Introduction
Hepatitis C is a liver disease caused by the hepatitis C virus (HCV). About 15–45% of infected individuals spontaneously clear the virus within 24 weeks without any treatment. The remaining 55–85% of individuals would develop chronic HCV infection ranging in severity from progressive liver fibrosis to subsequent liver cirrhosis and hepatocellular carcinoma [Reference Matsuura1].
To date, the mechanism of HCV chronic infection was not yet clear. HCV eradication included the host spontaneous clearance and the interferon (IFN)-mediated clearance [Reference Welzel2, Reference Kuiken3]. It is generally known that the virus clearance depends on the interaction of the virus and the host factor, whereas the genetic polymorphism on the immune-related pathway was a vital host factor. It has been reported that interleukin 28B (IL28B) gene and myxovirus resistance A (MxA) gene were widely studied [Reference Hijikata4]. Multiple genome-wide association study (GWAS) reported that single nucleotide polymorphisms (SNPs) of IL28B gene were identified to be associated with antiviral therapy among different HCV infection populations [Reference Tanaka5–Reference Ge7]. Since then, the association of multiple host gene polymorphisms with therapeutic effects has been explored in populations of different ethnic and different HCV genotypes [Reference Zheng8–Reference Par10]. Quite a few researchers have also found the association in Chinese Han population, but the results were not consistent in different studies.
Our previous studies have demonstrated that IFN-λ relevant gene polymorphisms may be associated with HCV spontaneous clearance [Reference Cui11]. Besides continuing to explore the association of IL28B gene and MxA gene with HCV spontaneous clearance, this study focused on the establishment of 231 chronic hepatitis C (CHC) patients using a prospective cohort treated by pegylated IFN and ribavirin (pegIFN-α/RBV), combined with molecular epidemiology laboratory tests, so as to study the predictive factors of sustained virological response (SVR) in patients and provide evidence for the evaluation of clinical antiviral therapy.
Methods
Participants
A total of 1090 subjects for investigating the factors of HCV spontaneous clearance were enrolled from the Nanjing compulsory detoxification centre from May to December 2006, nine hospital haemodialysis centres in southern China from October 2008 to January 2010, and a former remunerated blood donation population in Danyang City and six villages in Zhenjiang City from October 2008 to January 2013.
These 1090 subjects were categorised into two groups for analysis. The spontaneous clearance group (Clearance group) consisted of 428 spontaneous viral clearance cases who were anti-HCV seropositive, in which HCV-RNA detection was negative. The persistent HCV infection group (Chronic group) was composed of 662 persistent HCV infection cases who were anti-HCV seropositive and HCV-RNA positive continuously. All together 231 hospitalisation and outpatient hepatitis C treatment cases recruited from Jurong People's Hospital of Jiangsu Province from January 2011 to December 2016 were investigated to identify the factors influencing the therapeutic response.
This study was approved by the Ethics Committee of Nanjing Medical University, and all subjects provided voluntary informed consent to participate in the study. A standardised questionnaire was administered by well-trained interviewers to collect information on demographic data and environmental exposure history. After the interview, an approximately 5 ml venous blood sample was collected from each participant. The serum and white blood cells were separated and stored at −80 °C until assay.
Treatment and effect evaluation
All patients underwent weekly intramuscular injection of pegylated IFN-α2a (3MU IH QD) and ribavirin (RBV, 800–1200 mg/d) was combined orally for 48 weeks.
According to the manufacturers’ instructions, HCV-RNA was quantified in all patients at baseline and after 4, 12, 24 and 48 weeks of treatment and 24 weeks after cessation of treatment by Cobas Amplicor HCV Monitor Test, v2.0 (Roche, Basel, Switzerland). In this study, early virological response (EVR) was defined as ⩾2 log reduction in HCV-RNA level compared to baseline HCV-RNA level. SVR was defined as HCV-RNA negative 24 weeks after treatment-free follow-up, which meant that the desired therapeutic effect was achieved.
Genotyping
Rs12980275 near the IL28B, rs2071430 and rs17000900 on the MxA gene that may be related to HCV clearance were selected in this study. These three SNPs were predicted to have a role in the transcription factor-binding position on SNP info site (http://snpinfo.niehs.nih.gov/) and they were filtered with the criteria: minor allele frequency ⩾0.05.
