INTRODUCTION
The leatherback sea turtle (Dermochelys coriacea Vandelli, 1761) lives in ocean basins and shows high behavioural plasticity in habitat use and foraging strategies (Fossette et al., Reference Fossette, Girard, López-Mendilaharsu, Miller, Domingo, Evans, Kelle, Plot, Prosdocimi, Verhage, Gaspar and Georges2010). The species is widely distributed throughout tropical and subtropical regions of the world (López-Mendilaharsu et al., Reference López-Mendilaharsu, Rocha, Miller, Domingo and Prosdocimi2009), and can be found in temperate waters while feeding (Fossette et al., Reference Fossette, Girard, López-Mendilaharsu, Miller, Domingo, Evans, Kelle, Plot, Prosdocimi, Verhage, Gaspar and Georges2010; Stewart et al., Reference Stewart, James, Roden and Dutton2013). The global population of the leatherback turtle encompasses seven Regional Management Units (or subpopulations – IUCN, 2017) defined by nine mitochondrial DNA stocks and 652 rookeries among the Pacific, Atlantic and Indian Oceans (Wallace et al., Reference Wallace, DiMatteo, Hurley, Finkbeiner, Bolten, Chaloupka, Hutchinson, Abreu-Grobois, Amorocho, Bjorndal, Bourjea, Bowen, Duenas, Casale, Choudhury, Costa, Dutton, Fallabrino, Girard, Girondot, Godfrey, Hamann, López-Mendilaharsu, Marcovaldi, Mortimer, Musick, Nel, Pilcher, Seminoff, Troeng, Witherington and Mast2010, Reference Wallace, DiMatteo, Bolten, Chaloupka, Hutchinson, Abreu-Grobois, Mortimer, Seminoff, Amorocho, Bjorndal, Bourjea, Bowen, Briseno Duenas, Casale, Choudhury, Costa, Dutton, Fallabrino, Finkbeiner, Girard, Girondot, Hamann, Hurley, López-Mendilaharsu, Marcovaldi, Musick, Nel, Pilcher, Troeng, Witherington and Mast2011). Subadult and adult animals play an important role in maintaining ocean health by feeding on and controlling populations of jellyfish (Purcell et al., Reference Purcell, Uye and Lo2007).
Leatherback turtles exhibit complex behaviour, with long migrations between nesting and feeding areas. Most of the worldwide genetic studies of this species have focused on nesting areas (Dutton et al., Reference Dutton, Bowen, Owens, Barragan and Davis1999, Reference Dutton, Hitipeuw, Zein, Benson, Petro, Pita, Rei, Ambio and Bakarbessy2007, Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013; Carreras et al., Reference Carreras, Godley, Leon, Hawkes, Revuelta, Raga and Tomas2013; Molfetti et al., Reference Molfetti, Vilaça, Georges, Plot, Delcroix, Le Scao, Lavergne, Barrioz, dos Santos and de Thoisy2013). These studies analysed data collected from females when laying eggs in the sand, which poses fewer methodological challenges and is less expensive than sampling in oceanic areas. Only one study has focused on the genetic diversity of leatherback turtles at sea in the North Atlantic (Stewart et al., Reference Stewart, LaCasella, Roden, Jensen, Stokes, Epperly and Dutton2016) and three on turtles in the South Atlantic (Vargas et al., Reference Vargas, Araujo, Monteiro, Estima, Almeida, Soares and Santos2008, Reference Vargas, Molfetti, Vilaça, Monteiro, Estima, Soares, Almeida, Thoisy, Naro-Maciel and Santos2013; Prosdocimi et al., Reference Prosdocimi, Dutton, Albareda and Remis2014).
The South-west Atlantic Ocean leatherback subpopulation regularly nests only in Espírito Santo, south-eastern Brazil, between latitudes 19°40′S and 18°25′S (Thomé et al., Reference Thomé, Baptistotte, Moreira, Scalfoni, Almeida, Rieth and Barata2007). Occasional nests are recorded in other states, such as Piauí, Rio Grande do Norte, Bahia, Rio de Janeiro, São Paulo, Santa Catarina and Rio Grande do Sul (Barata & Fabiano, Reference Barata and Fabiano2002; Loebmann et al., Reference Loebmann, Alves Legat, Puchnick Legat, De Camargo, Erthal, Severo and De Goes2008; Bezerra et al., Reference Bezerra, Bondioli, Maistro and Ebert2014). The Espírito Santo rookery is characterized by its small population size, with the number of nests laid per year varying between 6 and 92, and an estimated 1 to 19 nesting females per year (Thomé et al., Reference Thomé, Baptistotte, Moreira, Scalfoni, Almeida, Rieth and Barata2007).
Additionally, leatherback turtles can be seen along the Brazilian coastal and offshore waters, and individuals are found stranded on beaches along the entire Brazilian coast (Barata & Fabiano, Reference Barata and Fabiano2002). Records from 1995 to 2014 show that 376 leatherback turtles stranded on beaches only in Rio Grande do Sul State, southern Brazil, along a 355 km stretch of beach (Monteiro et al., Reference Monteiro, Estima, Gandra, Silva, Bugoni, Swimmer, Seminoff and Secchi2016). For Espírito Santo State, in south-east Brazil, there are 27 records of animals stranded on beaches or found dead in nets from 1991 to 2014 (TAMAR/SITAMAR database). There have been less frequent records of stranded animals found in Parana (D'Amato, Reference D'Amato1991), Rio Grande do Norte (Sanches et al., Reference Sanches, Bellini and Silva Neto1999), Ceará, Bahia (Barata et al., Reference Barata, Lima, Borges-Martins, Scalfoni, Bellini and Siciliano2004), Rio de Janeiro and São Paulo (Lima et al., Reference Lima, Melo and Godfrey2007; TAMAR/SITAMAR database), Maranhão (Santana et al., Reference Santana, Lima-Junior, Aragão, Silva, Silva-Leite, Leite-Junior and Leite2007) and Pará States (Figueredo et al., Reference Figueredo, Carmo, Albuquerque and Pezutti2008). The large number of recorded strandings cannot be accounted for by the small number of females nesting per year in Brazil, and so might comprise animals from a number of different rookeries (Barata et al., Reference Barata, Lima, Borges-Martins, Scalfoni, Bellini and Siciliano2004; Sales et al., Reference Sales, Giffoni and Barata2008).
