INTRODUCTION
Enterococci are important nosocomial infection pathogens with Enterococcus faecalis and E. faecium being the most prevalent in humans. The spread of vancomycin-resistant enterococci (VRE), i.e. vanA- or vanB-positive E. faecalis or E. faecium, in the hospital environment may make it difficult to treat infections and leads to an increased risk of mortality and higher costs associated with prolonged stay of patients in hospitals [Reference Schouten1–Reference Werner3].
The motile enterococci, E. gallinarum and E. casseliflavus/flavescens, are characterized by the presence of the vanC gene cluster and show low-level resistance to vancomycin [Reference Vincent4, Reference Leclercq5]. However, acquisition of a vanA or vanB gene cluster results in high-level resistance to vancomycin [Reference Dutka-Malen6]. E. gallinarum and E. casseliflavus/flavescens have been shown to colonize the intestinal tracts of both hospitalized and non-hospitalized individuals [Reference Van Horn and Rodney7, Reference Toye8] but they are not considered to be important factors in nosocomial infection control. Since drug susceptibility testing is rarely implemented for isolates from non-sterile sites, they are sporadically detected during surveillance of VRE [Reference Patel9–Reference Biavasco12]. There are few reports of infections by vanA- or vanB-positive E. gallinarum and little data regarding their clinical importance. Two cases of sepsis due to vanA-positive E. gallinarum have been described [Reference Biavasco13, Reference Merquior14] and two outbreaks in Argentina and Brazil have been reported [Reference Corso15, Reference Neves16]. There are no epidemiological reports regarding regional spread of vanA- or vanB-positive E. gallinarum.
In 2005, an outbreak occurred in a hospital located in the southern district of Kyoto City in which more than 100 faecal carriers of vanA-positive E. faecium were detected. Following this outbreak, we began conducting regional surveillance of VRE annually and promoted screening of clinical faecal samples using selective agar culture in hospitals in Kyoto prefecture. Consequently vanA- or vanB-positive E. gallinarum were detected in multiple hospitals and long-term care facilities (LTCFs) in this region. This paper reports the results of the surveillance exercise and the molecular characterization of the isolates recovered.
MATERIALS AND METHODS
Regional surveillance and sample collection
Kyoto prefecture (population about 2·6 million) is located in the middle of Japan. The capital, Kyoto City, has a population of about 1·5 million inhabitants accounting for 58% of Kyoto prefecture.
Two types of VRE surveillance were performed and prospectively presumptive VRE samples were collected from hospitals and LTCFs in this region. The first was an annual surveillance from 2005 to 2008 in which about 100 hospitals and 40–60 LTCFs (accounting for more than 50% of prefectural hospitals and LTCFs) participated. The number of samples per facility was about 10% of capacity. The second approach was an enhanced VRE screening programme of clinical faecal samples through the collaboration of hospitals and clinical reference laboratories. This programme commenced in 2006 and was performed during the same period as the annual surveillance; about 60% of prefectural hospitals participated but LTCFs were not involved in this programme.
Patients who met more than one of the following criteria were selected: urinary and/or faecal incontinence; had nasogastric feeding tubes or had undergone gastrostomy; presence of urethral catheters; received antimicrobial chemotherapy within the previous 2 weeks, or had undergone surgery within 1 month. Each patient was assigned by facility personnel and not identified by name.
Microbiological methods
In the annual surveillance, rectal swabs or faecal samples were inoculated into 10 ml bile aesculin azide broth containing 15 μg/ml vancomycin (Nissui Pharmaceutical Co. Ltd, Japan). Preliminary experiments had confirmed that multiple strains of vanB-positive E. faecalis, with minimum inhibitory concentrations (MICs) of 4–8 μg/ml, grew well in this broth. After incubation at 35°C for 48 h, the broth samples with dark brown or black discoloration were streaked on VRE selective agar® (Nippon Becton, Dickinson and Company, Japan) containing 32 μg/ml vancomycin and vanA/vanB inducing agents [Reference Uzawa17]. Samples from the enhanced clinical surveillance were streaked directly on the VRE selective agar.
