Introduction
Norovirus is the leading cause of non-bacterial acute gastroenteritis (AGE) across all age groups. It causes about 18% of all cases of AGE worldwide [Reference Ahmed1]. In adults, most of the NoV infection cases manifest slight symptoms or no obvious symptoms at all. Although NoV-associated gastroenteritis is a self-limiting disease, it usually causes unneglectable disease burden. The previous study has shown that there is no difference between treating symptomatic and asymptomatic infections; the estimated virus excretion time is 60 days [Reference Teunis2]. Thus, the asymptomatic infections may have a great impact on the risk of disease spread. In a previous report, cross-sectional surveys in children <5 years of age showed a large variation ranging between 2% and 24% [Reference My3–Reference Huynen5]. Studies including the population of all age groups have been only reported in England [Reference Phillips4] and Netherlands [Reference de Wit and Kortbeek6]. In China, there are no related studies on asymptomatic NoV infection in a community population.
Oysters not only act as the transmission vector of NoV in the environment but also are an important reservoir of NoV [Reference Burkhardt and Calci7]. NoV outbreaks associated with oyster consumption have been well documented all over the world. The literature reported that approximately 90% of human NoV sequences were discovered in coastal regions of China [Reference Yu8]. The people living in a coastal area commonly consumed more shellfish compared with inland population. However, there is very little understanding of the norovirus infection prevalence in the coastal population. In the present preliminary study, we investigated the prevalence of norovirus contamination on one oyster farm area (Changsha Bay area) and found that the total norovirus contamination rate in oysters was 32.6% [Reference An-na9], which was higher compared with contamination level of 12% found in cultured oysters from China's coastal areas [Reference Ming10]. Therefore, a population survey addressing the norovirus contamination in villages surrounding oyster farms were conducted along with universal stool collection.
Epidemiological methods
Objective and case definition
Cases with asymptomatic norovirus infection were those cases which did not belong to AGE or where AGE symptoms were not present, but stool samples tested positive for infection (analysed using real time-qualitative polymerase chain reaction (RT-qPCR) and semi-nested PCR). The exposed population was people who are working on oyster farm sites, including their family members.
AGE was defined as diarrhoea of ⩾3 loose stools in a 24-h period, or as significant vomiting with at least one other symptom (abdominal pain/cramps, fever), but excluding cases with (a) Crohn's disease, irritable bowel syndrome, colitis, diverticulitis of large intestine, or another chronic illness with symptoms of diarrhoea or vomiting, or (b) pregnancy, consuming excess alcohol, undergoing chemotherapy/radiotherapy, drugs, or had food allergy.
Study design and sample collection
Changsha Bay, located in the southeast of Guangdong Province and regarded as one of the major marine product industries in China, was chosen as the main target area. Nine villages located around these three oyster farms, i.e. farm A, B and C were selected as survey spots (Fig. 1). These farming sites do not have a wastewater treatment plant, instead, the domestic sewage is discharged directly into Changsha Bay without purification. The villagers on average do not have high levels of education, they have poor health awareness and some have the habit of eating raw oysters.