Genomic DNA was extracted from leucocytes in peripheral blood by sodium dodecyl sulphate lysis and protease K digestion and followed by phenol–chloroform extraction and ethanol precipitation. Genotyping was performed by the TaqMan allelic discrimination assay on ABI PRISM 7900HT Sequence Detection system (Applied Biosystems, San Diego, CA, USA). PCR was performed in accordance with the recommended thermal profile: 50 °C for 2 min (preheating), 95 °C for 10 min (preincubation) followed by 40 cycles at 95 °C for 15 s (denaturation) and 60 °C for 1 min (annealing). The primers and probe sequences for the selected SNPs were shown in online Supplementary Table S1. Two blank samples as negative controls and five repeated samples as positive controls were assigned into each genotyping assay with a 100% accordance rate of each SNP for the repeated experiments of 10% random samples.
Statistical analysis
The database was established by EpiData3.10 (EpiData Association, Odense, Denmark). All statistical analyses were performed by 12.0 software (StataCorp LP, College Station, TX, USA) after the log check. Baseline clinical features between different treatment outcomes were calculated by the Student t test and the χ 2 test. Univariate and multivariate logistic regression analysis by the odds ratios (ORs) and 95% confidence intervals (CIs) were used to estimate the associations of SNPs with HCV spontaneous clearance, EVR and SVR. The life table was used to analyse the associations of selected SNPs with virological response time. The line graphs were used to observe the decrease in viral load at each follow-up point. All the statistical tests were a two-sided test, P < 0.05 was statistically significant.
Results
Demographic characteristics
In 428 subjects with spontaneous clearance and 662 patients with CHC, gender, age, alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were statistically significant (P < 0.05). The percentage of ALT and AST larger than 40 U/l in the chronic group was higher than the clearance group, as showed in Table 1. In the pegIFN-α/RBV treatment cohort, a total of 231 chronic-infected patients were included in the final analysis. The SVR rate was 52.0%. There was significant difference in the level of platelet, α fetoprotein (AFP) and blood glucose (GLU) between the two groups (SVR group and N-SVR group) (P < 0.05).
Table 1. Subjects characteristics
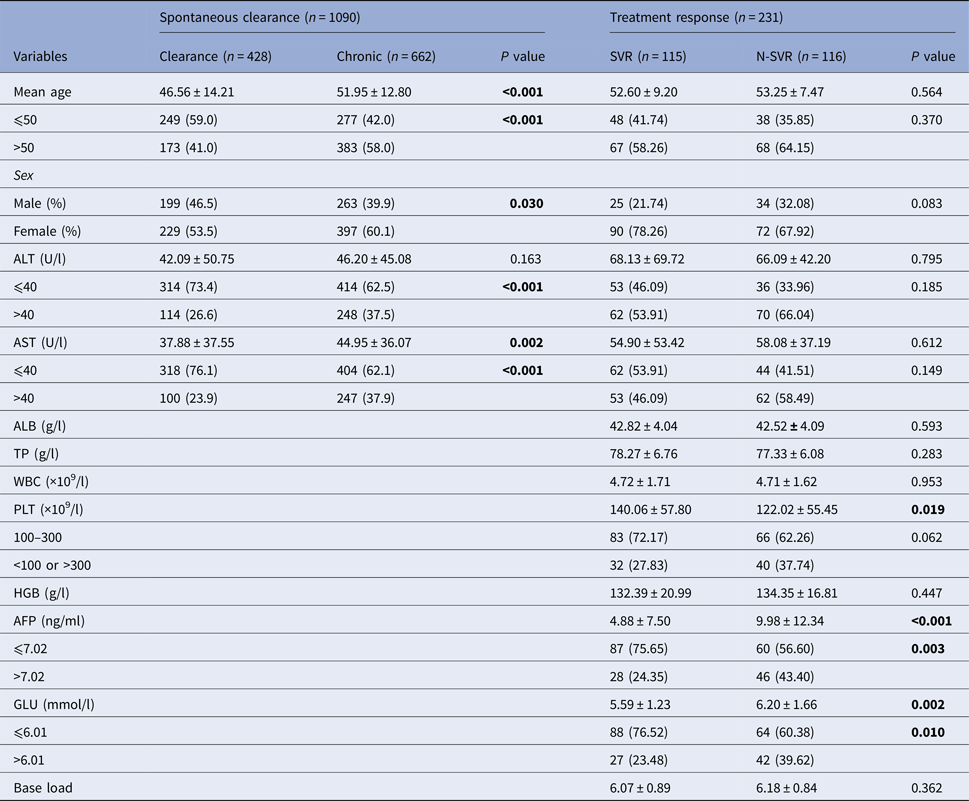
N-SVR, non-sustained virological response; SVR, sustained virological response; GGT, γ-glutamyl transpeptidase; TP, total protein; ALB, albumin; WBC, white blood cell.