Presumptive VRE isolates from the selective agar were subcultured to 5% sheep blood agar (Eiken Chemical Co. Ltd, Japan). Seven primer sets targeting the genes vanA, vanB, vanC1, vanC2/C3, E. faecalis-specific, E. faecium-specific, and rrs (16S ribosomal RNA) were used for multiplex PCR as previously described [Reference Kariyama18, Reference Elsayed19]. The species were identified by a motility test, production of a yellow pigment [Reference Leclercq5, Reference Cartwright20, Reference Clark21], and multiplex PCR. E. gallinarum was confirmed if an isolate was motile and vanC1-positive.
Pulsed-field gel electrophoresis (PFGE)
The first isolate from each facility was subjected to typing by PFGE using SmaI enzyme (New England Biolabs, USA) as previously described [Reference Turabelidze22, Reference Duck23]. Electrophoresis was performed in a Genepath System (Bio-Rad, USA) with pulse times increasing from 1·0 to 14·0 s for 18·5 h at 200 V (6 V/cm). Genetic relatedness was analysed with the aid of Gel Compar II (Applied Maths, Belgium). Dendrograms of percentage similarity were calculated using Pearson's correlation coefficient and represented by the unweighted pair-group method with mathematical averages algorithm. A cut-off of 92% similarity was set to cluster strains as belonging to the same clone, according to Morrison et al. [Reference Morrison24].
As a control for genetically related bacterial strains, six isolates of both vanA- and vanB-negative E. gallinarum detected during this surveillance period were used. Three isolates were recovered in Kyoto prefecture, and the others were each from different prefectures in Japan (Osaka, Miyagi, Fukuoka).
Antimicrobial susceptibility testing
Isolates were tested for susceptibility (MIC) to ampicillin, erythromycin, vancomycin and levofloxacin (Eiken Chemical Co. Ltd.) using a microdilution method according to Clinical and Laboratory Standards Institute (CLSI) guidelines [25].
RESULTS
Regional surveillance and bacterial strains
The number of samples collected during annual surveillance was 2872 in 2005, 2451 in 2006, 2406 in 2007, and 2735 in 2008; for the enhanced clinical laboratories screening programme the number of samples was 11 820 in 2006, 17 184 in 2007 and 14 748 in 2008.
Table 1 shows that 88 patients with vanA-positive E. gallinarum were found in 11 hospitals and one LTCF and 10 patients with vanB-positive E. gallinarum were detected in eight hospitals. Multiple patients were identified in ten hospitals: eight hospitals had 2–57 patients with vanA-positive E. gallinarum, and two hospitals each had two patients with vanB-positive E. gallinarum; both van genotypes were found in two hospitals. Seven of the 11 hospitals with vanA-positive E. gallinarum also harboured patients with vanA-positive E. faecium and in four of the eight hospitals with vanB-positive E. gallinarum, vanB-positive E. faecium was also detected concurrently during the study period.
Table 1. Number of isolates of vanA- or vanB-positive E. gallinarum in Kyoto prefecture
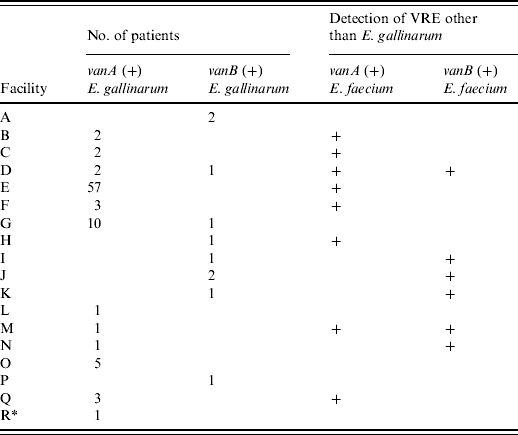
Numbers indicate the number of patients with vanA- or vanB-positive E. gallinarum.