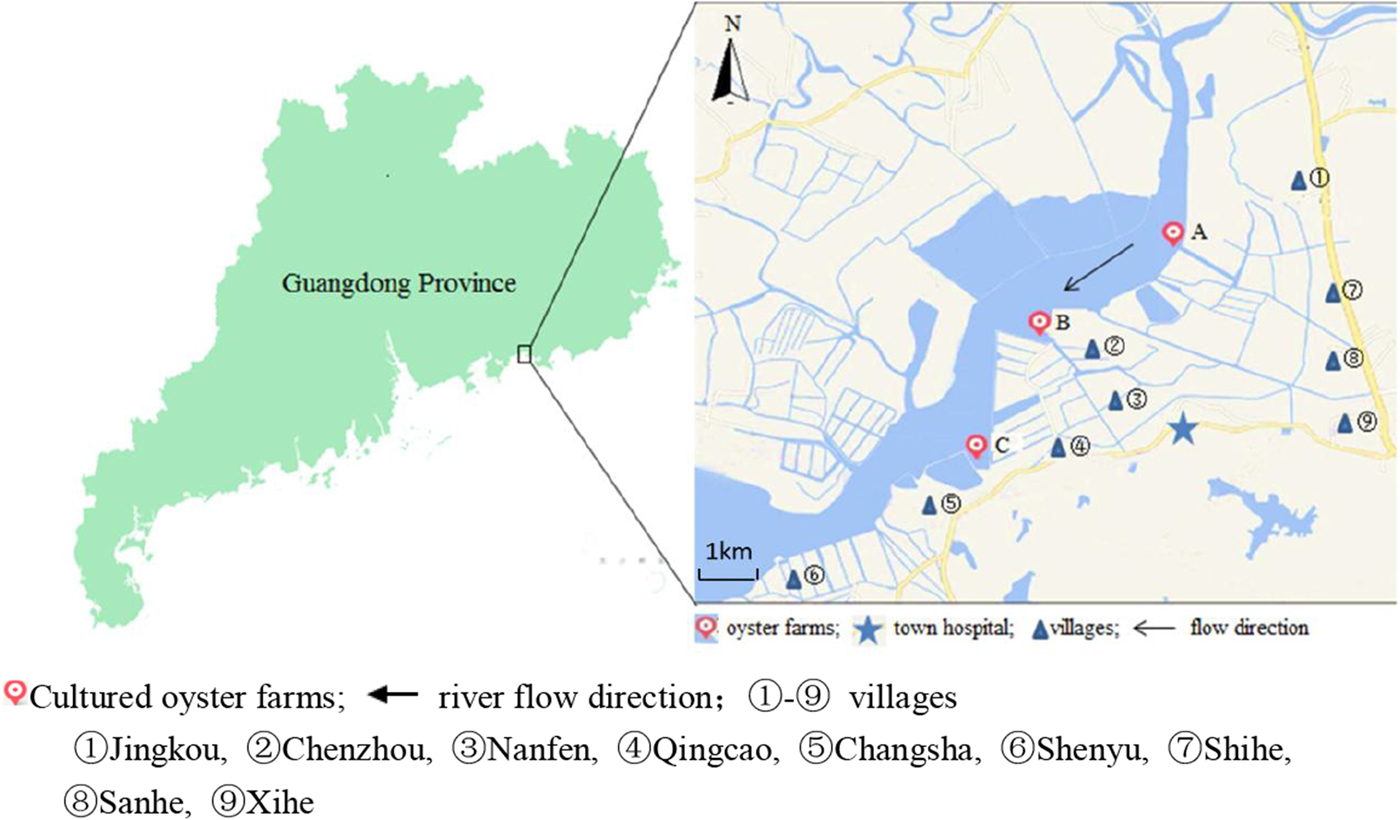
Fig. 1. Map of Changsha Bay and survey spots in the southeast of Guangdong province, China.
A cross-sectional study was conducted including all the residents around the oyster farms. The survey was evenly distributed within 6 months; two consecutive surveys were distributed between January and June 2014 and between July and December 2014. According to the household registration data, there were 19 800 residents in the area. A total of 10 415 stool samples were collected; the response rate was about 30%. Taking into account the representativeness and feasibility of samples, part of samples were randomly selected according to population proportion among nine villages. According to the living distance to the oyster farms, the nine villages were divided into two groups, <3 km and ⩾3 km group. The <3 km group included five villages (Jingkou, Chenzhou, Nanfen, Qingcao and Changsha), while the ⩾3 km group included four villages (Shihe, Sanhe, Xihe and Shenyu).
Household interviews were conducted on monthly basis by trained health workers using a standard questionnaire for AGE. The questionnaire was developed and validated specifically for the purposes of this study. Adults completed the questionnaire themselves, while a parent or guardian completed the questionnaire on behalf of children <18 years old. Each respondent was asked to provide a stool sample regardless if he/she was suffering from AGE. The samples were collected within 24 h after the interview. The stool samples were collected by the village health workers and were directly transferred to the town hospital and stored at −20 °C. During the last week of each month, all of the samples were transferred to the laboratory for testing.
The survey and stool sample collection were approved by the medical ethics committee and informed consent was obtained from all the participants.
Laboratory methods
RNA preparation and extraction
Aliquots of 0.1 g stools were re-suspended in 1 ml; and then every ten-stool suspension was mixed to one mixture sample, the mixture volume was 1 ml. The mixture samples were centrifuged at 8000 × g at 4 °C for 10 min. Viral RNA was extracted from the 200 µl of supernatants using the High Pure Viral RNA Kit (Roche, USA) according to the manufacturer's instructions. Viral RNA was then eluted into 50 µl of elution buffer and stored at −80 °C. Every single sample contained in the mixture sample was tested one by one when the mixture sample can get Ct value through RT-qPCR detection. The viral RNA was extracted as explained above.
RT-qPCR
RT-qPCR was performed on an CFX96 real-time PCR instrument (Bio-Rad, USA) and carried out using the One Step PrimeScript™ RT-PCR Kit (Perfect Real Time) (Takara, JPN). For NoV GI analysis, previously described [Reference Kageyama11] primers COG1F and COG1R and probes RING1(a)-TP and RING1(b)-TP were used. For NoV GII analysis, primers COG2F and COG2R and probe RING2-TP were used. The detection results were analysed according to the following criteria: (a) when the Ct value was ⩽35, the sample was considered positive; (b) when the Ct value was ⩾40, the sample was considered negative; (c) when the Ct value was between 30 and 40, the sample was suspected positive and was the re-tested using semi-nested PCR to confirm the result.