Relationship between MxA rs2071430 and HCV spontaneous clearance
Using the co-dominant model (wild type vs. homozygous type vs. hybrid type), the dominant model (homozygous type and hybrid type vs. wild type) and the additive model (homozygous type vs. wild type), the spontaneous clearance group and the chronic infection group were compared to estimate the impact of MxA and IL28B gene polymorphisms on HCV spontaneous clearance. Table 2 indicated that patients with MxA rs2071430 TT genotype were more likely to develop HCV infection chronicity than GG genotype (additive model: OR 1.22, 95% CI 1.01–1.48, P = 0.042) by logistic regression analysis after adjusting for age, gender, PLT, AFP, GLU and baseline HCV load.
Table 2. Relationship between selected polymorphisms and HCV spontaneous clearance
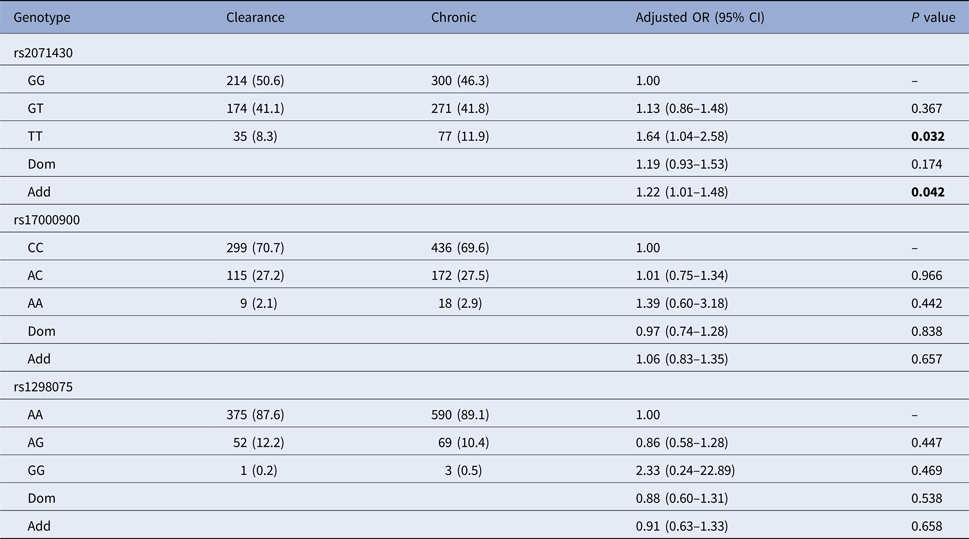
Clearance, the spontaneous clearance group; Chronic, the chronic infection group.
Association of MxA rs17000900 and IL28B rs12980275 with therapeutic response
After the covariate adjustment, rs12980275 analysis showed that the frequency of mutant genotype GG in SVR group was significantly lower than that in N-SVR group (additive model: OR 0.58, 95% CI 0.34–0.98, P = 0.040), which suggested that patients with GG genotype might not have good treatment response. Rs17000900 analysis showed that patients with CC wild genotype were more likely to achieve an SVR (additive model: OR 0.54, 95% CI 0.30–0.99, P = 0.048) (Table 3). It can be seen from online Supplementary Table S2 that patients with rs12980275 GG genotype were also unfavourable to EVR (additive model: OR 0.38, 95% CI 0.20–0.71, P = 0.003).
