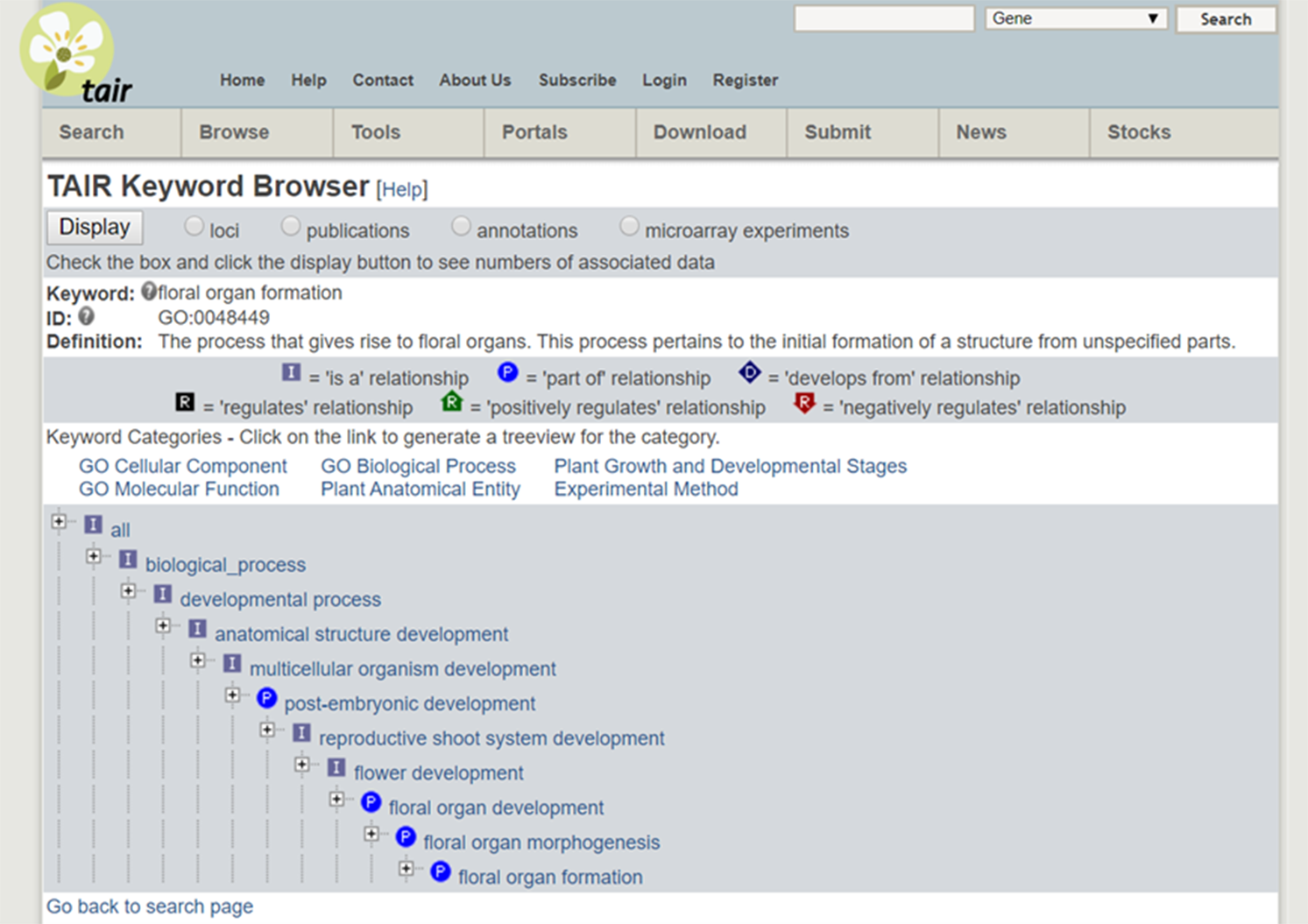
Figure 7. A “treeview” from the TAIR database.