Semi-nested PCR
The suspected positive samples and confirmed positive samples detected during the last step were subjected to gene amplification of partial NoV capsid gene to determine the genotype. The first round of semi-nested PCR was performed using PrimeScript One Step RT-PCR Kit (Takara, JPN), while the second round was performed using Premix Taq™ (Ex Taq™ Version 2.0) both in a 25 µl reaction. The primers used for PCR have been described previously [Reference Kojima12]. Negative controls were included to avoid false-positive results because of cross-contamination and false-positive PCR product has been excluded.
Sequencing and phylogenetic analysis
The second-round PCR products were separated by electrophoresis in a 1% agarose gel, then visualised under ultraviolet (UV) illumination after ethidium bromide staining and finally sent to the Sangon Biotechnology Company for sequencing. The genotype was defined by NoV Genotyping Tool (http://www.rivm.nl/mpf/norovirus/typingtool). The length of NoV gene region used was 291 bp and 303 bp for NoV GI and GII, respectively. The phylogenetic tree was constructed based on partial nucleotide sequences of the NoV capsid gene and according to the neighbour-joining method using MEGA 5.0 software. The reliability of the phylogenetic tree was assessed by a bootstrap sampling of 1000 replicates and the genetic distances were calculated by Kimura's two-parameter method.
Statistical analysis
EpiData version 3.1 was used for data entry. Statistical analysis was performed using Microsoft Excel 2010 and SPSS version 19.0. The comparison of positive rates was done using χ 2 test or Fisher's exact test. P-values <0.05 were considered statistically significant. Multivariate analysis of these variables and their interaction terms was done using manual forward, backward stepwise and best subsets logistic regression, with the 95% confidence interval being used to determine a variables' significance. Variables with a P-value <0.10 were included in the final model.
Results
The prevalence of NoV asymptomatic infection
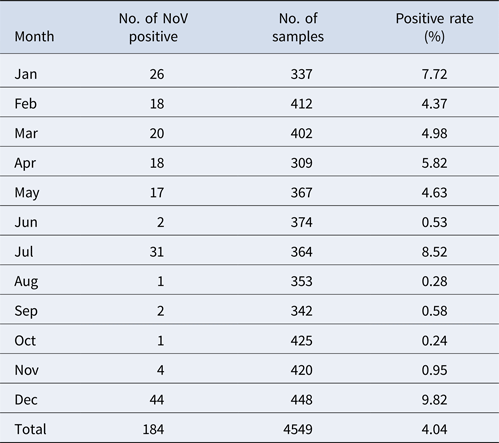
Stool samples were obtained from 10 415 respondents during a period between January and December 2014. Among those, 85 AGE cases were found; others were asymptomatic or healthy. Taking into account the representativeness and feasibility of samples, we further selected 4549 stool samples according to the population proportion from each village. Among these, 184 samples tested positive for NoV with the total asymptomatic infection rate of 4.04%. GI and GII were detected in 88 (47.8%) and 91 (49.4%) samples, respectively; while 5 (2.7%) samples were positive for both GI and GII.
Monthly distribution of NoV asymptomatic infection
The monthly NoV asymptomatic infection cases are shown in Table 1. The highest positive rate (9.82%) was observed in December and the lowest (1%) between August and November. Significantly higher detection rates were observed in winter/spring seasons (December–May) compared with summer/autumn seasons (June–November) (6.20% vs. 1.80%, χ 2 = 59.25, P < 0.05). It is worth noting that the rate in July was as high as 8.52%. For the genogroup distribution, the highest positive rate for GI was observed in July and for GII in December (Fig. 2).
