INTRODUCTION
Noroviruses are a group of RNA viruses in the family Caliciviridae that can cause gastrointestinal illness in humans. Transmission can be via the faecal–oral route, contaminated food or water or person-to-person transmission via airborne droplets [Reference Parashar1]. The low infectious dose and high transmissibility of noroviruses makes them a frequent cause of foodborne and waterborne outbreaks, usually with high attack rates. Waterborne norovirus outbreaks usually occur in the context of faecal contamination of drinking water, although outbreaks related to recreational waters have been described [Reference Gelting2–Reference Sartorius9]. Viral contamination of surface waters may also occur during heavy rainfall and include discharges of raw sewage or wash-off of animal manure. These outbreaks can be difficult to identify since a large number of cases is usually required for health staff and authorities to recognize water as a possible source of infection [Reference Maunula, Miettinen and von Bonsdorff10]. Moreover, detection of these viruses in water remains challenging despite improvements in the methodology used. As a result of this, noroviruses are not identified in water in most suspected norovirus-related waterborne outbreaks. In Sweden there have been norovirus-related waterborne outbreaks in recent years caused by contamination of drinking water or recreational water, but the virus was detected in water in only two of these outbreaks [Reference Carrique-Mas5, Reference Nygård6, Reference Sartorius9, Reference Lysen11]. The identification of norovirus in water to support the results of an epidemiological investigation can avoid delays in implementing appropriate control measures caused by lack of conviction of local authorities regarding the waterborne nature of the outbreak [Reference Carrique-Mas5]. We used epidemiological and molecular investigations to study a large waterborne outbreak caused by contaminated municipal drinking water.
METHODS
Outbreak description
On 15 April 2009, the Environmental Office (EO) in a municipality in Western Sweden was informed of several cases of vomiting and diarrhoea in a village during the Easter weekend. The EO contacted the County Medical Office (CMO) the same day. In the initial investigation, over 60 households reported at least one member ill with vomiting and/or diarrhoea. These households were distributed throughout the village, which is located in the mountains in Western Sweden and has a population of around 400.
On 16 April, the Swedish Institute for Infectious Disease Control (SMI) was informed. An outbreak control team was formed including epidemiologists and laboratory experts from SMI, the CMO, the municipal EO, the National Food Administration and the National Water Catastrophe Group (VAKA). Given the extent of the outbreak and the lack of an obvious common event, water was suspected to be the source of the outbreak. An investigation was started to assess the extent of the outbreak, identify the cause and confirm the mode and vehicle of transmission in order to guide the implementation of appropriate long-term control measures.
Epidemiological investigation
We decided to carry out a retrospective cohort study that would include all households in the village. A questionnaire was delivered by hand to all households on 18 April 2009. The questionnaire was divided into two sections: one with questions pertaining to the household as a whole, and a second section with questions to be answered individually by each household member.
A case was defined as an individual resident in the village and who developed vomiting and/or diarrhoea (defined as ⩾3 loose stools per day) from 7 to 17 April 2009. Although there were four cases with symptom onset in the week before 7 April (dates of onset 28–29 March and 1 April), they were considered to be background diarrhoea cases and were excluded from the study. There were also four cases reported after 17 April with date of onset from 19 to 22 April. These were excluded since the date of onset was after the questionnaire had been sent.
The exposure was defined as consumption of unboiled water in different locations (home, village community centre, work and school/day-care centre). The average number of glasses of water consumed per day in each location as well as data on the source of the household water (municipal water network or own well) was also recorded.
Analysis of the data was performed with Stata/IC 10 (Stata Corp., USA). Place- and source-specific attack rates (ARs), relative risks (RRs) and 95% confidence intervals (95% CIs) were calculated for the consumption and source of water in different locations.
Environmental investigation
The municipal EO started an investigation in order to detect possible anomalies in the water network that could have caused the contamination.
Microbiological investigation
Clinical microbiology
Stool samples from six ill individuals were collected and investigated for bacterial enteropathogens (including Salmonella, Shigella, Campylobacter, Yersinia) and intestinal parasites (Cryptosporidium, Giardia) at the county laboratory. Six stool samples were sent to SMI and were analysed for enteric viruses by electron microscopy (EM) [Reference Hedlund, Rubilar-Abreu and Svensson12] and for norovirus and sapovirus with single-round multiplex capsid RT–PCR.