Table 3. Association of selected polymorphisms with SVR
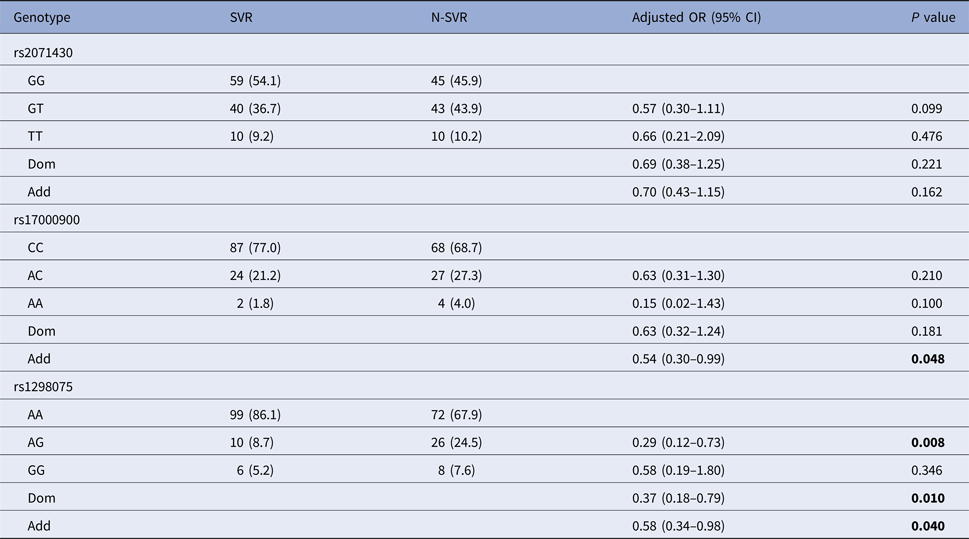
N-SVR, non-sustained virological response; SVR, sustained virological response.
Logistic regression analyses adjusted for age, gender, GLU, AFP, platelets.
Effect of IL28B rs12980275 on the virological response
The association of IL28B rs12980275 (additive model) in CHC patients achieving virological response time (after viral load reduction to 500 copies/ml) was analysed using the life table. The median time of rs12980275 AA, AG, GG genotype was 7.40, 13.56 and 6.76 weeks, respectively (Fig. 1). The time of viral response in patients with AG genotype was later than AA/GG genotype and the difference was statistically significant (AA vs. AG: P = 0.018, AG vs. GG: P = 0.097, AA vs. GG: P = 0.715).
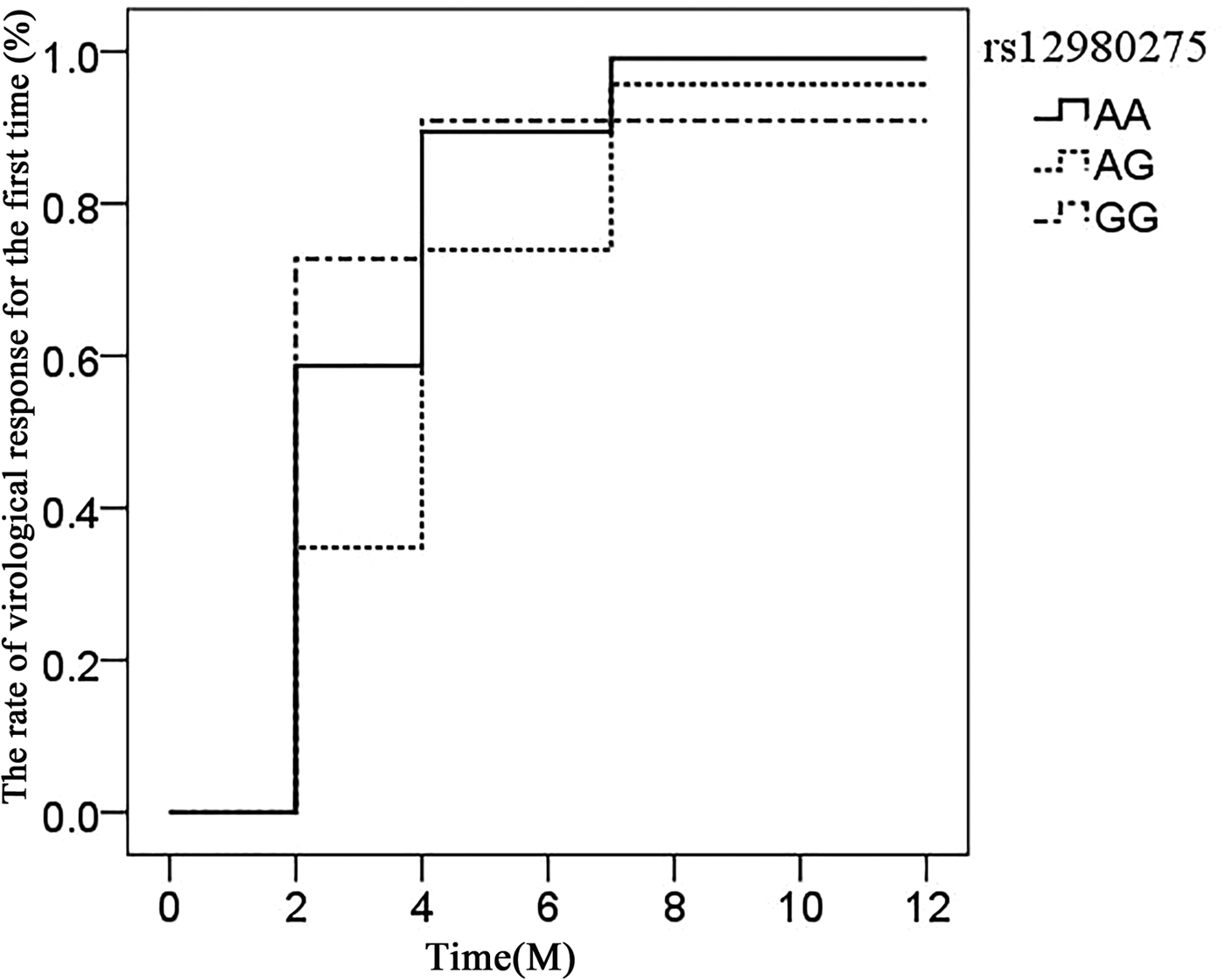
Fig. 1. Life expectancy chart of the virological response of different IL28B rs12980275 genotypes.
Viral load decline during treatment
By comparison with patients with IL28B rs12980275 AG/GG genotype, patients with AA genotype had a greater reduction in viral load in 4 weeks (W-4), 12 weeks (W-12), 24 weeks (W-24) and 48 weeks (W-48) after treatment (W-4: 6.21 ± 0.86 lg (copies/ml) vs. 