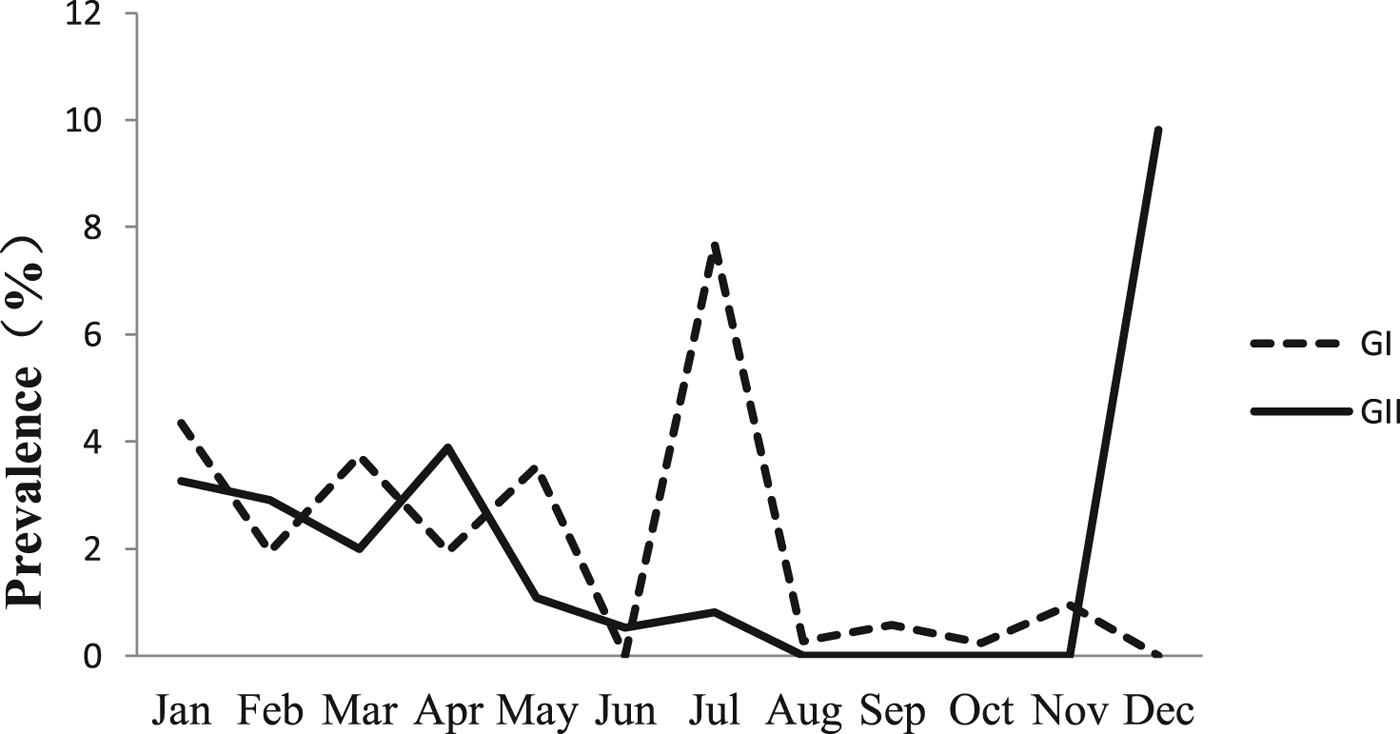
Fig. 2. Monthly prevalence of asymptomatic norovirus infection, by genogroups GI and GII, in Changsha Bay oyster farm sites, China.
Table 1. Monthly distribution of asymptomatic norovirus infection cases in Changsha Bay oyster farm sites, China from January to December 2014
Population distribution of NoV asymptomatic infection
NoV asymptomatic infection occurred in all age groups (Table 2). The highest positive rate was found in 0–5 age group (5.12%) and lowest in 6–17 age group (3.48%); nevertheless, no significant difference was observed between different age groups. Similarly, there was no significant difference in positive rates between male and female (3.89% vs. 4.24%, χ 2 = 0.38, P > 0.05). With reference to occupation, the NoV positive rate in oyster farming population was higher compared with other occupations (5.20% vs. 3.65%, χ 2 = 5.49, P < 0.05). Moreover, considering the prevalence of high positive rate in children under 5 years, we further conducted a comparative analysis and found that there is still a significant difference in positive rates (5.08% vs. 3.53%, χ 2 = 5.23, P < 0.05).
Table 2. Characteristics of asymptomatic norovirus infection cases in Changsha Bay oyster farm sites, China from January to December 2014
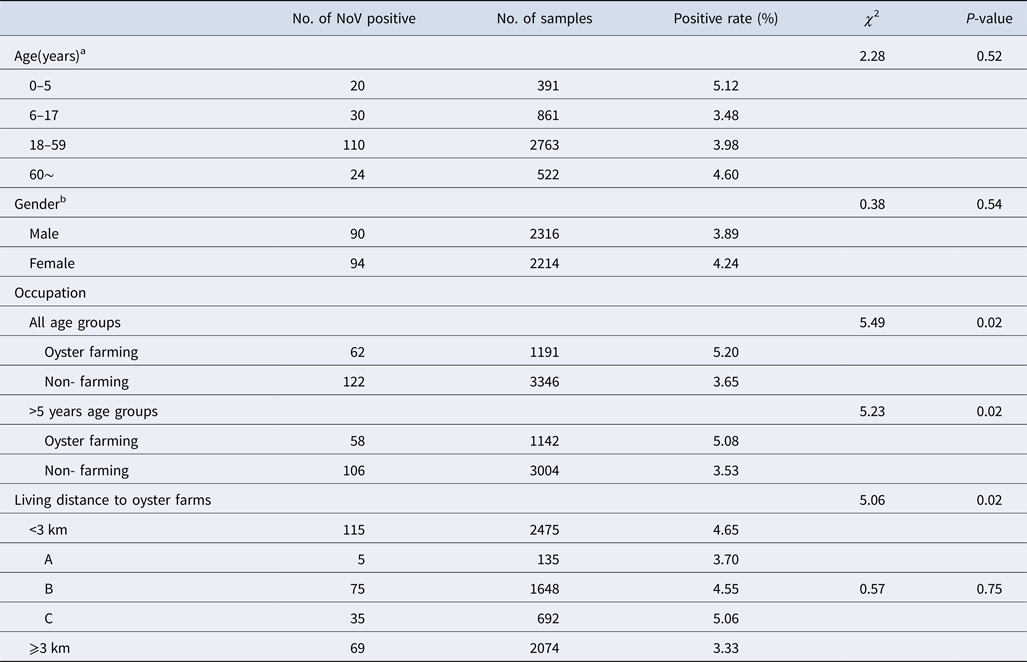
a Delete 12 missing value, n = 4537.