The origins of individuals found outside the nesting areas are largely unknown for leatherback turtles. There have been only three studies analysing samples from by-catch and stranded animals in the South Atlantic Ocean: in southern Brazil and Buenos Aires province, Argentina (Vargas et al., Reference Vargas, Araujo, Monteiro, Estima, Almeida, Soares and Santos2008, Reference Vargas, Molfetti, Vilaça, Monteiro, Estima, Soares, Almeida, Thoisy, Naro-Maciel and Santos2013; Prosdocimi et al., Reference Prosdocimi, Dutton, Albareda and Remis2014). Following the first of these studies, which used mixed-stock analysis to examine the natal origins of the sampled individuals (Vargas et al., Reference Vargas, Araujo, Monteiro, Estima, Almeida, Soares and Santos2008), there have been other studies analysing samples from nesting areas. These studies involved further source populations, larger sample sizes and longer mitochondrial DNA sequences (Carreras et al., Reference Carreras, Godley, Leon, Hawkes, Revuelta, Raga and Tomas2013; Dutton et al., Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013; Molfetti et al., Reference Molfetti, Vilaça, Georges, Plot, Delcroix, Le Scao, Lavergne, Barrioz, dos Santos and de Thoisy2013). The growing database of DNA sequences from leatherback turtles has been important for improving the power of mixed-stock analyses and for informing conservation programmes for this species.
Leatherback turtles are considered critically endangered in Brazil (Almeida et al., Reference Almeida, Eckert, Bruno, Scalfoni, Giffoni, López-Mendilaharsu and Thomé2011). The long-term conservation work carried out by Projeto Tamar (the Brazilian Sea Turtle Conservation Programme) increased nest numbers in recent years, with the nearly complete cessation of egg harvesting and turtle take, together with nest protection, management and enlargement of protected areas. New threats are emerging, however, such as the high levels of incidental captures in fisheries in Ceará, Espírito Santo, São Paulo, Santa Catarina and Rio Grande do Sul States (Sales et al., Reference Sales, Giffoni and Barata2008; Fiedler et al., Reference Fiedler, Sales, Giffoni, Monteiro-Filho, Secchi and Bugoni2012), and also the coastal development seen in Espírito Santo (Almeida et al., Reference Almeida, Eckert, Bruno, Scalfoni, Giffoni, López-Mendilaharsu and Thomé2011; da Silva et al., Reference da Silva, Mitraud, Ferraz, Lima, Melo, Santos, da Silva, de Castilhos, Batista, Lopez, Tognin, Thomé, Baptistotte, da Silva, Becker, Wanderline, de Vasconcellos Pegas, Róstan, dei Marcovaldi and dei Marcovaldi2016).
Here we present a genetic analysis of leatherback turtles in the Atlantic Ocean, with a focus on the Brazilian rookery. We aim to: (i) describe the genetic diversity of the Brazilian rookery and the southern Brazil pelagic aggregate; (ii) identify the natal origin of the samples from this aggregate; (iii) compare the natal origin of animals from Brazil with those from a nearby region (Buenos Aires, Argentina); (iv) infer migration patterns and connections among nesting and feeding areas for the species; (v) understand how the genetic diversity of the species is distributed throughout the Atlantic Ocean; and (vi) contribute to better-designed conservation management strategies for the species.
MATERIALS AND METHODS
Data collection
This study was approved by the Instituto Chico Mendes de Conservação da Biodiversidade, and conducted under SISBIO and Centro TAMAR-ICMBio licences #14122-10 and # 15962-1. Tissue samples from females nesting at the Espírito Santo rookery (Comboios and Povoação beaches) were collected during the 2008/2009 nesting season, which runs in the region during the austral summer (N = 2), building up to the ones (N = 11) reported by Vargas et al. (Reference Vargas, Araujo, Monteiro, Estima, Almeida, Soares and Santos2008). We took precautions to guarantee that living animals were sampled only once by tagging them with Inconel flipper tags.
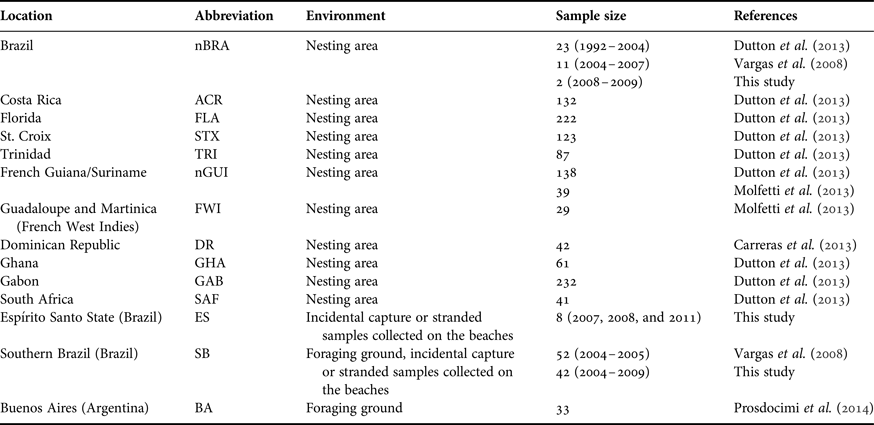
Additionally, samples were collected throughout the year, from animals that were either incidentally caught by fishnets or stranded on the beach. Eight samples were collected in Espírito Santo State, including five stranded on the beaches and three incidentally caught in fisheries’ nets. Another 42 samples were collected in south-eastern and southern Brazilian waters between January 2004 and November 2009. Of these, 40 were from individuals stranded on beaches and two were incidentally caught in fishing nets. We combined the new sequences with those published by Vargas et al. (Reference Vargas, Araujo, Monteiro, Estima, Almeida, Soares and Santos2008) (collected after 2004) and Dutton et al. (Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013) (collected before 2004). The total number of samples was 36 from nesting females and 94 from foraging grounds (Table 1 and Figure 1).