Access to such databases helps enhance the value of the data by decontextualizing it in ways that enable it to travel and later be recontextualized by researchers interested in an entirely different organism.Footnote 9 By using widely known phenomena terms to categorize the data, researchers with disparate backgrounds are able to navigate the data architecture. Synonyms are provided for users when it is known that various communities refer to the same or similar phenomena using different expressions. Furthermore, the database interface suggests paths for finding data that users might have been unaware of when designing their original query, by, for example, pointing toward similarly annotated genes from other organisms. This method of using a relational database helps enhance the value of data across contexts.
Employing phenomena-laden databases for integration in this way promotes certain methodological developments. Particularly important for our analysis is the work performed by dedicated data curation teams. Data curators shoulder the responsibility for the decisions surrounding database creation and management, including their ontological structure and selection of data to include in the databases. In model organism research, curators are generally professionals with strong backgrounds in both biology—often a doctorate—and in information technology. They typically work for consortia—often funded by organizations like the National Science Foundation or National Institutes of Healths—established for the purpose of managing and disseminating biological data donated by a myriad of researcher groups. They do not engage in fundamental biology research, but rather they are the key packagers of data for travel. These curators work with the community to establish data standards and serve as gatekeepers that enforce and lead revisions of those standards. In collaboration with the research communities they serve, curators choose the data architecture to be employed (e.g., the primary keys and valid codes), the criteria by which submitted data will be judged for inclusion in the database, design the workflows to handle submitted data, and lead efforts to revise the database architecture. In essence, data-centrism in this form aims to help enhance the value of data across research contexts, relying on heavily curated relational databases to do so. The curators address big data velocity by including in the databases only data they consider valuable. They address big data variety by transforming formats and metadata for increased consistency and especially by ontologizing the data into phenomena that are meaningful for researchers across specialties.
The practices in regional climate modeling constitute a different variety of data-centrism generated specifically in response to challenges arising from data volume; neither velocity nor variety are of primary concern. There are very few producers of regional climate-model data because of the computational requirements for producing such data; a supercomputer with massive storage capabilities is practically essential. Datasets are thus a relatively rare commodity. Such datasets are also relatively low in variety, due to the standardized netCDF format used for data management, the relatively consistent sets of variables of interest, and the similarity amongst models.
While scientists in a variety of subspecialties use RCM output datasets in downstream research activities, sometimes beyond those envisioned by the modelers, that variety—in the scientific users and uses rather than the data—also does not end up posing a significant challenge for use of big data.Footnote 10 The broad community of climate scientists is more epistemically unified (than that of biologists) in an important sense: their training, background knowledge, methods, objects of study, and so on, are similar enough to provide sufficient epistemic cohesion to alleviate the need for extensive decontextualization.Footnote 11 Climate scientists who are not modelers understand RCMs and their output to a level needed to identify relevance, evaluate provenance, acquire, and make use of output datasets in the context of their production, without curators identifying phenomena.Footnote 12 For example, consider the Navier–Stokes equations: Virtually every climate scientist knows the role they play in climate models, whereas there really is no comparable law grounding data-intensive model organism research. While modelers may occasionally worry about how nuanced some downstream researchers’ appreciation of details may be, the ecosystem of climate science research functions successfully in this manner (Edwards Reference Edwards2010), reinforcing that specializations in climate science are not as epistemically disparate as those in biology.
Phenomena-agnostic data-centrism therefore seeks to overcome challenges regarding data volume when packaging data to ensure its travel. The general strategies are to divide, parse, and limit data—as well as distribute labor—for efficient data distribution. These strategies lead to “lean” information architectures and efficient data management tools that facilitate storage, processing, and dissemination of massive files. Simply saving the files requires extensive storage space and manipulating and editing them during the packaging process is computationally expensive.
The information architecture employed in packaging RCM data—as exemplified by the netCDF framework—helps minimize storage requirements and maintain computational tractability, even when compared to the data architectures deployed in model organism research. The netCDF framework consists of simple arrays of values, with each element in each array standing alone. This simplicity stands in contrast to relational databases, like those used by the bio-ontologies, which are organized into records (the rows in tables) with the columns linked together (as an n-tuple by record) and additional relationships between tables. In a relational database system, referential integrity must be maintained when a record’s value is changed. Such a change prompts cascading alterations that update other dependent values and ensure references to the changed object remain consistent. Such computational complexity is avoided with netCDF because there is no referential integrity to maintain. The format simplifies the database management actions needed when working with data, which helps maintain computational tractability. A relational database likewise must store not only the data but also additional information about relationships, valid codes, and so forth, multiplying the size of stores. This is unnecessary in the netCDF framework. The methods of data handling, afforded by netCDF, help further the goal of making the bulky datasets from regional climate modeling packageable for travel.