b Delete 19 missing value, n = 4530.
The living distance factor was also associated with NoV infection. Briefly, the infection rate was significantly higher in residents who are living <3 km from the farm compared with those living ⩾3 km (4.65% vs. 3.33%, χ 2 = 5.06, P < 0.05). Yet, for people living <3 km, no significant differences were observed in the infection rates of villages located near three oyster farms (χ 2 = 0.57, P > 0.05).
Multivariable analysis
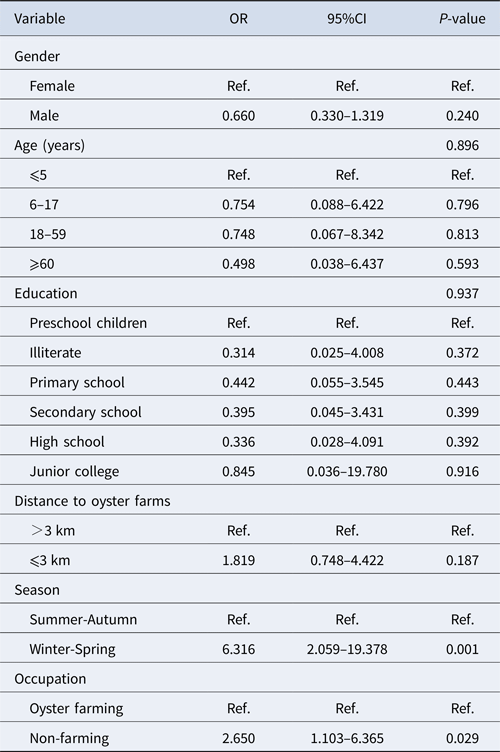
Among 184 positive samples, 93 were GI and 96 were GII. Multivariable analysis of the prevalence of genogroup was undertaken with gender, age group, level of education, residence distance, season and occupation as explanatory variables (Table 3). Variables with a P-value <0.10 were kept in the model. The significant variables in the final model were season (P < 0.001) and occupation (P = 0.029). Winter/spring season and individuals related to oyster culture were more likely associated with GII infection.
Table 3. Multivariable analysis of risk factors associated with GI and GII norovirus infection in Changsha Bay oyster farm sites, China
Genotyping analysis
A total of 184 NoV positive samples were identified by RT-qPCR and semi-nested PCR. Among these, 136 sequences were obtained, including 66 GI and 70 GII sequences. GI strains were clustered into six genotypes, namely GI.2, GI.3, GI.5, GI.6, GI.8, GI.9. GII strains were clustered into eight genotypes, namely GII.2, GII.3, GII.4, GII.5, GII.6, GII.8, GII.13 and GII.17. The genetic diversity was found to be highest in February, including eight genotypes. The largest numbers of strains were detected in December. GI.9 was the most prevalent genotype in GI strains; GI.9 was distributed over 5 months (January, February, April, May and July) and the highest number of strains (18) were observed in July. In addition, GII.17 was the predominant genotype in GII strains; GII.17 was distributed over 5 months (February, March, April, May and June) and the highest number of strains (42) were observed in December (Table 4).