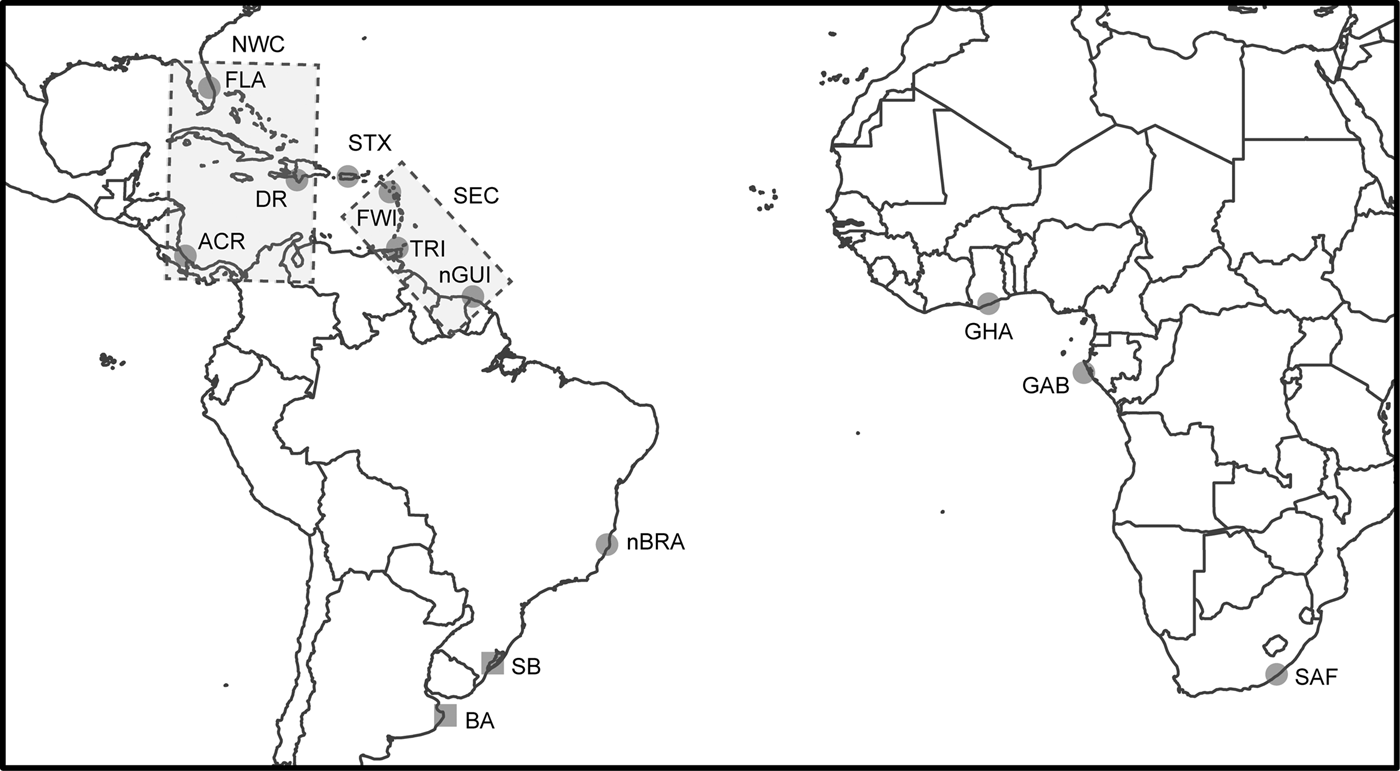
Fig. 1. Sampling locations of leatherback turtle rookeries (circles) and feeding grounds (squares) in the Atlantic and South-west Indian Oceans and the seven identified genetic stocks. Rectangles represent the genetic stocks involving multiple locations: North-west Caribbean (NWC) genetic stock including ACR, DR and FLA rookeries and South-east Caribbean (SEC) genetic stock including FWI, TRI and nGUI rookeries. The other five genetic stocks are represented by the same name as the rookery. Rookery abbreviations are defined in Table 1.
Table 1. Details of samples of leatherback turtles (Dermochelys coriacea) analysed in this study.
Fresh skin biopsies were taken from the individuals incidentally caught in fishing nets or from the anterior flipper of females during oviposition. Biopsies from skin and other tissues were also collected from dead individuals found on beaches. All samples were stored in 98% ethanol at room temperature, and total DNA was extracted as described by Lara-Ruiz et al. (Reference Lara-Ruiz, Lopez, Santos and Soares2006).
The primers LCM 15382, H950 (Abreu-Grobois et al., Reference Abreu-Grobois, Horrocks, Formia, Dutton, LeRoux, Vélez-Zuazo, Soares and Meylan2006) and HDCM1 (Allard et al., Reference Allard, Miyamoto, Bjorndal, Bolten and Bowen1994) were used to amplify the mitochondrial control region. Polymerase chain reaction (PCR) mixes of 12.5 ml included 2 µl of genomic DNA (~40 ng), 1 U of Taq polymerase (Phoneutria, Belo Horizonte, Brazil), 200 µM of dNTP, 1 × Tris-KCl buffer with 1.5 mM MgCl2 (Phoneutria), and 0.5 µM of each primer. The amplification programme consisted of 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 51°C, 1 min at 72°C, and a final extension step of 9 min at 72°C. PCR products were then processed and sequenced as previously described (Lara-Ruiz et al., Reference Lara-Ruiz, Lopez, Santos and Soares2006). We used Geneious 6.1.7 (Drummond et al., Reference Drummond, Ashton, Buxton, Cheung, Cooper, Duran, Field, Heled, Kearse and Markowitz2010) to align the sequences using a global alignment with free end gaps, and the consensus sequences were checked manually.
The data used for the population genetics and mixed-stock analysis regarding rookeries and feeding grounds in the Atlantic Ocean from areas outside Brazil were collected from the literature (Table 1; Molfetti et al., Reference Molfetti, Vilaça, Georges, Plot, Delcroix, Le Scao, Lavergne, Barrioz, dos Santos and de Thoisy2013 – GenBank accession numbers JX629739.1–JX629672.1; Dutton et al., Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013 – nos. HM452343.1–HM452352.1 and HM452364.1; Carreras et al., Reference Carreras, Godley, Leon, Hawkes, Revuelta, Raga and Tomas2013 – no. EF513272; Vargas et al., Reference Vargas, Araujo, Monteiro, Estima, Almeida, Soares and Santos2008; Prosdocimi et al., Reference Prosdocimi, Dutton, Albareda and Remis2014 – nos. EF513272.1–EF513278.1). Consequently, we used an alignment of the 695 base pair sequences (GenBank accession numbers: MF346872–MF346884) in further analyses to maintain compatibility with previous data.
Mixed-stock analysis
To quantify the relative contributions of the different genetic stocks in the Atlantic Ocean to the foraging grounds in Brazil and Argentina (southern Brazil + Buenos Aires, N = 127), we performed a mixed-stock analysis. We used a Bayesian algorithm with Markov chain Monte Carlo (MCMC) sampling and the ‘many-to-many’ hierarchical model (Bolker, Reference Bolker2012 – mixstock R package), which uses population sizes as priors. A total of 11 Markov chains were run for 20,000 steps each, with the first 10,000 steps treated as burn-in. To check for convergence we used Gelman-Rubin diagnostic implemented in the R package CODA (Plummer et al., Reference Plummer, Best, Cowles and Vines2006) and values below 1.1 were considered converged.
