Introduction
Some members of the genus Acinetobacter are important healthcare-associated pathogens, being closely linked to outbreaks worldwide [Reference Peleg, Seifert and Paterson1]. The most common species isolated from clinical samples is A. baumannii, however, with improved laboratory identification methods, an increasing number of other Acinetobacter species are now recognised as being clinically significant particularly due to their association with nosocomial outbreaks and antibiotic resistance [Reference Yamamoto2]. A. calcoaceticus, A. baumannii, A. pittii and A. nosocomialis are closely related species that are phenotypically indistinguishable by routine laboratory technologies, hence the term ‘A. calcoaceticus–A. baumannii (Acb) complex’. Recently, A. seifertii and A. dijkshoorniae have been included in this complex [Reference Nemec3, Reference Cosgaya4].
Acinetobacter spp. are widespread in the hospital environment, especially A. baumannii strains belonging to worldwide distributed clones such as European clones I, II and III. These are defined by multilocus sequence typing and assigned according to the Pasteur Institute scheme (MLST-IP) to clonal complexes (CCs) 1, 2 and 3, respectively [Reference Peleg, Seifert and Paterson1]. More recently, the isolation of multidrug-resistant (MDR) strains of A. pittii and A. nosocomialis of various sequence types (STs) in healthcare facilities has been described around the world [Reference Al Atrouni5]. Carbapenems are the drug of choice against infections caused by MDR Acinetobacter and the production of oxacillinase (OXA)-type carbapenemases is the main resistance mechanism to these antimicrobials. These comprise four main groups, namely, OXA-23-like; OXA-24/40-like; OXA-58-like; OXA-51-like (intrinsic in A. baumannii); OXA-143-like and OXA-235-like are quite rare [Reference Higgins6].
Patients with cystic fibrosis (CF) are frequently admitted to hospitals for the treatment of acute pulmonary exacerbations and severe complications due to a limited range of bacterial pathogens, primarily Pseudomonas aeruginosa, Staphylococcus aureus, among others, but Acinetobacter spp. are seldom detected in these patients [Reference Coenye7]. We report here the characterisation of a series of isolates of Acinetobacter spp. recovered from the sputum of Brazilian patients with CF with regard to species distribution, CC and antimicrobial resistance mechanisms.
Materials and methods
We conducted a retrospective study of Acinetobacter spp. isolated from CF patients from June 2005 to February 2014 attending the Instituto Nacional da Saúde da Mulher, da Criança e do Adolescente Fernandes Figueira (IFF-FIOCRUZ), and Hospital Universitário Pedro Ernesto (HUPE-UERJ), two reference centres for paediatric and adult patients with CF, respectively, in Rio de Janeiro, Brazil. Respiratory samples were cultured in accordance with standard bacteriological protocols for CF samples [Reference Gilligan, Kiska and Appleman8] and presumptive Acinetobacter spp. isolates were confirmed by phenotypic tests and assigned to species level by PCR amplification and partial sequencing of the rpoB gene as described previously [Reference Gundi9].
Sequence typing
All isolates classified as members of the Acb complex were subjected to MLST, by PCR amplification and sequence analysis of seven housekeeping genes (cpn60, fusA, gltA, pyrG, recA, rplB and rpoB) in accordance with the MLST-IP scheme [Reference Diancourt10]. CCs were identified by the eBURST algorithm (http://eburst.mlst.net/), and defined as a cluster formed by the founder ST linked to other STs sharing six identical (single locus variants – SLVs) allele profiles at each of the seven MLST loci [Reference Feil11].
Antimicrobial susceptibility
Isolates were tested for antimicrobial susceptibility by the disk diffusion method according to the Clinical and Laboratory Standards Institute guidelines [12] with the following agents: amikacin, ampicillin/sulbactam, cefepime, cefotaxime, ceftazidime, ciprofloxacin, gentamicin, imipenem, meropenem, piperacillin/tazobactam and tobramycin (Oxoid Ltd Basingstoke, UK). P. aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as quality controls. Strains that were resistant to at least one agent in three classes of antimicrobials were defined as MDR. Multiplex PCR for the detection of genes encoding OXA bla OXA23-like, bla OXA24-like, bla OXA51-like, bla OXA58-like and bla OXA143-like was performed as previously described [Reference Woodford13, Reference Higgins, Lehmann and Seifert14].
Results
Fifty-three Acinetobacter spp. strains were isolated from 32 paediatric and seven adults (Table 1). In seven paediatric patients, more than one Acinetobacter strain defined by species, ST and/or resistance markers was isolated. The mean age of first isolation of Acinetobacter spp. was 6 and 26 years for paediatric and adults, respectively, and in four patients (one child and three adults) colonisation was first detected on hospitalisation. Twenty of the 39 patients were also colonised with pathogens commonly found in CF, with P. aeruginosa being the most frequent.