Prior to RT–PCR, RNA was extracted from a 10% stool suspension in PBS on a Biorobot M48 (GenoVision, Qiagen, Germany) using the MagAttract Viral RNA M48 kit (Qiagen) according to the manufacturer's instructions. The Single-round multiplex capsid RT–PCR was performed as previously described [Reference Yan13] including sapovirus, norovirus GI and norovirus GII but excluding astrovirus detection. Briefly 2·5 μl cDNA was added to 22·5 μl of a mix containing 0·4 μl from 33 μm each of forward primers: G1-SKF, G2-SKF, SLV5317 and the reverse primers G1-SKR, COG2F, SLV5749, in 2·5 μl of 25 mm MgCl2, 0·5 μl of 10 mm dNTP, 2·5 μl of 10×PCR buffer (pH 8·3) and 1·25 U of AmpliTaq Gold enzyme (Invitrogen, USA). The PCR program used was: 8 min denaturation at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, 30 s at 72°C with a final extension at 72°C for 7 min. DNA was separated on a 1·5% agarose gel and fragments of interest were cut out and purified with GEL-M kit (Viogene, USA). For sequencing oligonucleotides used in the PCR reaction described above corresponding to the fragment of interest were used. Sequencing was performed with BigDye 3 (Invitrogen) and loaded onto an ABI 3130xl Genetic Analyzer sequencer (Applied Biosystems, USA) according to the manufacturer's instructions. Sequence data was analysed with SeqMan, Lasergene 6, and nucleotide alignment search was performed at the Rijksinstituut voor Volksgezondheid en Milieu (RIVM, The Netherlands), and/or the National Centre for Biotechnology Information (NCBI, USA) nucleotide databases. The received sequence was compared to the same part of the genetic material obtained from the norovirus-positive water sample.
Environmental microbiology
Sixty-seven water samples taken in different locations and at different time points were obtained and investigated for E. coli, coliforms, intestinal enterococci and Clostridium perfringens at the county laboratory.
Seven water samples were sent to SMI for investigation of the presence of somatic coliphages and norovirus. For virus concentration, volumes of ~1 litre were filtered on Zetapor 47 mm, positively charged with pore size 0·45 μm (Cuno, USA). Following filtration, the filters were incubated with 3 ml of 50 mm glycine 1% beef extract buffer (pH 9·5) at room temperature for 30 min with agitation at 300 rpm. Thereafter the pH was adjusted to 8 and the sample was transferred to Amicon Ultra-4, Centrifugal Filter Devices 100K (Millipore, USA) and concentrated to 140 μl. The concentrated sample was subsequently used for RNA extraction using QIAamp Viral RNA Mini kit (Qiagen) according to the manufacturer's instructions.
To generate cDNA, 7·5 μl RNA was added to a 7·5 μl mix containing 0·5 μl (200 U/μl) SuperScript II (Invitrogen), 2·05 μl of 5x first strand buffer, 2·7 μl of 2·5 μm random primers, 0·75 μl of 10 mm DDT, 0·25 μl RNAse free water, 0·5 μl (40 U/μl) RNAse out (Invitrogen) and 0·75 μl of 10 mm dNTP (Invitrogen). The reaction was run for 10 min at 25°C, 60 min at 42°C and 15 min at 70°C. For detection of norovirus GI and GII two in-house semi-nested RT–PCRs based on primers described earlier [Reference Yan13, Reference Gallimore14] were used. For norovirus GI the first round of PCR was run with 2·5 μl cDNA added to a 22·5 μl mix containing 0·2 μl from each of three forward primers: GIFF-1, GIFF-2 and GIFF-3 in concentrations of 20 μm and 0·4 μl of the reverse primer G1-SKR in a concentration of 33 μm, 5 μl of 25 mm MgCl2, 1 μl of 10 mm dNTP (Invitrogen), 2·5 μl of 10×PCR buffer (pH 8·3) and 1·25 U of AmpliTaq Gold enzyme (Invitrogen). The PCR program used was: 2 min denaturation at 95°C followed by 40 cycles of 30 s at 95°C, 30 s at 55°C, 2 min at 72°C with a final extension at 72°C for 5 min. The second round of PCR was performed by adding 2·5 μl product from first-round PCR to a 22·5 μl mix containing 0·4 μl of forward primer GI-SKF and reverse primer GI-SKR in a concentration of 33 μm, in 4 μl of 25 mm MgCl2, 1 μl of 10 mm dNTP (Invitrogen), 2·5 μl of 10× PCR buffer (pH 8·3) and 1·25 U of AmpliTaq Gold enzyme (Invitrogen). The PCR program used was the same as for the single-round multiplex capsid RT–PCR described above.