Table 4. Monthly distribution of norovirus genotypes detected from asymptomatic norovirus infection cases in Changsha Bay oyster farm sites, China
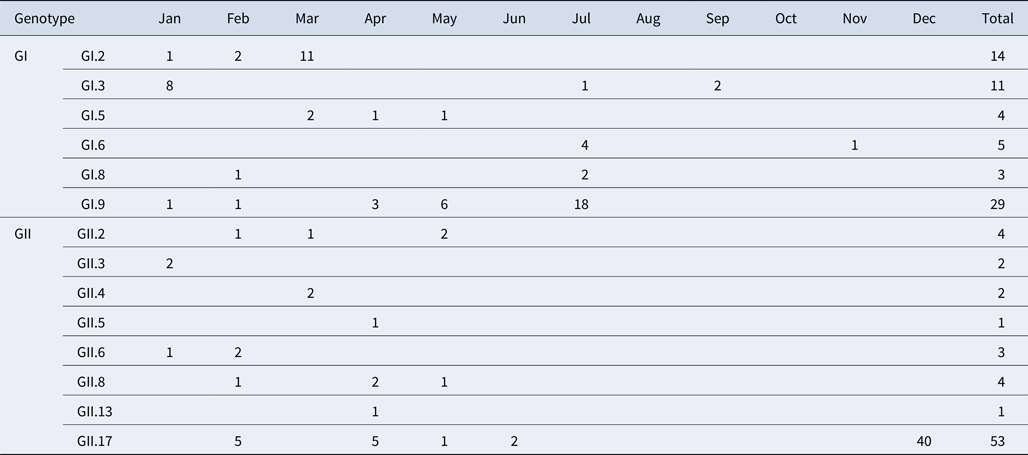
The number in the table represents the numbers of strains in each genotype per month.
Phylogenetic analysis
The phylogenetic analysis was conducted using all 138 sequences identified from asymptomatic norovirus infection cases. GI strains were clustered into six major clusters; GI.9 were divided into two minor clusters (Fig. 3). The GI.5 sequences were highly similar to the sequences isolated from Korea clam (KT438802) and an oyster-related outbreak in Shanghai (KP325650). In addition, the GI.8 sequences were closely related to the strain isolated from an outbreak on a cruise ship in Nanjing (KP753284). The GI.9 was the most frequently detected genotype; a total of 29 sequences shared 89.6% to 100% nucleotide sequence identity to strains found in Korea (KT383939), New Zealand (KT151005) and China (KP753287, KF586509). GII sequences included eight major clusters among which GII.17 was divided into two minor clusters (Fig. 4). The two GII.4 strains belonged to the Sydney 2012 variant. The GII.5 sequence was highly similar to the strain from the waterborne outbreak in Korea (DQ004642) and the sewage in Nanning (KM246934). GII.17 was the predominant genotype and the strains were grouped into two minor cluster, cluster A and B. In the cluster A, the sequences shared 70.9% to 100% nucleotide sequence identity to strains from Japan (AB983218, LC043305), Korea (KF773972), Kenya (KF916584) and China (KR107599). In the cluster B, the sequences shared 92.5% to 99.3% nucleotide sequence identity to the strains from Hong Kong (KT589391), Korea (KT384029) and China (KT716724); whereas the high range of genetic distance was observed between the sequences isolated before 2012 and after 2012. On the basis of the phylogenetic analysis, all of the GII.17 sequences identified in this study belonged to the novel GII.17 variant, which emerged and caused an increased number of related outbreaks at the end of 2014–2015 in China.
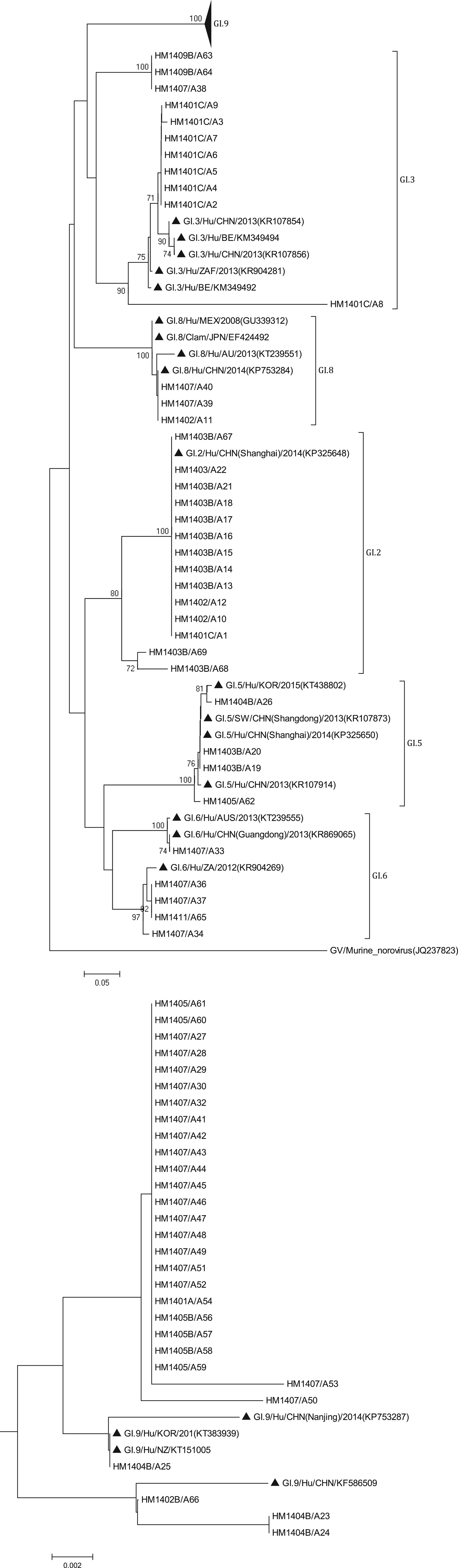
Fig. 3. Phylogenetic tree based on partial nucleotide sequences (291bp)of the capsid gene (ORF2) of total of 68 NoV GI strains identified in this study, and constructed by using Mega 5.0 software. The distance was calculated by Kimura's two-parameter method, and the tree was plotted by the neighbour-joining method. Numbers at each branch indicate bootstrap values for the clusters supported by that branch, and GV murine norovirus strains (JQ237823) was used as outgroup. The GenBank and strains accession numbers of the reference were as follows: GI.3, KR107854, KM349494, KR107856, KR904281, KM349492; GI.8, GU339312, EF424492, KT 239551, KP753284; GI.2, KP325648; GI.5, KT438802, KR107873, KP325650; KR107914; GI.6 KT239555, KR869065, KR904269; GI.7 KT150981; KF395222; GI.9, KP753287, KT383939, KT151005, KF586509.