+ Indicates the facilities in which vanA- or vanB-positive E. faecium were concurrently detected during the study period.
* Indicates long-term care facility (facility R).
Figure 1 shows that during the 3 years following an outbreak of vanA-positive E. faecium at facility E in 2005, carriers of vanA- or vanB-positive E. gallinarum were found in multiple hospitals. The latter genotyopes were not detected in 2005, but in 2006, they began to be recovered in the South district of Kyoto City which includes the hospital where the first outbreak of VRE occurred. Spread of these genotypes was evident through the northern district of Kyoto City in 2007, and subsequently outside the city.
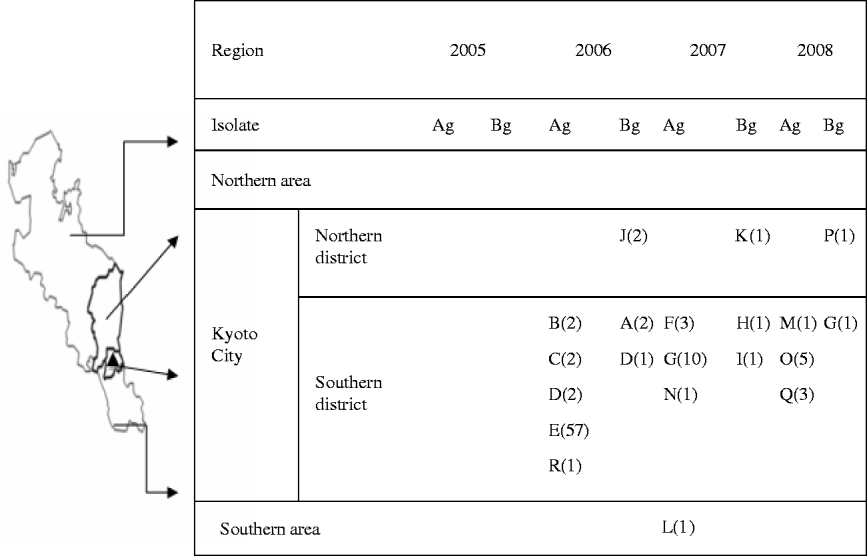
Fig. 1. Regional spread of vanA- or vanB-positive E. gallinarum after first outbreak of vanA-positive E. faecium. ▴, The first outbreak of vanA-positive E. faecium occurred at facility E in 2005. A–R indicates the number of facilities; values in parentheses indicate the number of patients with vanA- or vanB-positive E. gallinarum; Ag, vanA-positive E. gallinarum; Bg, vanB-positive E. gallinarum.
PFGE typing and antimicrobial susceptibility
Figure 2 shows the PFGE profiles of 23 isolates from Kyoto facilities and three controls of vanA- or vanB-positive E. gallinarum from each facility; 12 isolates were vanA-positive, eight vanB-positive and six were negative for both elements. Eleven clones were distinguished with the largest single group consisting of 14 isolates (11 vanA-positive, three vanB-positive). The other five vanB-positive isolates fell into four clones. These clones along with three vanA- and vanB-negative isolates (P, S, N) were clearly distinguishable from the predominant clone; two of the latter isolates clustered together on the dendrogram. The control vanA- and vanB-negative isolates from other prefectures were distinct from the major clone.

Fig. 2. PFGE profiles and antimicrobial resistance patterns of Enterococcus gallinarum isolates included in the study. * Other regions in Japan. AMP, Ampicillin; ERY, erythromycin; LVF, levofloxacin.
Eighteen of 20, i.e. all vanA-positive E. gallinarum isolates and six of eight vanB-positive E. gallinarum isolates, showed characteristic susceptibility patterns being susceptible to ampicillin and resistant to both erythromycin and levofloxacin. All vanA- or vanB-positive E. gallinarum isolates were resistant to vancomycin.