For norovirus GII the first round of PCR was performed by adding 2·5 μl cDNA into a 22·5 μl mix containing 0·2 μl from each of three forward primers: GIIFB-1, GIIFB-2 and GIIFB-3 in concentrations of 20 μm and the reverse primer G2-SKR in a concentration of 33 μm, 5 μl of 25 mm MgCl2, 0·5 μl of 10 mm dNTP (Invitrogen), 2·5 μl of 10× PCR buffer (pH 8·3) and 1·25 U of AmpliTaq Gold enzyme (Invitrogen). The PCR program used was: 2 min denaturation at 95°C followed by 40 cycles of 30 s at 95°C, 30 s at 50°C, 2 min at 72°C with a final extension at 72°C for 5 min. The second round of PCR was performed by adding 2·5 μl product from first-round PCR to a mix containing 0·2 μl of forward primer COG2F and reverse primer G2-SKR in a concentration of 33 μl, in 3 μl of 25 mm MgCl2, 0·5 μl of 10 mm dNTP (Invitrogen), 2·5 μl of 10× PCR buffer (pH 8·3) and 1·25 U of AmpliTaq Gold enzyme (Invitrogen). The PCR program used was the same as for the single-round multiplex capsid RT–PCR described above. All PCRs were run on a 2720 Thermal Cycler (Applied Biosystems).
DNA fragments of interest were handled as described above to obtain genetic sequence to compare to the results obtained from the stool samples.
RESULTS
Epidemiological investigation
Out of a total of 155 households, 116 returned the completed questionnaire (75% response rate). The questionnaires contained information about 270 individuals (including guests) which constituted our study cohort. Of these, 173 individuals fulfilled the case definition, giving an AR of 64%.
The median household size was 3 (range 1–7). Ninety-five (82%) households reported having at least one member ill and 27 (23%) reported having had guests who became ill during the same time period. All those guests had consumed water at the house during their stay. In total, 104 (90%) households reported being connected to the public water network.
Figure 1 shows the case distribution by date of symptom onset. The profile corresponds to a point-source outbreak, with the first cases appearing on 7 April and the number of cases increasing progressively and peaking on 14 April, decreasing thereafter with the last case registered on 17 April.
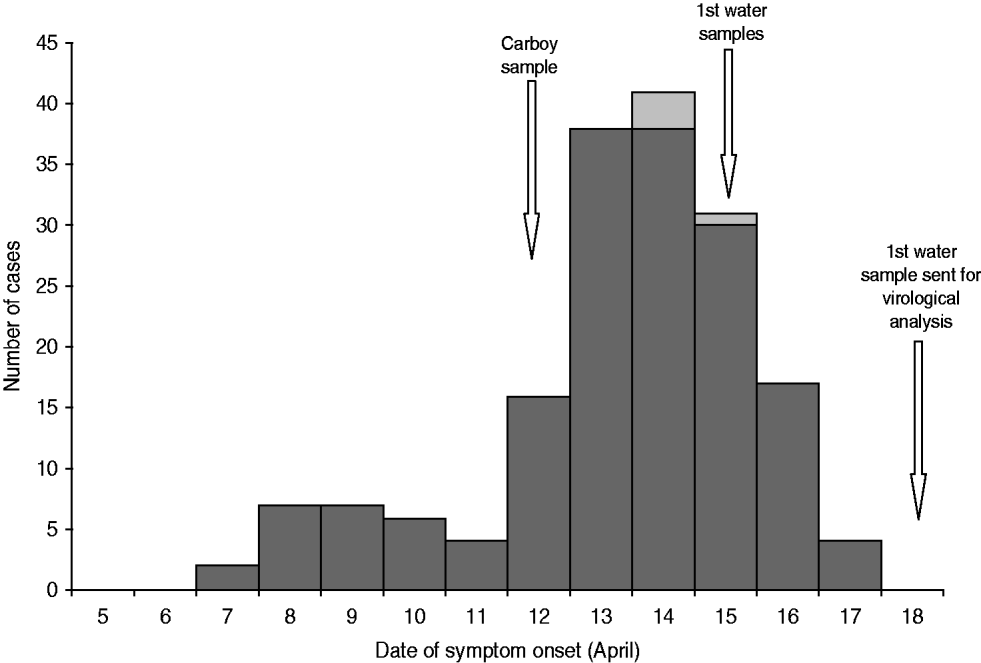
Fig. 1. Distribution of cases of gastrointestinal illness by date of symptom onset, April 2009.