5.81 ± 0.80 lg (copies/ml), P = 0.004; W-12: 6.21 ± 0.88 lg (copies/ml) vs. 5.83 ± 0.81 lg (copies/ml), P = 0.008; W-24: 6.22 ± 0.84 lg (copies/ml) vs. 5.81 ± 0.81 lg (copies/ml), P = 0.003; W-48: 6.22 ± 0.86 lg (copies/ml) vs. 5.79 ± 0.93 lg (copies/ml), P = 0.027). There was a significant difference in the viral load reduction between rs12980275 AA group and AG/GG group at each time point.
Taking into account the mixed effects of baseline HCV-RNA viral load levels, the treatment population was further divided into four groups: baseline HCV-RNA <105, 105–106, 106–107, ⩾107 copies/ml. In patients with HCV-RNA <105 and105–106 copies/ml, IL28B rs12980275 had no effect on the decrease in viral load during the treatment (all P > 0.05). In patients with HCV-RNA 106–107 copies/ml, better treatment effect could be achieved among patients with IL28B rs12980275 AA genotypes (W-4: 6.62 ± 0.30 lg (copies/ml) vs. 6.29 ± 0.26 lg (copies/ml), P = 0.002; W-12: 6.51 ± 0.31 lg (copies/ml) vs. 6.31 ± 0.27 lg (copies/ml), P = 0.006; W-24: 6.50 ± 0.30 lg (copies/ml) vs. 6.29 ± 0.28 lg (copies/ml), P = 0.005) (Fig. 2). The sample size of HCV-RNA ⩾107 copies/ml group was too small to be allowed for stratified analysis.
