Cholera remains a serious and significant bacterial disease in developing countries, including many African countries. Eight pandemics of cholera have been reported since 1817 [Reference Sack1]. The ongoing seventh pandemic started in 1961 in Asia and eventually reached the continent of Africa in 1970 [Reference Sack1]. Cholera then quickly spread throughout Africa and it is now endemic over a vast area of the continent. Africa has in recent years contributed more than 80% of the total cholera cases worldwide [Reference Naidoo and Patric2]. Angola and Namibia are countries located along the South-western coastline of Africa. To date, there has been no published data of cholera occurring in Namibia. The opposite situation exists in Angola, which is Namibia's immediate northern neighbour. Angola has a large public health problem due to cholera, with continuous cholera outbreaks occurring. As at 19 June 2006, Angola had reported a total of 46758 cases of cholera that included a daily incidence of 125 cases being reported [3]. In the present study, we report on the first outbreak of cholera in Namibia. We report on the serotype, biotype and antimicrobial susceptibility profile of the Vibrio cholerae isolates and investigate the genotypic relatedness of the strains.
From December 2006 to February 2007, Namibia experienced an outbreak of cholera in the North-western parts of the country, within the Omusati and Kunene provinces, which immediately borders the country of Angola. Unfortunately very little information is available regarding the outbreak. We do know that more than 250 cases of cholera were reported amongst Namibian and Angolan citizens living in the area, which included more than seven deaths. During this outbreak period, the Enteric Diseases Reference Unit (EDRU) of the National Institute for Communicable Diseases (NICD) in South Africa received nine bacterial isolates for confirmation of cholera diagnosis. The isolates were cultured from stool specimens of patients (aged between 2 and 55 years) who all presented with symptoms of diarrhoea. Bacterial isolates were confirmed as V. cholerae using standard microbiological identification techniques [Reference Winn4]. Serogrouping and serotyping were performed by the slide agglutination method with polyvalent antisera and mono-specific Inaba and Ogawa antisera, according to the manufacturer's instructions (Murex Biotech Ltd, Dartford, UK). Determination of biotype and detection of the cholera enterotoxin was performed by polymerase chain reaction (PCR) using a previously described method [Reference Keasler and Hall5]. For this PCR, we used primers as described in the above method and employed 28 cycles of 95°C for 1 min, 60°C for 1 min and 72°C for 1 min. The template for PCR was purified genomic DNA and was prepared using a previously described method [Reference Smith6]. Susceptibility testing to antimicrobial agents (ampicillin, augmentin, trimethoprim, sulfamethoxazole, chloramphenicol, streptomycin, erythromycin, tetracycline, kanamycin, imipenem, nalidixic acid, ciprofloxacin, ceftriaxone and ceftazidime) was determined by the E-test (AB Biodisk, Solna, Sweden), according to the manufacturer's instructions. The genotypic relatedness of the outbreak strains was investigated by pulsed-field gel electrophoresis (PFGE) analysis of digested genomic DNA using a PulseNet standardized protocol [Reference Cooper7]. For PFGE analysis, we performed separate digestion and analysis with SfiI and NotI restriction enzymes. The CHEF-DR III system (Bio-Rad Laboratories Inc., Hercules, CA, USA) was used for PFGE analysis, with an electrophoresis gradient of 6 V/cm and a 2-block run programmed as follows. Block 1 included an initial switch time (IST) of 2 s to a final switch time (FST) of 10 s, over a run time of 13 h. Block 2 included an IST of 20 s to a FST of 25 s, over a run time of 7 h.