The median age of cases was 48 years (range 0–85 years) and 50% of the cases were male. There were no significant differences in age and sex between cases and non-cases. Of the 173 cases, 88% presented with diarrhoea, 84% with abdominal pain, 75% with vomiting and 50% with fever. The median duration of illness was 46 h (range 2–154 h). Ninety-eight per cent of the cases (169/173) reported living in a house connected to the municipal water network, compared to 81% of non-cases (78/96).
Table 1 describes the ARs and RRs with 95% CIs for all the exposures studied. Residents living in households connected to the public water network were at an increased risk of developing disease (RR 4·80, 95% CI 1·68–13·73) compared to those with no connection to the public network. There was also an increased risk of developing disease for those residents who drank unboiled water at home but it did not reach statistical significance (RR 1·76, 95% CI 0·60–5·18). We did not find an association between the development of disease and other exposures related to the different locations where the residents drank water.
Table 1. Attack rates (AR) and relative risks (RR) with 95% confidence intervals (95% CI)
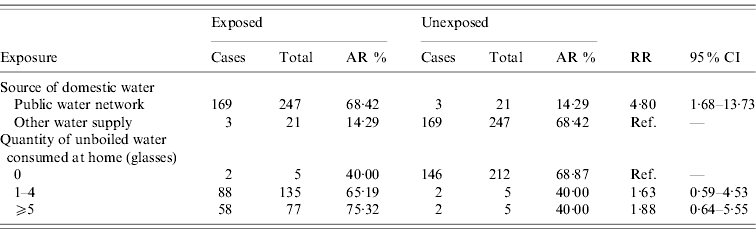
Information about the number of glasses of water consumed at home was available for 217 residents (148 cases, 69 non-cases). The median number of glasses consumed at home for the cohort was three (range 0–25) with no difference between cases and non-cases. We found the risk of developing disease to be higher the more glasses of unboiled water that were consumed at home, although it did not reach statistical significance.
Environmental investigation
The village was served by two different water sources. Source A was a borehole situated 2 km from the village. The water was treated in an on-site water plant through rapid sand filtration and UV-light disinfection. The inspection of this source did not reveal any irregularities and no obvious source of contamination was identified. The water from this source had last been tested on 7 April (samples were taken from raw and treated water at the plant, as well as from the house of a user) without any alterations in the water quality being reported.
Source B was a well situated on the outskirts of the village. The water was treated in an on-site water plant with the same methods as for source A. This source was intended for use at times of increased demand in the area and was not used during 2008. Source B was only put to use just before the annual Vasa cross-country ski competition took place (18 February to 1 March 2009), in anticipation of an increased demand for water in the area. On inspection, water source B was found to have inadequate physical barrier protection. The water from this source had not been tested for over a year since it had last been used. No samples were tested before this source was reconnected to the municipal water network. During the time period of the outbreak there was abundant snow melting in the area.
The village also had a water reservoir with two tanks situated uphill from the village. This reservoir was served by the same water pipe network as the village. When the water demand was not met by the two water sources, the water from the reservoir would discharge into the water pipe network to meet this demand.