We included four new source populations that were not previously included in the dataset of Prosdocimi et al. (Reference Prosdocimi, Dutton, Albareda and Remis2014): French West Indies (FWI) population, which includes Guadeloupe and Martinique from Molfetti et al. (Reference Molfetti, Vilaça, Georges, Plot, Delcroix, Le Scao, Lavergne, Barrioz, dos Santos and de Thoisy2013); the new French Guiana/Suriname (nGUI) rookery, which includes the samples from Awala-Yalimapo beach (Dutton et al., Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013; Molfetti et al., Reference Molfetti, Vilaça, Georges, Plot, Delcroix, Le Scao, Lavergne, Barrioz, dos Santos and de Thoisy2013), from Matapica beach in Suriname (Dutton et al., Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013), and from Cayenne in East French Guiana (Molfetti et al., Reference Molfetti, Vilaça, Georges, Plot, Delcroix, Le Scao, Lavergne, Barrioz, dos Santos and de Thoisy2013); the new Brazilian (nBRA) rookery, which includes samples collected in Brazil before 2004 (Dutton et al., Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013) or after 2004 (Vargas et al., Reference Vargas, Araujo, Monteiro, Estima, Almeida, Soares and Santos2008), plus the two new samples collected during the 2008/2009 and 2009/2010 nesting seasons (this study); and the rookery from Dominican Republic (Carreras et al., Reference Carreras, Godley, Leon, Hawkes, Revuelta, Raga and Tomas2013).
The F ST values found among ACR, FLA and DR populations and among TRI, nGUI and FWI populations indicated genetic homogeneity (Table 3). Consequently, we combined these populations as North-west Caribbean (NWC) and South-west Caribbean (SEC) genetic stocks, respectively. This follows the approach used by Prosdocimi et al. (Reference Prosdocimi, Dutton, Albareda and Remis2014), but with the addition of one more rookery for each genetic stock. The number of females nesting per year in each population was determined from the literature, and this information was used to specify the prior (Table 2; Revuelta et al., Reference Revuelta, León, Feliz, Godley, Raga and Tomás2012; Dutton et al., Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013; Molfetti et al., Reference Molfetti, Vilaça, Georges, Plot, Delcroix, Le Scao, Lavergne, Barrioz, dos Santos and de Thoisy2013; Fossette et al., Reference Fossette, Witt, Miller, Nalovic, Albareda, Almeida, Broderick, Chacon-Chaverri, Coyne, Domingo, Eckert, Evans, Fallabrino, Ferraroli, Formia, Giffoni, Hays, Hughes, Kelle, Leslie, López-Mendilaharsu, Luschi, Prosdocimi, Rodriguez-Heredia, Turny, Verhage and Godley2014; Prosdocimi et al., Reference Prosdocimi, Dutton, Albareda and Remis2014). To maintain compatibility with DNA sequences from other studies, our analyses were based on 695 bp of the mitochondrial control region. We chose to exclude the haplotype Dc9.1 (GenBank accession #HM452364.1) from our analysis because information about its frequency and the source population(s) was unavailable.
Table 2. Mitochondrial diversity of leatherback turtles in the Atlantic Ocean, based on 695 bp of the mitochondrial control region (haplotype designations correspond to those from Dutton et al., Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013).
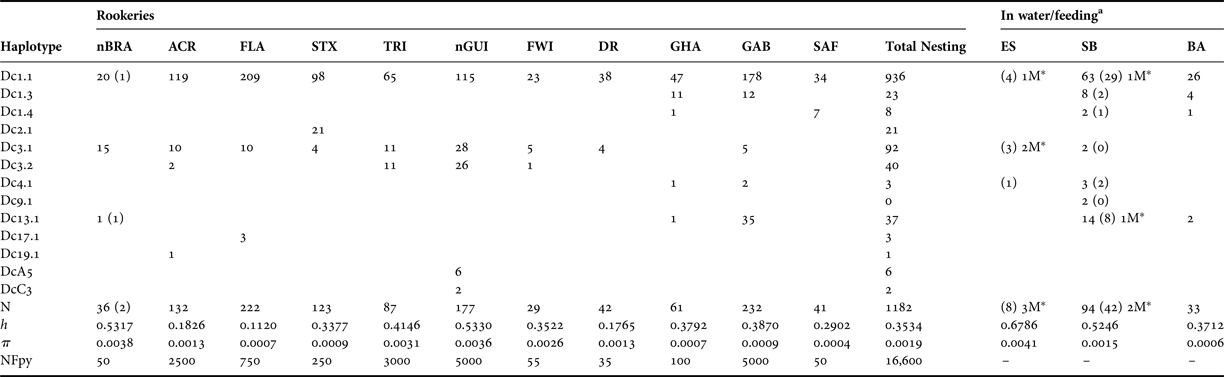
Standard and molecular diversity indices are given for rookeries and in-water/feeding samples from the Atlantic. Values are given for sample number (N), haplotype diversity (h), nucleotide diversity (π) and nesting females per year (NFpy). Numbers in parentheses are samples collected in this study (see Table 1 for details).
a The number of samples identified as males in this study is denoted by M*.
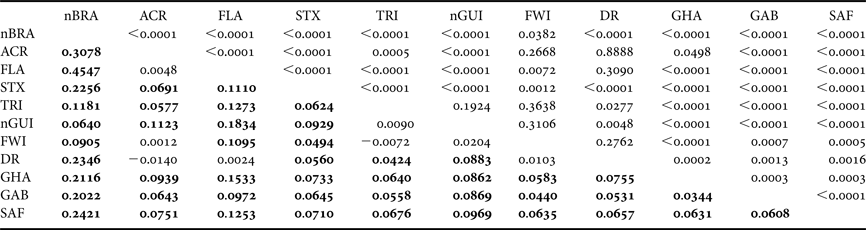
Table 3. Population pairwise F ST based on haplotype frequencies (below the diagonal) between Atlantic rookeries of the leatherback turtle (significant F ST values are shown in bold) and P-values of exact tests of population differentiation method (above the diagonal). Rookery abbreviations are defined in Table 1.