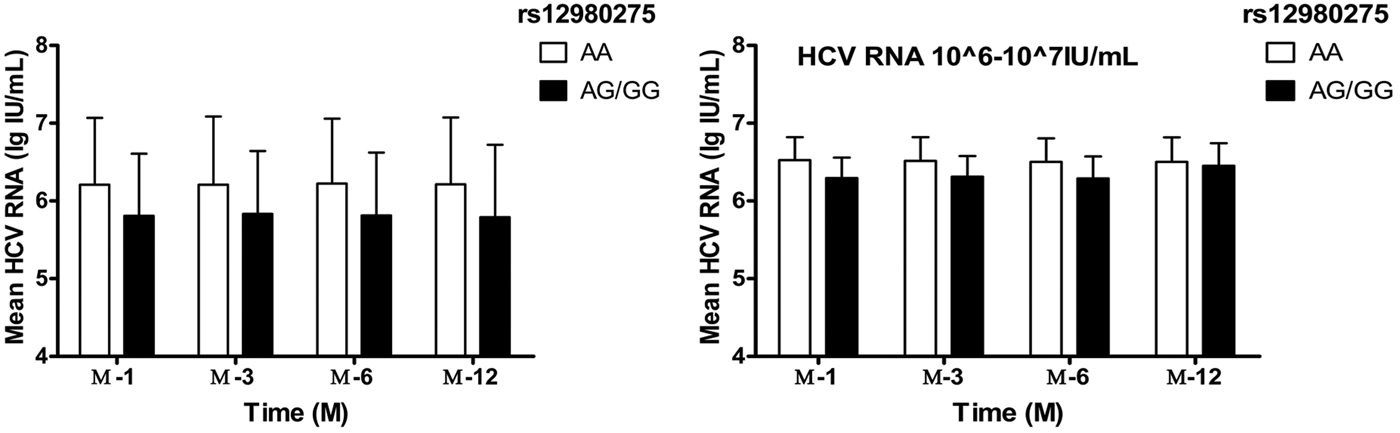
Fig. 2. The viral load decline IL28B rs12980275 AA and AG/GG group. W-4, W-12, W-24 and W-48 represented the mean logarithm of the difference between baseline viral load and 4, 12, 24 and 48 weeks after initiation of treatment.
Compared with patients carrying MxA rs17000900 AC/AA genotype, patients with CC genotype had a greater reduction in viral load in 4 weeks (W-4), 12 weeks (W-12), 24 weeks (W-24) and 48 weeks (W-48) after treatment (W-4: 6.21 ± 0.81 lg (copies/ml) vs. 5.90 ± 0.94 lg (copies/ml), P = 0.023; W-12: 6.21 ± 0.81 lg (copies/ml) vs. 5.91 ± 1.00 lg (copies/ml), P = 0.050; W-24: 6.19 ± 0.82 lg (copies/ml) vs. 5.99 ± 0.90 lg (copies/ml), P = 0.136; W-48: 6.22 ± 0.83 lg (copies/ml) vs. 5.79 ± 1.02 lg (copies/ml), P = 0.021). There was a significant difference in the viral load reduction between rs17000900 CC group and AC/AA only at 4 weeks (Fig. 3).

Fig. 3. The viral load decline MxA rs17000900 CC and AC/AA group. M-1, M-3, M-6 and M-12 represented the mean logarithm of the difference between baseline viral load and 4, 12, 24 and 48 weeks after initiation of treatment.
Discussion
HCV infection was one of the leading causes of advanced liver disease and hepatocellular carcinoma in the world [Reference Omata12]. CHC virus infection clearly posed a challenge to public health. With the development of molecular biology, more and more researches focus on the relationship between gene polymorphism and hepatitis C infection [Reference Xu13, Reference Domagalski14]. The present study investigated the association of IL28B gene and MxA gene polymorphisms with HCV clearance among Chinese Han population. The results indicated that patients with MxA rs2071430 mutant genotype exhibited a worse chance of infection chronicity. IL28B rs1298075 and MxA rs17000900 were also analysed to play a role in the prediction of SVR in CHC patients with pegIFN-α/RBV treatment.
In recent years, sofosbuvir, daclatasvir and the sofosbuvir/ledipasvir combination are part of the preferred regimens in the WHO guidelines, demonstrating cure rates higher than 90% [Reference Polo15]. These direct acting antiviral (DAA) medicines were much more effective, safer and better tolerated than the IFN-based treatment regimens [Reference Welzel16–Reference Bersoff-Matcha18]. Meanwhile, there remains a limited role for pegIFN-α/RBV in certain scenarios where currently data are limited in supporting DAA-only therapies [19, Reference Michelin20]. In addition, the DAA medicines remain very expensive in many high- and middle-income countries, such as China. Therefore, pegIFN-α/RBV is still used as the main treatment in China [Reference Wu21]. IFN-λ relevant genes were in the antiviral treatment pathway, not only influenced HCV spontaneous clearance, but also affected the effectiveness of antiviral treatment.