On 10 April, the engineer on duty at the municipal water network was notified of an interruption in the water service caused by a fall in water pressure which affected households in the upper part of the village. A leak in the water pipe network was suspected to be the cause. It was also noted that the reservoir was not working correctly. One of the tanks did not have a brim and with the fall in pressure all the water was discharged into the water pipe network.
The whole water pipe system in the village was examined and several leaks were found. No other abnormalities were detected in the system.
Microbiological investigation
Clinical microbiology
All stool samples were negative for enteropathogenic bacteria, Cryptosporidium and Giardia. Norovirus were identified by EM in 5/6 stool samples and norovirus RNA was detected in all six samples by RT–PCR. Nucleotide sequencing of the capsid identified the microorganism as norovirus GI.3 when compared with sequences from the GenBank database in the six samples.
Environmental microbiology
From 15 April to 7 May, 67 water samples were taken for analysis: nine from water source A (eight raw water, one treated water), 10 from water source B (nine raw water, one treated water), two samples from the community centre, two samples from the day-care centre and 44 samples from 19 households connected to the public water network where cases had occurred (15 households were sampled once and four households were sampled repeatedly with a total of 29 samples). The analysis of a sample of treated water from water source B on 15 April had 5 c.f.u./100 ml coliforms and no E. coli. A water sample taken the same day from the home of a user had 12 c.f.u./100 ml coliforms and 2 c.f.u./100 ml E. coli. Samples taken after 15 April were mostly negative for coliforms except for four raw water samples from water source B (2–6 c.f.u./100 ml).
Initially, six water samples from five different locations, two samples each with raw water collected on different occasions from the two waterworks and two tap-water samples from users, were sent to SMI to investigate for the presence of norovirus. All six samples were negative for norovirus RNA. On 12 April, a 25-litre carboy was filled with water at a private home connected to the village public water network. The existence of the carboy was brought to our attention on 12 May and a sample sent to SMI for analysis. This sample tested positive for norovirus GI.3 and the analysed part of the capsid sequence was identical to the findings in the stool samples.
Control measures
Since water was the suspected source of the outbreak from the start, a boiling recommendation had already been issued on 16 April. Source B was disconnected from the public water network the same day. As a result of the investigation, the leaks found in the water pipe network were repaired and the municipal water reservoir was emptied and cleaned 1 week after the fall in pressure. Since norovirus had not been identified in water, source B was reconnected to the public water network on 17 May. After identification of the virus in the carboy sample, water source B was finally closed down on 17 June. The boiling water advice was continued until 26 June, after repeated testing of water source A had revealed no abnormalities. However, just before the advice was discontinued, it was decided to start chlorination of the water until the whole water pipe system could be flushed and cleaned. A new water source was identified to supply the municipal water network replacing both sources.
DISCUSSION
The objective of this investigation was to identify the source of the outbreak. The epidemiological, environmental and laboratory investigations supported the initial hypothesis indicating water as the source of the outbreak. The epidemiological curve showed a point-source outbreak indicating that contamination of the source was probably present for a limited period of time. The curve starts slowly with few cases on the first days and then increases sharply. This may have been because of increased person-to-person transmission in the second part of the outbreak. However, it was impossible to establish the extent of person-to-person transmission within this outbreak since distinction between primary and secondary cases was not possible because of the high rate of exposure to municipal drinking water in the cohort.
We found a statistically significant RR of 4·80 (95% CI 1·68–13·73) for residents connected to the public water network compared to residents not connected to the water network. However, the wide CI reflects the low numbers unexposed to the municipal water network in our cohort. We found a high RR for those residents who stated drinking unboiled water at home (RR 1·76, 95% CI 0·60–5·18) but it did not reach statistical significance, probably because of the low number of respondents to this question among the unexposed group. It was not possible to compare between the two water sources since both are connected to the same network and therefore all households in the village are served by both sources.
We failed to demonstrate a dose–response effect for the quantity of unboiled water consumed at home as has been reported in other norovirus waterborne outbreaks [Reference Carrique-Mas5]. However, lack of association between the amount of water consumed and disease in norovirus-related outbreaks is not always clear [Reference Werber4] and recall bias of the number of glasses of water consumed during the outbreak period may also influence the outcome.