Genetic diversity, structure and demography
To characterize patterns of genetic variation, we used the program Arlequin 3.5 (Excoffier & Lischer, Reference Excoffier and Lischer2010) to calculate several measures of diversity: haplotype diversity (h), nucleotide diversity (π), the average number of nucleotide differences (k), and number of variable sites. To describe the genetic structure of the Atlantic populations, we also conducted exact tests of population differentiation and determined population pairwise F ST values (based on haplotype frequencies). We tested for a correlation between genetic distance (F ST/[1 − F ST]) and geographic distance (logarithm of distance in km) matrices using Mantel non-parametric permutation (N = 10,000) tests. The shortest distances (km) between rookeries (accounting barriers like continents) were calculated using the ruler tool of Google Earth (LeRoux et al., Reference LeRoux, Dutton, Abreu-Grobois, Lagueux, Campbell, Delcroix, Chevalier, Horrocks, Hillis-Starr, Troëng, Harrison and Stapleton2012).
Analyses of molecular variance (AMOVA; Excoffier & Lischer, Reference Excoffier and Lischer2010) among a hierarchical grouping of populations were carried out in Arlequin to test for regional genetic structure. We performed hierarchical AMOVAs based on the haplotype frequencies using a reduced alignment of 695 bp to maintain compatibility with the published sequences that were included in the dataset. This analysis considered the variation from four to eight groups, until we got the highest significant F CT value: (1) four groups (Atlantic North-west ACR, FLA, STX, TRI, nGUI, DR, and FWI; South-west Atlantic nBRA; South-east Atlantic GHA and GAB; South-west Indian Ocean SAF) based on the Regional Management Units from Wallace et al. (Reference Wallace, DiMatteo, Hurley, Finkbeiner, Bolten, Chaloupka, Hutchinson, Abreu-Grobois, Amorocho, Bjorndal, Bourjea, Bowen, Duenas, Casale, Choudhury, Costa, Dutton, Fallabrino, Girard, Girondot, Godfrey, Hamann, López-Mendilaharsu, Marcovaldi, Mortimer, Musick, Nel, Pilcher, Seminoff, Troeng, Witherington and Mast2010), (2) five groups (STX;ACR, DR, FLA, TRI, nGUI and FWI; nBRA; GHA and GAB; SAF); (3) six groups (STX; ACR, DR and FLA; TRI, nGUI and FWI; nBRA; GHA and GAB; SAF); (4) seven groups (STX; ACR, DR and FLA; TRI, nGUI and FWI; nBRA; GHA; GAB; SAF); and (5) eight groups (STX; FLA; ACR and DR; TRI, nGUI and FWI; nBRA; GHA; GAB; SAF). The results of hierarchical AMOVA should be verified with caution. For two groups, this analysis appears to require at least six populations per group to detect among group structure (Fitzpatrick, Reference Fitzpatrick2009).
We conducted neutrality tests (Tajima's D and Fu's Fs) using Arlequin to detect evidence of demographic expansion for the populations studied here. Using DnaSP 5 (Librado & Rozas, Reference Librado and Rozas2009), we tested for changes in population size using the R 2 test (Ramos-Onsins & Rozas, Reference Ramos-Onsins and Rozas2002) and mismatch distribution (Rogers & Harpending, Reference Rogers and Harpending1992).
To characterize temporal patterns of mitochondrial DNA variation of the leatherback turtles collected in Brazilian waters/feeding grounds, we divided the samples in four temporal groups: 2004, 2005, 2006, and all samples obtained after 2006 (>2006). We used the script TempNet (Prost & Anderson, Reference Prost and Anderson2011) in R (Team, Reference Team2013) to construct a three-dimensional statistical parsimony network using the sequence data.
RESULTS
Genetic diversity
From the new samples of leatherback turtles analysed in this study (N = 52), we sequenced 695 base pairs of the mitochondrial control region. We found six different haplotypes when considering the samples from nesting females in Espírito Santo (nBRA; N = 2) and the samples from incidental captures in fisheries and strandings in southern Brazil (SB; N = 42) and Espírito Santo (ES; N = 8; Table 1). A previously unrecorded haplotype for the Brazilian rookery, Dc13.1, was found in one of the samples collected from the nesting females. Until now, this haplotype had only been reported from nesting females from GAB and GHA rookeries (Dutton et al., Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013). We found low haplotype and nucleotide diversities across the Atlantic population of leatherback turtles as a whole, however there was considerable variation across rookeries (Table 2).
Genetic structure and demography
The overall single group AMOVA F ST was 0.092 (P < 0.01) and the pairwise F ST and exact tests showed non-significant (P > 0.05) differences between eight pairs of populations that were compared (Table 3). The hierarchical AMOVA based on the haplotype frequencies showed low and very low but significant differences among groups (F CT) and among population within groups (F ST) respectively, only when it considered six or more groups, with the association of seven groups exhibiting the highest significant F CT. The variation within populations was highest in all five cases (Table 4).
Table 4. Results from the analyses of molecular variance of Atlantic Ocean populations of the leatherback turtle.
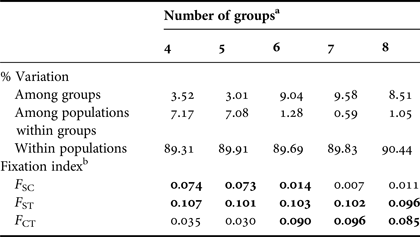
a Groups considering the 11 designated rookeries = ACR, FLA, STX, TRI, GUI, DR, FWI, BRA, GHA, GAB and SAF.
b F-statistics were used to estimate the proportion of genetic variability among groups (F CT), among populations within groups (F SC) and among populations (F ST). Values in bold are significant (P < 0.001).
The Mantel test for isolation by distance among Atlantic populations did not indicate a positive correlation between genetic and geographic distance (r = 0.029, P = 0.418), and the relationship (r 2) was weak and only accounted for 1.5% of the variation in the data. When accounting for temporal sampling (2004, 2005, 2006 and >2006), the comparison among the samples from the in water/feeding grounds in Brazil showed significant pairwise F ST values (data not shown) as well as evidence of different recruitment among years.
We found significant negative value of Tajima's D and significant value of R 2 for GHA, suggesting demographic expansion (Table 5). There are no signs of demographic growth for the other populations studied or for the combined Atlantic Ocean population. On the other hand, population bottlenecks are potentially indicated by the non-significant positive values of Tajima's D associated with multimodal mismatch distributions (data not shown) found for nBRA and SEC.
Table 5. Results of neutrality tests and population-size changes in leatherback turtle genetic stocks (rookeries or groups of rookeries) from the Atlantic Ocean, including Tajima's D, Fu's F S and R 2.