During acute HCV infection phase, many IFN-stimulating genes (ISGs) were involved in the viral eradication process. The level of ISGs expression was related to IL28B genotype, where carrying the good variant of IL28B could express the lower level of intrahepatic ISG. During the pegIFN-α/RBV treatment, high expression of ISGs was obtained and high SVR rate was achieved [Reference Urban22]. MxA could inhibit the HCV virus replication and protein synthesis by affecting the early transcription of HCV nucleic acid in cytoplasm after binding viral nucleoprotein [Reference Hijikata4, Reference O'Connor23].
Previous meta-analysis results demonstrated that MxA gene was a strong SVR predictor for HCV patients treated with pegIFN-α/RBV in Chinese Han population, which was consistently found in populations of Egyptian [Reference Chen24, Reference Shaker25]. Another study pointed out that MxA gene mutation may be linked with the outcomes of HCV infection [Reference Hu26]. In this study, patients with CC wild genotype were more likely to obtain an SVR, which was first discovered and the stability of the results needed to be validated in different infected individuals (drug addiction, dialysis and other racial groups) or by expanding the sample size.
GWAS in populations of different races have provided valuable clues that there was a strong association between IL28B rs12980275 and HCV treatment response [Reference Suppiah6, Reference Ge7, Reference Liu27]. Consistent with these findings, CHC patients carrying the rs12980275 AA genotype received a better virologic response rate after pegIFN-α/RBV treatment in Chinese Han population. While studying the infection outcome, the sample size of rs12980275 AA mutant patients was too low to be statistically analysed. It was precisely because the mutation frequency of AA genotype was too low in Chinese Han population, and the association between IL28B rs12980275 and HCV treatment outcome was rarely studied. IL28B rs12980275 affected the SVR rate representing the end of treatment, as well as the independent factors of EVR. Moreover, compared with patients with genotype AG/GG, patients carrying the AA genotype could achieve virologic response earlier. The functional importance may be explained by another hepatitis B virus (HBV) infection study, which suggested that serum IL28B levels of HBV patients with AA genotype were higher than other genotypes, which was conducive to reducing HBV load and reduce liver inflammation [Reference Li28].
During the treatment, the decline in viral load was different in different genotypes of IL28B rs12980275. However, the results of stratified analysis of baseline viral load showed that HCV-RNA <106 copies/ml was not significantly affected by IL28B rs12980275, and the lower the viral load, the better the therapeutic effect. By contrast, the influence of MxA rs17000900 on viral load decline was much weaker. The viral load in patients with CC genotype decreased significantly only after 12 weeks treatment.
There were some strengths in our study. First, in prior similar researches, the part of inclusion population was not only infected with HCV, but also with advanced liver diseases, human immunodeficiency virus (HIV) and other diseases [Reference Lunge29–Reference Prokunina-Olsson32]. In our study, all subjects were only suffering from HCV infection without co-infection of HBV or HIV, or without other liver-related diseases. This advantage allowed us to study the HCV clearance more convincingly. Second, we conducted genetic polymorphisms study in the population of HCV spontaneous clearance, and simultaneously investigate the IFN-mediated viral clearance in the treatment cohort. Finally, the sample size of 1090 cases for studying HCV spontaneous clearance was large enough to evaluate the influence of IL28B and MxA gene polymorphisms on HCV clearance. Certainly, the study has some deficiencies. First of all, all patients of the treatment cohort for studying the IFN-mediated virus clearance were from the same hospital in eastern China, which might cause selection bias to a certain extent. Second, although we founded the association of IL28 and MxA genes with HCV clearance, it remains to be explored whether those loci have functional polymorphisms. Frankly speaking, future studies that involve more functional loci are essential for exploring the mechanism of HCV clearance.
To summarise, CHC patients carrying IL28 rs12980275 AA genotype were easier to get a better virological response rate after pegIFN-α/RBV treatment in Chinese Han population. MxA rs2071430 may be the new loci associated with CHC, which could be further studied and verified.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268817002928
Acknowledgements
The current study was supported in part by National Natural Science Foundation of China (No. 81703273, 81473029, 81502853), Natural Science Foundation of Jiangsu Province (BK20171054, BK20151026), the Science and Technology Development Fund Key Project of Nanjing Medical University (2016NJMUZD012), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).