Fig. 4. Phylogenetic tree based on partial nucleotide sequences (303bp) of the capsid gene (ORF2) of total of 79 NoV GII strains identified in this study, and constructed by using Mega 5.0 software. For NoV GII.17 sequences, a separate phylogenetic tree was built. The distance was calculated by Kimura's two-parameter method, and the tree was plotted by the neighbour-joining method. Numbers at each branch indicate bootstrap values for the clusters supported by that branch, and GV murine norovirus strains (JQ237823) was used as outgroup. The GenBank and strains accession numbers of the reference were as follows: GII.2, KF495143, GII.5, DQ004642, KM346934; GII.13, KR706460, KR706455; GII.8, KP963765, JF292510, JF802498, JF973385; GII.6, KT151026, KT151027, KM036377; GII.3, KC820420, LC089390, KC464495; GII.4, KJ716357, KJ938996, KJ533133,KF177432, KR107566; GII.17, KT384021, KP718638, AB983218, KF773972, LC043305, KF916584, KR107599, LC037415, KT716724, KT589391, KT384029,GQ266696, KC495680, JQ944348, DQ438972, KC915021.
Discussion
Norovirus is considered the leading cause of non-bacterial AGE worldwide. This study investigated the prevalence of asymptomatic norovirus infections in the population living around oyster farms. During the period from January to December 2014, a total of 184 out of 4549 stool samples were identified and positively screened for norovirus; the total asymptomatic infection rate was 4.04% among all age groups. The highest infection rate was detected in January (7.72%), July (8.52%) and December (9.82%); these results very similar with data from Netherlands (5.2%) [Reference de Wit and Kortbeek6], somewhat higher compared with data from Korea (2.3%) [Reference Koo13] and significantly lower compared with data from Japan (12%) [Reference Okabayashi14] and England (12%) [Reference Phillips4]. The study carried out in Netherlands was based on community population; studies in Korea and Japan were based on food handlers, while the study in England investigated all age groups between 1993 and 1996. Considering the fact that the prevalence of norovirus could be influenced by population composition, study design and norovirus epidemic season, the data that show a range of fluctuations are still acceptable [Reference Matthews15]. Compared with other studies which have reported that infection rate of norovirus-related AGE increased in October, our data indicated that the infection rate was significantly increased in December, while it was low (<1%) in November. Guangdong province is located in a tropical region, thus the temperatures during the late November are still warm compared with northern countries. Meanwhile, we found that the positive rate of gastroenteritis was only 0.10 times/person-years [Reference Lin16], which is significantly lower compared with 0.56 times/person-years in China [Reference Chen17]. Although the incidence of AGE cases was in low level, the asymptomatic infection rate was high. This suggested that the population living in the oyster farming area might have immune protection, which needs to be further investigated.