The viral pathogen involved was identified early in the investigation in stool samples from six cases. Evidence of faecal contamination was found in one sample of treated water from source B and one sample taken from a household connected to the municipal water network, as well as in 4/9 samples of raw water from source B. The pathogen responsible was identified as norovirus GI.3 and was found in both the six stool samples and in the water sample from a carboy stored since 12 April in one household. The water from the carboy had been used for drinking and one of the two consumers in the household became ill after they had left the village to return to their usual place of residence. The first water samples in relation to the outbreak were taken on 15 April, on the same day as the outbreak alert. By then, the peak of the outbreak was over and therefore the virus could have already disappeared from the water or been present at much diluted concentrations if it was a transient contamination. This may explain why no norovirus was detected in the water samples collected on 18 and 23 April and only detected in the water sample from the carboy collected on 12 April. The sensitivity of the semi-nested RT–PCR used in this study for detection of norovirus GI and GII in water has not been fully evaluated. The method is tested with end-point titration using water spiked with human norovirus from faecal samples. However, the semi-nested RT–PCR was compared to the established single-round multiplex capsid RT–PCR [Reference Yan13] used for clinical samples and it was found to increase sensitivity by 10- to 100-fold without loss of specificity (data not shown).
Norovirus-related waterborne outbreaks are not rare in Sweden, with at least 13 outbreaks reported since 2002. GI strains were identified as the most frequent cause of norovirus-related waterborne outbreaks in Sweden, while GII strains were more common in norovirus-related foodborne outbreaks and healthcare settings [Reference Lysen11]. This association between GI strains and waterborne outbreaks has also been seen in Finland [Reference Maunula, Miettinen and von Bonsdorff10]. The reason for this association is not known but it has been suggested that GI strains could be more stable in water than GII strains [Reference Maunula, Miettinen and von Bonsdorff10, Reference Lysen11].
Water source B was identified as the possible cause of the outbreak. Five samples showed evidence of faecal contamination and the source lacked adequate protection barriers. No abnormalities were identified in the water treatment process. However the UV-light disinfection could have been negatively affected by humic material in the water. The water was not chlorinated. In Sweden chlorination is not used on a regular basis in water treatment processes. In the week prior to the outbreak there had been considerable amounts of snow melting that could have overflowed into the system. The contamination of both the raw water source and the water pipe network could have been caused through the identified leaks. Noroviruses are also known to be able to migrate through the ground and contaminate groundwater [Reference Lawson7]. The fall in water pressure could have amplified this contamination of the water pipe network. This could explain the sudden increase in the number of cases after the fall in pressure. In addition, residents reported the occurrence of clusters of cases of diarrhoea in the village in previous years around the same time of the year, which would be consistent with melting snow causing or contributing to the contamination of the drinking water.
Concerning control measures, a boiling advice was issued shortly after the outbreak alert. As the number of cases was already declining by then it is difficult to establish whether this measure was effective in controlling the outbreak. Water source B was initially disconnected from the public water network but reconnected a month later because of lack of microbiological evidence of water as the source of the outbreak. The water pipe network was repaired promptly, and the cleaning of the water reservoir was done within a week of the outbreak alert. Identification of norovirus in a water sample supported the results of the epidemiological investigation and dispelled any remaining doubts that water was the source of the outbreak. This finding enabled the local authorities to take the decision to close down water source B.
This is the first time that noroviruses have been isolated both in patients and drinking water in connection with a waterborne outbreak in a municipal drinking-water supply in Sweden. This outbreak investigation illustrates the importance of inter-agency collaboration and the added value of using molecular methods in outbreak investigations in order to prompt adequate and timely control measures.
ACKNOWLEDGEMENTS
The authors thank Lena Eriksson, Edita Platbardis and Per-Ove Brandt (Älvdalen municipality), Torbjörn Lindberg (National Food Administration) and Peter Norberg (VAKA) for their contribution to the outbreak investigation and revision of the manuscript.
DECLARATION OF INTEREST
The work of Margarita Riera-Montes was supported by the European Programme for Intervention Epidemiology Training (EPIET), funded by the European Centre for Disease Prevention and Control (ECDC).