Beyond the information architecture, various methods are employed to help ameliorate the difficulties with large data volumes when packaging and disseminating data. While the RCMs run with relatively fine-grained time-steps, only certain variables are made available at these fine resolutions; for most variables only daily and longer time resolutions are published. Likewise, climate scientists have decided to only publish datasets for certain variables, with publication of some variables dependent on resources. For example, the NA-CORDEXFootnote 13 archive categorizes variables into tiers as essential (always archived), high priority (archived if at all possible), and aspirational (archived if time and resources allow) (McGinnis, Mearns, and Gutowski Reference McGinnis, Mearns and Gutowski2016). The data archives often provide a feature that allows users to “subset” the data to a region of interest,Footnote 14 as well as selecting only variables of interest. In this way, users can reduce volumes to a level where they can download and work with the files.
The roles curators play and the methods they employ in the RCM context reflect the concern over handling large data volumes and differ from those of curators in model organism research. RCM curators are normally members of modeling groups, not members of consortia working on dedicated data-curation projects. Their multidisciplinary backgrounds in science and information systems make them unique members of these groups. Because data volume prohibits these “data wranglers” from postprocessing data in commercial software tools, they must be able to write programs in low-resource languages, such as R or Python. Bias correction requires serious data science chops, too. Moving RCM output datasets to a centralized location and processing them en masse would be costly in terms of computational and network requirements due to data volume. It is also advantageous for data curators to have access to detailed knowledge of the model when postprocessing the data, for example in helping them understand the original grid system, which may be idiosyncratic, or having a feel for model biases to correct. For these reasons, regional modeling groups have incentives to hire or develop in-house data curators who prepare and publish their team’s data rather than relying on third parties to compile and integrate data from multiple contributors.
The general approach to phenomena-agnostic data-centrism found in RCM research is to eschew, rather than embrace, decontextualization. Curators do not organize data in terms of phenomena. To do so is likely to be unwieldy, unnecessary, and add to the already significant computational costs. Thus, data are preserved in a form much closer to the context of their production as they travel, which makes sense given the goal is distribution of massive datasets within an ecosystem of researchers with overlapping expertise. Not only can users identify phenomena of interest using criteria relevant for their work but also curators could not possibly do so for many of the applications RCM data serves. Identifying hurricanes, for example, requires visualizing the data and looking at it play out over time, a labor-intensive process. This lack of concern for decontextualization is reflected in the organization of curatorial practices in regional climate modeling: Rather than installed as part of a dedicated team, data curators are often individuals that support groups of climate modelers. That the identification of phenomena within data is left almost entirely to end users is one noteworthy aspect in which work is distributed in this form of data-centrism. The goal in this form of data-centrism, which is supported by the chosen information architecture, is one of providing phenomena-agnostic data in context, not decontextualizing it by recourse to phenomena.
6. Trade-offs of the varieties of data-centrism
While they are each suited to the circumstances in which they are deployed, there are trade-offs involved in implementing either form of data-centrism. In particular, the extensive decontextualization that makes the phenomena-laden approach useful for researchers working across contexts also has downstream impacts that are sometimes disadvantageous. The lack of decontextualization in phenomena-agnostic data-centrism helps the approach avoid these potentially pernicious downstream impacts, but it also may prevent data from traveling to and being recontextualized by certain interested stakeholders.