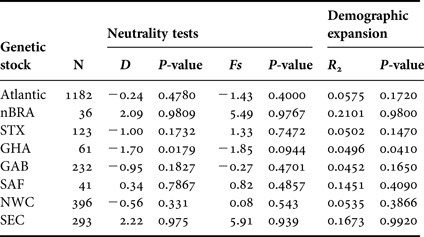
Genetic stocks including single rookery are defined with the same abbreviations of the individual rookeries (Table 1) and genetic stocks including multiple rookeries are defined as: North-west Caribbean (NWC = FLA + ACR + DR) and South-east Caribbean (SEC = nGUI + TRI + FWI).
Mixed-stock analysis and natal origin of non-nesting Brazilian samples
For the new samples collected in Brazilian waters or stranded on beaches (SB and ES), we found six out of the seven haplotypes identified by Vargas et al. (Reference Vargas, Araujo, Monteiro, Estima, Almeida, Soares and Santos2008), with the exception of the haplotype Dc9.1 (Table 2). The most common haplotype found in rookeries across the Atlantic Ocean, Dc1.1, is also very frequent in ES and SB and in BA (Argentina). It was found in 79.3% of the samples from nesting sites and in 69.7% of the samples from SB. This haplotype is spread across all the rookeries and feeding grounds that have been described so far from the Atlantic (Vargas et al., Reference Vargas, Araujo, Monteiro, Estima, Almeida, Soares and Santos2008; Dutton et al., Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013).
We did not find any differentiation between the samples from SB and BA (F ST: 0.005, P = 0.2698); however, SB had higher haplotype and nucleotide diversities (Table 2). The temporal network highlighted that the samples from SB and ES comprised different haplotypes and frequencies across years, with samples from 2005 being the most diverse (Figure 2).
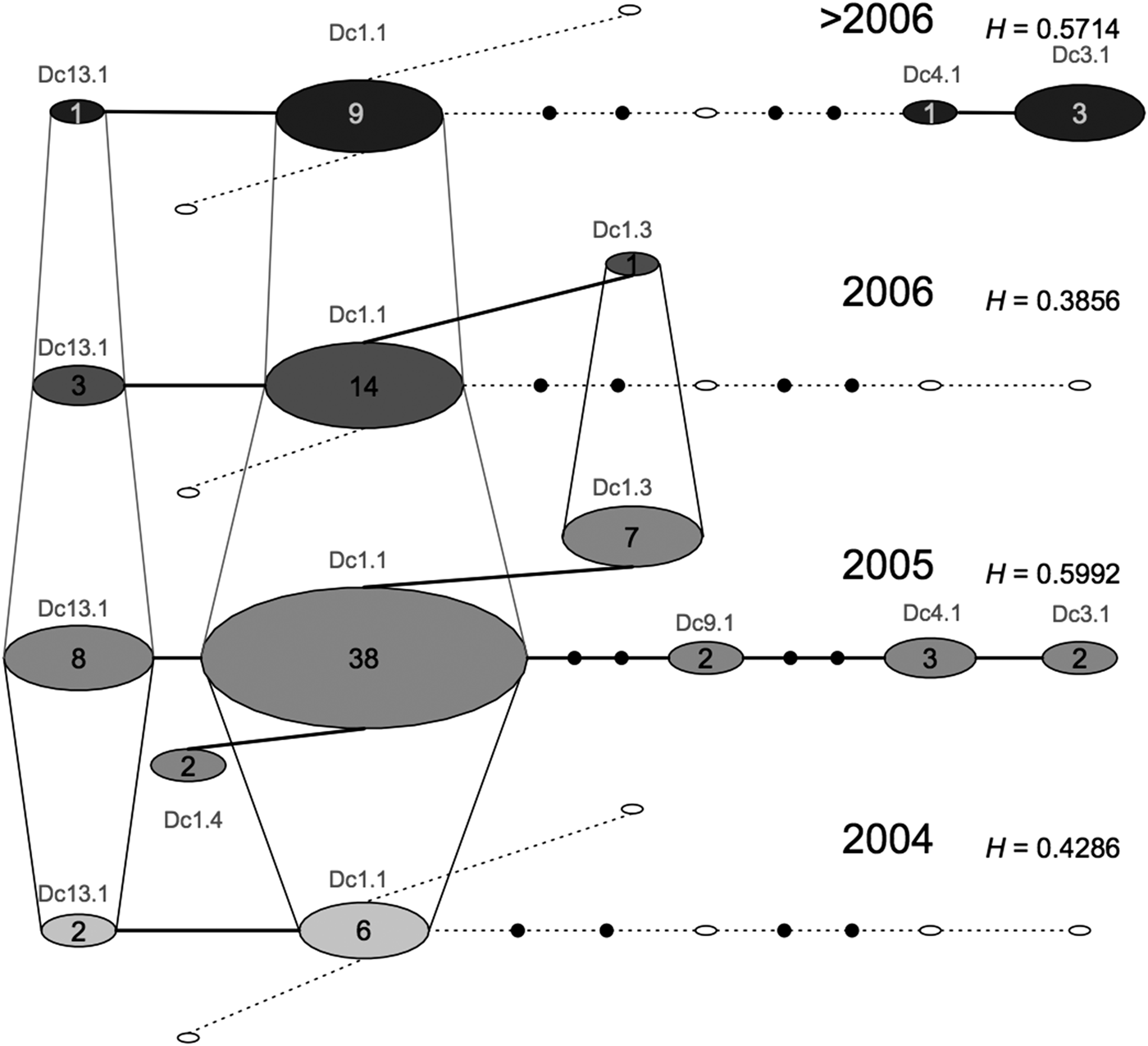
Fig. 2. Median temporal network of the seven mitochondrial control region (695 bp) haplotypes found in Brazilian samples of leatherback turtles from foraging ground, incidental capture, or stranded on the beaches (ES and SB). Layers represent four different sampling times. Each circle represents one haplotype. Small filled black circles represent hypothesized median vectors. Empty circles represent haplotypes that are missing from one particular sampling time but that are present at another sampling time. Sizes of circles are proportional to haplotype frequencies.
The mixed-stock analysis indicated that the samples from SB + BA primarily represent animals from West Africa (84.3%; estimated mean GAB = 82.4% and GHA = 1.9%). North Atlantic genetic stocks also accounted for the diversity in the SB + BA samples (overall estimated mean of 13.7%). The stocks from BRA and SAF accounted for 0.7 and 1.3%, respectively (Figure 3).