DISCUSSION
E. gallinarum organisms carrying vanA or vanB genes have been detected in various environments (e.g. soil, water) and animals (e.g. chickens, other poultry, pigs) [Reference Biavasco12, Reference Messi26, Reference Roberts27]. Only a few isolates of E. gallinarum carrying vanA or vanB genes have been identified in human faecal samples during and/or after outbreak surveillance of non-motile VRE [Reference Patel9–Reference Biavasco12]. To our knowledge, two cases of sepsis due to vanA-positive E. gallinarum [Reference Biavasco13, Reference Merquior14] and two possible outbreaks with this genotype have been reported [Reference Corso15, Reference Neves16]. One described 15 isolates from an intensive care unit (ICU) of a hospital in Argentina comprising two clonal types defined by SmaI PFGE [Reference Corso15] and the other described seven isolates of the same clone from faecal carriers over 3 months in a university hospital in Brazil [Reference Neves16].
This report is to our knowledge the first to document regional spread of vanA- or vanB-positive E. gallinarum. This was an unexpected finding following an outbreak of vanA-positive E. faecium. Despite there being no evidence of gene transfer in this region, previous reports have described vanA genes as being transmissible between E. faecium and E. gallinarum [Reference Corso15, Reference Foglia28].
We have no evidence of the movements of individual VRE-positive patients, since we were not able to follow the course of each individual VRE carrier in this study. However, circumstantial evidence suggests that the spread of VRE from facility E, one of the core hospitals in Kyoto City had occurred as the transfer of patients to or from this facility is routine. Further, in multiple hospitals or LTCFs included in this study, some VRE-positive patients were confirmed to have been transferred from facility E. We acted as infection control consultants in facility E and advised intensive screening and hygienic precautions. However, large-scale, ongoing patient movements in this hospital made it difficult and time-consuming to successfully prevent or even curtail the movements of VRE carriers, resulting in regional spread from this centre during the surveillance period.
Since E. gallinarum has been shown to colonize the intestinal tract, drug susceptibility testing is rarely conducted for isolates from non-sterile sites. The vanC1 gene is intrinsic to E. gallinarum in which it mediates low-level resistance to vancomycin [Reference Leclercq5]. Even when vancomycin-resistant E. gallinarum is detected, we usually do not conduct susceptibility testing or PCR for of resistance genes and therefore may overlook vanA- and/or vanB-resistance elements in this species.
The motile enterococci account for 3–8% of all enterococcal bacteraemia cases [Reference Reid, Cockerill and Patel29, Reference Choi30] and severe infections of various body sites due to E. gallinarum have been reported, and vanA- and vanB-positive strains can be relatively difficult to treat [Reference Reid, Cockerill and Patel29–Reference Koganemaru and Hitomi33]. Infection control measures are required to stem the spread of these genotypes. Moreover, as E. gallinarum may act as a reservoir for the vanA or vanB gene, motile enterococci should be screened for the presence of these genes, especially in regions or facilities where VRE is endemic.
This study has limitations. We collected faecal samples from anonymous patients, and thus obtained no information about their clinical backgrounds, such as whether any of the cases were carriers or had symptomatic infections. We also could not follow patient transfers between hospitals or LTCFs. Therefore, we were unable to determine whether or not vanA- or vanB-positive E. gallinarum were spread by direct transfer. Despite these limitations, this study confirms the regional spread of these organisms and emphasizes the need for their surveillance.
ACKNOWLEDGEMENTS
We express our appreciation to Dr Haruyoshi Tomita, Dr Shuhei Fujimoto, and Dr Yasuyoshi Ike, Gunma University Graduate School of Medicine for helpful advice and discussions. We also express our appreciation to the following individuals for providing the bacterial strains used in this study: Dr Hiroyuki Kunishima, Tohoku University Hospital; Dr Masayuki Murata, Kyushu University Hospital; Mr Nobuyoshi Tamagawa, Osaka City General Hospital. This study was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology (Kiban B, 18390172).
DECLARATION OF INTEREST
None.