All nine bacterial isolates from this diarrhoeal disease outbreak were phenotypically identified as V. cholerae O1 serotype Inaba. They were further identified as the El Tor biotype, using a multiplex PCR technique. This biotyping PCR targets the tcpA gene of V. cholerae, which encodes the toxin co-regulated pilus. Nucleotide sequence differences exist in the tcpA gene from classical and El Tor strains and the PCR exploits these sequence differences in order to diagnose a biotype. The presence of cholera enterotoxin amongst the isolates was confirmed using a PCR technique which determined the presence of the ctxA gene. Susceptibility testing to antimicrobial agents revealed the following results. Isolates were susceptible to [minimum inhibitory concentrations (MICs) given in parentheses]: ampicillin (2–4 μg/ml), augmentin (2–4 μg/ml), chloramphenicol (8 μg/ml), nalidixic acid (0.38–0.75 μg/ml), ciprofloxacin (0.002 μg/ml), tetracycline (0.38–0.75 μg/ml), kanamycin (4–6 μg/ml), imipenem (0.75 μg/ml), ceftriaxone (0.016 μg/ml) and ceftazidime (0.19–0.125 μg/ml). Isolates were non-susceptible to erythromycin (0.75–1 μg/ml). Isolates were resistant to: trimethoprim (>32 μg/ml), sulfamethoxazole (>1024 μg/ml) and streptomycin (64–96 μg/ml). PFGE analysis incorporating SfiI digestion revealed an identical pattern for all nine outbreak strains (Fig.). Analysis with a second restriction enzyme (NotI) also revealed an identical pattern for all strains. These genotypic fingerprint data, together with results indicating identical antimicrobial susceptibility profiles for all isolates, determined that the outbreak was caused by a single strain of V. cholerae O1 serotype Inaba biotype El Tor.
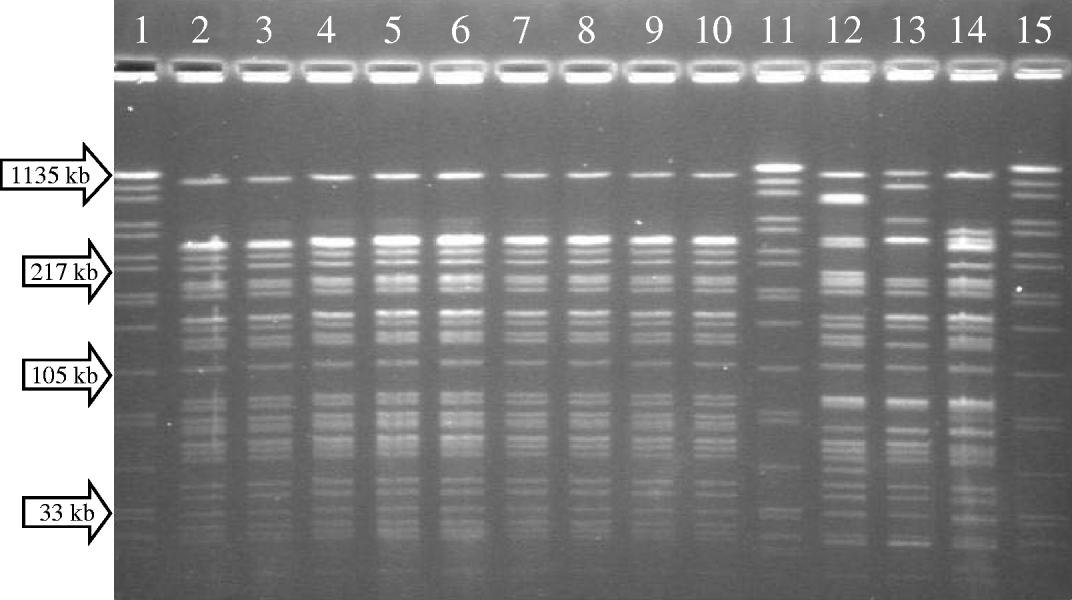
Fig. PFGE fingerprint patterns (SfiI digestion) of cholera isolates. Lanes 2–10, outbreak isolates from Namibia; lanes 12–14, unrelated isolates from South Africa; lanes 1, 11 and 15, reference standard (Salmonella Braenderup strain H9812 digested with XbaI).
To the best of our knowledge, this is the first published report of cholera from Namibia. The seventh pandemic of cholera has now reached Namibia. This outbreak occurred in an area of Namibia which immediately borders Angola. Initial cases were reported amongst Angolan citizens (L. de Wee, personal communication). The first case was reported from an Angolan citizen who was treated at the border post clinic of Otjimuhaka which serves people from Angola and Namibia [8]. It is common practice for Angolan citizens living near the border to receive treatment at Namibian clinics, as there are no medical facilities in the Angolan border region. Cholera is endemic within Angola and there are continuous reports of cholera from this country [3]. These data suggest that the Namibian cholera outbreak may have been imported from Angola. Unfortunately, this conclusion cannot be supported by scientific evidence as no Angolan isolates were available for analysis.
ACKNOWLEDGEMENTS
This work was financially supported by the National Health Laboratory Service.
DECLARATION OF INTEREST
None.