The complexities of managing large relational databases with extensive phenomena-laden ontologies seemingly promotes a centralization of data management, as is noted in model organism research (Leonelli Reference Leonelli, Atkinson, Glasner and Lock2009, Reference Leonelli2016, chap. 2). Not only does this centralization tend to result in dedicated teams of curators that specialize in data management—creating an efficient division of scientific labor—but it also promotes cooperation between various stakeholders, including academia, government agencies, and industry. This cooperation is important in model organism research in light of the special properties that bio-data can have. The commodification and enormous resulting financial value of model organism data, for example, in biomedicine or biofuel development, introduces intellectual property issues. Additionally, some data used in model organism research comes with privacy concerns, such as medical datasets containing protected health information or other forms of personally identifiable information. The centralization of databases that store this data helps implement uniform data-handling procedures that accord with the relevant regulations adhered to by various stakeholders.
But the success of phenomena-laden data-centrism does not come without its problems. As Leonelli (Reference Leonelli2016, chap. 6) details, database architecture and management practices can have downstream consequences that might be viewed as pernicious. For example, data that is not easily machine-readable is laborious to integrate into a large relational data architecture and thus is often excluded from databases all together. This creates a preference for a certain kind of data, and marginalizes researchers working with certain methods of analysis or outside the omics fields (e.g., DNA, RNA, and protein sequence databases). Analyses that focus on photos, films, field notes, focal animal observation data, or whole animal morphology and physiology are already being sidelined as databases become a dominant research tool.
These are not the only problems that may result from phenomena-laden data-centrism. The standards for data inclusion in a database may be articulated in ways that exclude the work of laboratories that lack adequate funding, particularly those in the Global South, further marginalizing some already marginalized research teams. Furthermore, as databases become the standard tool for organizing information, scientists become reliant on research methods that are promoted by database use at the expense of developing new methods. As Leonelli explains, “Such unequal participation in data journeys has epistemologically significant implications. Perhaps the most important of those is that online data collections tend to be extremely partial in the data that they include and package for travel” (Leonelli Reference Leonelli2016, 163). In short, the success of phenomena-laden data centrism may reduce epistemic pluralism while promoting a kind of scientific conservatism and bias.
One advantage of phenomena-agnostic data-centrism is that it does not seem to promote these pernicious downstream effects. Because the extent of decontextualization is minimal, phenomena-agnostic data-centrism does not face the myriad of decisions required to ontologize data to promote information integration. Thus, the management of data, or the packaging of large datasets in an economical way, can be distributed among various research groups. This allows a kind of flexibility. In regional climate modeling, the netCDF architecture can be used by anyone; it is not a resource with a gatekeeper who enforces standards for access or admission. It is unclear how the netCDF data architecture and the decisions made in its administration could be used in exclusionary ways; the pernicious downstream effects are seemingly minimal.
However, because phenomena-agnostic data-centrism minimizes decontextualization and assumes a high level of knowledge among its users, certain stakeholders face difficult hurdles when utilizing data. An example is that nonexpert users, when they successfully access data, often fail to properly interpret models, believing that output values are forecasts (predictions) rather than projections, or worse, that there is a single “right” model. More concretely, a city planner might desire to know the exact number of days the city will experience more than 90°F twenty years in the future, for example, and might access model data assuming it can give them that information. RCMs today cannot provide such precise predictions; rather they give ranges of values that support only inferences about means and trends. Even less-savvy users face other problems: Despite being freely available, the relative simplicity of RCM data archives may result in novice users struggling to search for relevant datasets and accessing the necessary meta-information to properly interpret what they find. Climate data, in part because its ontology and structure are not focused on integration across a diverse community, has less epistemic value for certain untrained users.
That certain stakeholders might struggle to use data can be seen as a shortcoming of phenomena-agnostic data-centrism: The lack of integration puts some users at a disadvantage. For example, climate scientists recognize that particular stakeholders may struggle to access and properly use climate data; this “usability gap” (Lemos, Kirchhoff, and Ramprasad Reference Lemos, Kirchhoff and Ramprasad2012) has led to the emergence of a subdiscipline called “climate services.” Climate service providers aim to work with certain stakeholders—often decision makers—to tailor climate data—often from RCMs—to the users’ needs (Parker and Lusk Reference Parker and Lusk2019; Lusk Reference Lusk2020). They thereby recontextualize data for a particular stakeholder problem on a case-by-case basis. Part of the work of these specialists may be using the data to identify and quantify phenomena, for example, droughts, for which stakeholders need to plan. Some of the work that is done by curators on phenomena-laden approaches may still need to be performed, but that work is divided and accomplished piecemeal under the phenomena-agnostic approach. The path data need to travel to become useful for certain stakeholder projects might be significantly longer under phenomena-agnostic data-centrism. While each variety of data-centrism demonstrates a scientific focus on data management, the reasons for this focus, and the resulting paths of data travel, are significantly different.