Fig. 3. Results of the mixed-stock analysis (many-to-many) showing mean proportional contribution of nesting to the feeding ground (SB + BA). Circles denote the means and bars represent 95% confidence intervals. Abbreviations of the names of genetic stocks are given in Table 1. The ACR, DR and FLA rookeries were combined as North-west Caribbean (NWC) and FWI, TRI and nGUI as South-east Caribbean (SEC) because they showed genetic homogeneity.
DISCUSSION
Genetic diversity and demography
Our results suggest that the high estimated genetic diversities (nucleotide and haplotype) found among leatherback turtles in the Brazilian rookery are not directly proportional to the current population size recorded at this rookery. For instance, this population has a very small number of nesting females per year (between 1 and 19; Thomé et al., Reference Thomé, Baptistotte, Moreira, Scalfoni, Almeida, Rieth and Barata2007); however, it presents the second-highest haplotype diversity among all described Atlantic rookeries. It is even higher than that reported for the Gabon rookery, which is considered the largest population of leatherback turtles in the world (Witt et al., Reference Witt, Baert, Broderick, Formia, Fretey, Gibudi, Mounguengui, Moussounda, Ngouessono, Parnell, Roumet, Sounguet, Verhage, Zogo and Godley2009).
The Brazilian rookery is likely to comprise a blend of native animals and migrants from eastern Atlantic and from northern Atlantic populations. This hypothesis is supported by the fact that the second most frequent haplotype found in Brazil (Dc3.1, N = 15) has a greater frequency (78%) among North Atlantic rookeries and very low frequencies in the African rookeries (Gabon and Ghana). There is evidence that this rookery might be a link between the North Atlantic and East Atlantic rookeries, highlighting its importance for species conservation.
Evidence for gene flow between the Eastern Atlantic (Gabon and Ghana) and the Brazilian rookery is provided by one individual, which was found off Espírito Santo State in September 2011, bearing the very low-frequency haplotype Dc4.1. The turtle was found in an advanced stage of decomposition, and its sex could not be determined. Until now, this haplotype had only been described as present in nesting females from Gabon and Ghana (Dutton et al., Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013). This finding reveals the occurrence of gene flow between these rookeries, since there are no known foraging areas for leatherback turtles in the waters of Espírito Santo.
Three adult males (curved carapace length from 144 to 157 cm) bearing the haplotypes Dc1.1 and Dc3.1 were found dead in fishing nets in Espírito Santo State (Brazil) in 2011. This finding suggests that males might be vulnerable to threats when in waters adjacent to nesting beaches (James et al., Reference James, Eckert and Myers2005), highlighting the importance of in-water conservation actions. This lends further support to the conservation importance of this rookery, which represents a significant contribution to the genetic diversity of the species in the Atlantic Ocean.
No signs of demographic growth for the combined Atlantic Ocean population were found, but the GHA rookery exhibits signals of demographic expansion that can be associated with recent colonization events. The non-significant and positive Tajima's D values coupled with multimodal mismatch distributions (data not shown) found for nBRA and SEC might indicate population bottlenecks in the life history of these stocks. This has been recorded for French Guiana and French West Indies populations of leatherback turtles (Molfetti et al., Reference Molfetti, Vilaça, Georges, Plot, Delcroix, Le Scao, Lavergne, Barrioz, dos Santos and de Thoisy2013).
Mixed-stock analysis and feeding grounds
Our data suggest that samples from the feeding grounds in the South Atlantic are more closely related to the West African rookeries than to the Brazilian rookery. We also have found little evidence of trans-equatorial migration in the West Atlantic for leatherback turtles found in feeding grounds. This result is contradictory because telemetry and genetic studies have shown that movements between northern and southern hemispheres are very rare (Stewart et al., Reference Stewart, James, Roden and Dutton2013; Fossette et al., Reference Fossette, Witt, Miller, Nalovic, Albareda, Almeida, Broderick, Chacon-Chaverri, Coyne, Domingo, Eckert, Evans, Fallabrino, Ferraroli, Formia, Giffoni, Hays, Hughes, Kelle, Leslie, López-Mendilaharsu, Luschi, Prosdocimi, Rodriguez-Heredia, Turny, Verhage and Godley2014). Given that the Atlantic leatherback turtles have a high degree of behavioural plasticity (Fossette et al., Reference Fossette, Girard, López-Mendilaharsu, Miller, Domingo, Evans, Kelle, Plot, Prosdocimi, Verhage, Gaspar and Georges2010), movements from the north to the south might lead to an increase in genetic diversity if some animals occasionally mate or nest in southern areas (as possibly found for the Brazilian rookery). This would also increase the possibility of animals reaching southern waters to forage.
Our mitochondrial data do not provide sufficient resolution to allow more detailed inferences about the north-to-south movements. Instead, additional investigations using multiple nuclear markers (as done for the Canadian foraging ground; Stewart et al., Reference Stewart, James, Roden and Dutton2013) are needed to test this hypothesis. Another possible explanation is that the contribution of animals from some nesting sites along the West African coast have not yet been characterized (Prosdocimi et al., Reference Prosdocimi, Dutton, Albareda and Remis2014), suggesting that further sampling would be beneficial to elucidate this question.
We found a minor contribution from Northern Atlantic rookeries to the South-west Atlantic feeding grounds, but possibly with a higher connection between them and the Brazilian rookery. The connection between the South Atlantic and the Indian Ocean also needs to be taken into account, although this needs to be confirmed by further work. There has been evidence of migration between the South Atlantic and South Africa from broad tag and telemetry studies (Luschi et al., Reference Luschi, Lutjeharms, Lambardi, Mencacci, Hughes and Hays2006; Fossette et al., Reference Fossette, Witt, Miller, Nalovic, Albareda, Almeida, Broderick, Chacon-Chaverri, Coyne, Domingo, Eckert, Evans, Fallabrino, Ferraroli, Formia, Giffoni, Hays, Hughes, Kelle, Leslie, López-Mendilaharsu, Luschi, Prosdocimi, Rodriguez-Heredia, Turny, Verhage and Godley2014), but the connection between South Africa and South America (South Brazil and Argentina) needs more investigation. Further studies using multiple nuclear markers are needed to test this hypothesis and to guide conservation policies in the Southern Atlantic Ocean. Considering the current levels of at-sea mortality and the likelihood of interactions with fisheries in the region (Fossette et al., Reference Fossette, Girard, López-Mendilaharsu, Miller, Domingo, Evans, Kelle, Plot, Prosdocimi, Verhage, Gaspar and Georges2010), coordinated efforts among relevant countries are needed to ensure the conservation of the leatherback turtle in this region.