Bivalve molluscs, especially oysters, are recognised as high-risk food associated with norovirus-related gastroenteritis outbreaks worldwide. Over recent years, UK, France, Norway, Sweden and Denmark reported approximately 70 oyster-related norovirus gastroenteritis outbreaks, with more than 400 cases [Reference Westrell18, Reference McIntyre19]. Our preliminary study showed that the norovirus contamination rate in local oysters in the Changsha Bay is significantly higher compared with other coastal areas of China. In the present study, population working on oyster farms and those living <3 km near the farms had a significantly higher percentage of norovirus infection compared with other groups. Meanwhile, the multivariable analysis results showed that norovirus GII infection is more likely associated with population working on oyster farm sites. We found that the people who live near the farms are highly exposed to infection; farmers usually eat raw or salted oysters, or engage in processes such as washing, opening, airing and transportation of oysters and thus are exposed to direct contact which may cause them to inhale aerosol and increase the chance of infection. Moreover, GII strains tend to spread by oyster farm workers to their family members and other healthy villagers and thus pose a potential outbreak threat.
Furthermore, we conducted a phylogenetic analysis of nucleotide sequences isolated from local patients, cultured oysters and sewage in the survey area [Reference An-na9]. Our data showed that the sequences in these three types of samples shared a high nucleotide identity and a close genetic distance. The norovirus infection in the population and oyster contamination should be interconnected. Because there is no sewage treatment in the villages, the domestic dirt is discharged directly into Changsha Bay without purification. Therefore, we speculated that there is a circulating transmission route between the environment and population by the oysters considered as the transmission vector. To sum up, the infected people could excrete the virus from their body, thus contaminating the oysters in the water. Therefore, it is necessary to control the contamination of norovirus in aquaculture water and shellfish to decrease infection. The sewage treatment plant construction, the education about hygiene and healthy eating habits could be useful control measures.
In this study, the norovirus strains detected in asymptomatic infected persons were genetically diverse. A total of 14 different GI and GII genotypes were identified, namely GI.2, GI.3, GI.5, GI.6, GI.8, GI.9 and GII.2, GII.3, GII.4, GII.5, GII.6, GII.8, GII.13, GII.17. Genogroups GI and GII accounted for about 50%, which was somewhat unusual since GII is typically the most prevalent genotype in norovirus outbreaks. A systematic review showed that GI strains were associated with waterborne and foodborne outbreaks, while GII.4 strains were associated with person-to-person outbreaks [Reference Verhoef20]. This genotype was even detected from an outbreak that occurred on a cruise ship. Besides, there have been few case reports referring to GI.9. Nevertheless, in our study, we found many asymptomatic infected cases which reminded us that the risk caused by GI.9 genotype should not be neglected. Also, a recent study on norovirus outbreaks in the USA [Reference Verhoef20] indicated that GI.3, GI.6, GII.3 and GII.6 are the norovirus genotypes most often associated with foodborne outbreaks. Based on the phylogenetic analysis, our data suggested that norovirus infection is associated with environment pollution. There might exist a widespread contamination of norovirus in the local environment. Therefore, this study calls for immediate long-term and systematic surveillance of symptomatic and asymptomatic norovirus infected cases so as to trace and control outbreaks. Furthermore, it is essential to improve environmental sanitation.
GII.17 strains were detected over 6 months; the earliest strain was detected in February, while the largest number, i.e. 42 strains were detected in December. It is worth noting that GII.17 has replaced GII.4 and become the predominant genotype in the oysters farming area; while all of the GII.17 strains were blasted to be novel GII.17 variants. In China, the novel GII.17 has been firstly reported in the environmental surveillance report in Shandong Province [Reference Tao21], but not as the dominant genotype. Subsequently, the novel GII.17 variant caused a sharp increase in the number of AGE outbreaks in Asia, including Jiangsu [Reference Fu22] and Guangdong province [Reference Lu23] in 2014–2015 winter seasons. The strains have also been reported to spread broadly in America, Africa, as well as in European and Australian regions [Reference De24]. The recent data have shown that the novel GII.17 become the dominant genotype in AGE cases in China in 2015 [Reference Wang25]. In our study, GII.17 was first detected from an asymptomatic person in February 2014, which is much earlier than the AGE cases reported in Jiangsu and Guangdong provinces at the end of 2014–2015. These findings suggested that the continuous surveillance can reveal the emergence of novel variant strains. The prevalence of asymptomatic infection in population reflects the dynamic change and it is useful for identifying the outbreaks at the early stage.
In conclusion, we found that norovirus asymptomatic infection exists in the residents living around oyster farms and the infection was significantly present in the oyster farming population. The identified norovirus genotypes had a high genetic diversity and the GII.17 replaced GII.4 as the dominant genotype in this area. Long-term and systematic surveillance of the asymptomatic cases needs to be performed so as to predict the emergence of new variants and related outbreaks. Improving environmental sanitation, educating the population and avoiding eating raw food could serve as useful control measures.
Acknowledgements
We appreciated the assistance from the Hongcao and Magong village clinics staff. This study was supported by the National Science Foundation of China (No. 81502859).
Conflict of interest
The authors declare no conflict of interest.