7. Prescriptive recommendations for identifying phenomena in climate data
Data-centric practices are evolving as some tools for data analysis, such as ML and AI, become more useful. In this section, we detail a new concern for phenomena-agnostic data-centrism that it had thus far largely avoided, in contrast to the phenomena-laden variety, but which arises from this quite different cause.
Leonelli maps out some risks of the influence of certain groups, individuals, and institutions to determine the field’s ontologies, categories, and theories. These frameworks serve to organize big data and thus determine much about downstream research. On Leonelli’s account, in biology, these downstream researchers, who are typically other biologists, are often tightly restricted by those ontologies in their investigations into nature, steered only down certain pathways, and forbidden down others, with some negative effects on both the science and its overall fairness, as we discussed in the preceding text. Biologists have conferences and workshops to get users, theoreticians, and curators all together to discuss the ontologies, to both spread the word and mitigate any harms that might arise from them. Nevertheless, there are significant risks that remain, argues Leonelli.
A possible parallel risk applies to data-centrism in climate science, especially in analysis of climate model outputs. Clearly, the outputs of numerical experiments are rather different from the outputs of biological experiments. And currently, data managers and curators of, for example, regional model outputs are not in the habit of imposing much ontology, classification, or interpretation, that is, identifying phenomena and events within the dataset, and so forth, on their output data beyond the identification of variables, parameters, and parameterizations, as well as key instructions to make sense of the model output.
In other words, if a downstream user of the data wants to know what the atmospheric rivers are doing, or how many hurricanes occurred in the history of the system over a span of time, the user must identify phenomena in the data; the data curators do not define them. Thus, data curators are doing little of the sort of ontologizing that Leonelli has defined as socially, politically, or scientifically risky and biased or partial.
But there are potential risks bearing down on climate science in the future. As big data tools are developed and implemented, and it becomes easier to pick out entities and define ontologies through AI and ML, there will be ever-increasing risk of imposing just the kinds of error, bias, and partiality warned against by Leonelli in the biological setting.
For instance, take the recent developments and advancements in the detection of hurricanes from climate model data. ML algorithms are being developed to detect hurricanes from climate model output, a much desired and valued goal of modelers and end users.
Why is this change so desirable? Partly because it expands the evidential value of data, because users with lower levels of training or lack of access to theoretical models or tools can access the data and apply them to their own projects at the civic or environmental project level. ML algorithms are advantageous in that such discoveries would reduce the time required to find these phenomena in the large datasets climate models produce, in some cases dramatically. One does not have to take the time to find the events of interest, one can just leap ahead to asking questions about those events.
We can picture automated identification of heat waves, droughts, and atmospheric rivers in climate model outputs as ML is adopted in climate science. But taking note from Leonelli’s analysis, that ease comes with risks: Who will define the parameters of what counts as a drought, heat wave, or atmospheric river? Which datasets will be used to train the algorithms, and what consequences does that have for real-world applications? It makes a difference whether an algorithm comes up with a heat wave, drought, or atmospheric river in a particular context or contexts, especially for adaptation purposes. There is also the problem of users taking some of the models at face value; they might not present an accurate or good representation of surface rainfall, but the user sees such representation as “realistic,” and takes it as such.
Taking responsibility for mitigating the scientific, social, and political risks and trade-offs, detailed in the preceding text, that might be associated with faulty or biased definitions of climate phenomena falls with the community that is building the big data processing plus its users, both professional and downstream. In general, it would seem that the more centralized and standardized the organization of data using phenomena is, the greater the chance that potentially valuable minority views or practices may be excluded from the research conversation. Centralization and standardization, however, may also afford additional efficiencies. A more distributed and egalitarian approach to phenomena identification might bring epistemic advantages associated with a diversity of views, but it may not be as efficient and possibly could result in confusion or inconsistency when results are presented.