The temporal network of samples from the Brazilian in water/feeding grounds shows that there is considerable variation in mitochondrial diversity through time. Samples from 2005 appear to be more genetically diverse (presence of seven haplotypes) than those from other years, suggesting that these turtles might be using other areas when environmental conditions are not ideal. An important point to note is that an unexplained high mortality of leatherback turtles was seen in South Brazil in 2005 (Vargas et al., Reference Vargas, Araujo, Monteiro, Estima, Almeida, Soares and Santos2008), which resulted in a larger sample size for that year; this might have influenced the results from mitochondrial DNA recruitment. Further studies integrating multiple approaches, and considering ecological parameters, such as oceanographic data on currents and distribution of food resources, would be valuable to understand population dynamics in the region. Combining these different sources of information would help us to determine whether changes in the proportion of recruits are related to intrinsic complex behaviours or to transient responses to ecological factors.
Genetic structure and Management Units
The inclusion of new samples in the nGUI and nBRA rookeries did not change the overall pairwise F ST results found by Dutton et al. (Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013) among the Atlantic populations sampled (Table 3). However, the inclusion of FWI and DR rookeries in the analysis reveals a more complex and closer connection among the Northern Atlantic rookeries compared with those of the Southern Atlantic. This can be associated with the smaller geographic distances among the northern rookeries, which cover a much smaller area (less than half) than the rookeries from the Southern Atlantic and from the South-west Indian Ocean.
The results of our AMOVAs showed that there was a greater genetic variation among groups when the seven groups based on Management Units proposed by Dutton et al. (Reference Dutton, Roden, Stewart, LaCasella, Tiwari, Formia, Thomé, Livingstone, Eckert, Chacon-Chaverri, Rivalan and Allman2013) were considered for the Atlantic and South-west Indian Ocean. However, we can note that the greater amount of variation for the Atlantic leatherback turtles is within populations, rather than among populations within groups, or among groups. In addition, our F ST results indicate that the populations from western Africa, South Africa and Brazil might have divergent origins and should be considered as three different Regional Management Units, as proposed also by Wallace et al. (Reference Wallace, DiMatteo, Hurley, Finkbeiner, Bolten, Chaloupka, Hutchinson, Abreu-Grobois, Amorocho, Bjorndal, Bourjea, Bowen, Duenas, Casale, Choudhury, Costa, Dutton, Fallabrino, Girard, Girondot, Godfrey, Hamann, López-Mendilaharsu, Marcovaldi, Mortimer, Musick, Nel, Pilcher, Seminoff, Troeng, Witherington and Mast2010). Future studies using larger sample sizes and including populations from other ocean basins will help us to understand the possible origins of these populations. These will also shed light on the connections among nesting and foraging populations from the Atlantic and Indian Oceans along the Antarctic/African Corridor.
CONCLUSIONS
We have found evidence of a very complex genetic distribution among leatherback turtles in the South-west Atlantic Ocean. Because this species is considered to be less philopatric than other species of marine turtle (Rivalan et al., Reference Rivalan, Dutton, Baudry, Roden and Girondot2006), some degree of interchange between rookeries is expected, and genetic diversity can be hidden in populations that have not yet been sampled. In Brazil, occasional nests can be found along the northern and southern coasts (Barata & Fabiano, Reference Barata and Fabiano2002; Loebmann et al., Reference Loebmann, Alves Legat, Puchnick Legat, De Camargo, Erthal, Severo and De Goes2008; TAMAR/SITAMAR Database), in addition to their regular distribution in Espírito Santo. However, it was not possible to collect samples from these animals due to logistical and financial limitations.
The mixed-stock analysis done with the large sample size obtained here from foraging grounds shows evidence of migration from the North to the South Atlantic. This was an unexpected result, given that tracking studies indicate that these movements are rare (Fossette et al., Reference Fossette, Witt, Miller, Nalovic, Albareda, Almeida, Broderick, Chacon-Chaverri, Coyne, Domingo, Eckert, Evans, Fallabrino, Ferraroli, Formia, Giffoni, Hays, Hughes, Kelle, Leslie, López-Mendilaharsu, Luschi, Prosdocimi, Rodriguez-Heredia, Turny, Verhage and Godley2014). In view of the limited resolution in some of our analyses, we suggest that efforts should be directed towards obtaining longer DNA sequences from worldwide samples (including the Indo-Pacific Ocean populations). These data will help to improve the reliability of the inferences from phylogeographic and mixed-stock analyses and to understand the contribution of Indian and Pacific Ocean animals to the feeding grounds in the southern Atlantic Ocean. There has been evidence of migration between nesting and foraging grounds, and between different ocean basins, but further investigation is needed to understand these complex population dynamics.
Genetic analyses have determined the existence of seven independent genetic stocks (Management Units) of leatherback turtles that nest in the Atlantic Ocean. Leatherback turtles are considered vulnerable globally (IUCN, 2017), but distinct subpopulations make the assessment more complex because they vary in population size, demographic trends and geographic range. Understanding the dynamics of individual populations is key to assessing threats and designing effective conservation strategies. In highly migratory animals, such as the leatherback turtle, assessing threats is a difficult task. When in foraging grounds along the coast, turtles are susceptible to a variety of threats, such as fisheries by-catch, ship strikes, natural predation and entanglement in or ingestion of marine debris.
Using genetic studies, we were able to better characterize the Brazilian rookery, and evaluate the composition of turtles found along the Brazilian coast throughout their range in the South-west Atlantic. The Brazilian rookery deserves special attention for conservation programmes because it is possible that the Espírito Santo coast is being used as a nesting refuge in the Atlantic Ocean. Despite its small size, this rookery has the potential to experience population recovery if, in addition to the full protection of nesting grounds, effective management actions are developed to tackle the current high levels of at-sea mortality.
This information provided by our analyses is valuable for improving management actions and in highlighting where mitigation may be required. In this context, Brazil has an important role as the only country in the South-west Atlantic that includes both known nesting and foraging areas for the leatherback turtle, with these presenting high levels of genetic diversity compared with others in the South Atlantic.
ACKNOWLEDGEMENTS
The authors thank the fieldwork team involved in collecting samples in Espírito Santo and Rio Grande do Sul States.
FINANCIAL SUPPORT
This work was supported by CNPq–Conselho Nacional de Desenvolvimento Científico e Tecnológico (Post-Doctoral scholarship number 201968/2014-2 to S.M.V.).