The strategy in model organism research to address this problem was to convene conferences and workshops to come to consensus on definitions (and synonyms) of fundamental concepts that would be used in data ontologies (Leonelli Reference Leonelli2016, chap. 1). These conferences included users and institutional representatives to develop responsible, responsive ontologies and working definitions that can periodically be reviewed and updated as needed. We applaud this kind of effort, and a similar level of inclusiveness should characterize efforts in regional climate modeling.
The Atmospheric River Tracking Model Inter-comparison Project (ARTMIP), coorganized by researchers at NCAR (National Center for Atmospheric Research) and NOAA (National Oceanic and Atmospheric Administration) (UCAR 2019), may be taken as an exemplar of just the sort of thing we have in mind. It might be that a variety of definitions should be used and tested in a variety of settings, using a variety of techniques or algorithms, before a single standard is settled on; it will be important to ensure that the training data is available to the users for these purposes. These are issues that could be discussed at length at the planning workshops. What is most urgent is vigilance and awareness concerning these various risks, and an avoidance of a naïve and “gee whiz” or “Wild West” approach to big data processing, AI, and ML, despite its power—or rather, because of it.
8. Conclusion
The dominant narrative about big data is that it represents a new step in the evolution of science. Though there is nothing new about a concern for data in science, new tools and capabilities have increased the value of data and made data-centric science viable. But as we have shown, data-centrism is not monolithic.
Building on Leonelli’s framework, we argued that understanding how big data is harnessed by science requires investigating the information architectures of systems that organize, store, and disseminate data. Applying this method, we identified two data-centric approaches—phenomena-laden and phenomena-agnostic—as distinct species. This insight adds to our understanding of the role of phenomena in science: In one species, categorizing the data in terms of phenomena helps it travel to new contexts, while, in the other, phenomena are absent, and travel is catalyzed by other means.
Each of these species of data-centrism entails trade-offs. On one hand, the phenomena-laden approach requires curators to engage in extensive ontologizing to decontextualize data. Diverse users can thus easily access information, but there is a risk that curators implicitly steer downstream research. On the other hand, users are expected to search datasets in phenomena-agnostic approaches more directly, relying on shared knowledge to select and apply them, including identifying phenomena after dataset acquisition. Nonexpert users may struggle to employ data, but the risks associated with the steering of downstream research are diminished. Thus, the success of data-centric science, and who can easily utilize its products, is heavily shaped by the data practices employed. There is no single “big data” or “data-centrism,” and no single narrative to describe the expanded reliance on data in science.
Varieties of data-centric science exist because the opportunities and challenges presented by big data vary across scientific domains. Information architectures and data-packaging practices designed for model organism biology, in which many datasets are produced (velocity) with significant heterogeneity (variety), are not optimal for regional climate modeling, in which there are few and relatively homogenous datasets each of massive scale (volume). Harnessing the advantages of big data and facilitating the sharing of data needs to be done in domain-specific ways that address the specific conditions that scientists and other stakeholders face.
Looking forward, we expect data-centric science to further evolve through advancements in AI and ML. We briefly examined the potential use of these tools in climate science, noting that the looming implementation of ML models for identifying phenomena could introduce new vulnerabilities as well as capabilities and suggested some amelioration strategies. We believe there is more work to be done to understand the vulnerabilities of different forms of data-centrism and how they can be overcome. The identification of distinct types of data-centrism is a step forward in illuminating both the benefits and risks of big data in science.
Acknowledgments
This paper is based on research that was funded by the National Science Foundation (NSF) Award Number 1754740: A Methodological Study of Big Data and Atmospheric Science. We would also like to thank the National Center for Atmospheric Research [NCAR] Affiliate Scientist program for their support. We would like to thank the Mearns research group at NCAR in Boulder, Colorado for hosting us and contributing their space, time, and ideas. Thanks are also due to Ryan O’Loughlin, research assistant on the project, the philosophers of science at Michigan State, as well as Monica Morrison and Suzanne Kawamleh for their feedback.