Table 1. General features of Acinetobacter spp. strains isolated from Brazilian patients with CF
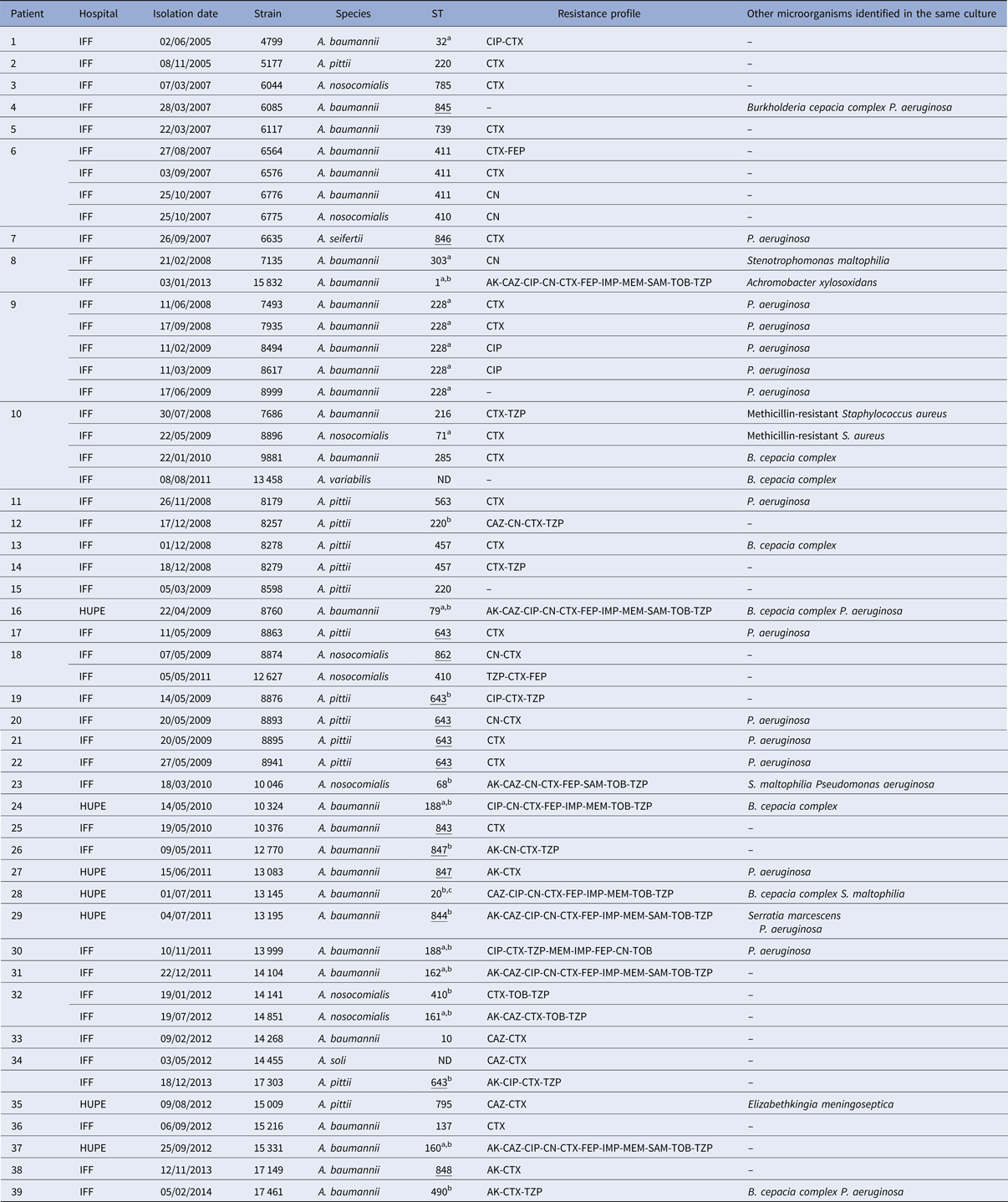
ND, MLST not determined.
Novel ST deposited in PubMLST from this study.
a Also isolated from non-CF patients in Rio de Janeiro state [Reference Grosso25].
b MDR strain.
c Also isolated from non-CF patients in São Paulo state [Reference Chagas26].
AK, amikacin; CAZ, ceftazidime; CIP, ciprofloxacin; CN, gentamicin; CTX, cefotaxime; FEP, cefepime; IMP, imipenem; MEM, meropenem; SAM, ampicillin/sulbactam; TOB, tobramycin; TZP, piperacillin/tazobactam.
Partial rpoB gene sequencing identified the species distribution as A. baumannii (n = 29), A. pittii (n = 13), A. nosocomialis (n = 8), and one each of A. seifertii, A. soli and A. variabilis.
Although most strains showed susceptibility to the majority of the antimicrobials tested, with the exception of cefotaxime (Table 1), 16 (30%) were classified as MDR (10 A. baumannii, three A. pittii and three A. nosocomialis). Eight of the MDR A. baumannii were resistant to carbapenems (meropenem and imipenem) and these were the only isolates harbouring the bla OXA23-like gene (Table 1). None of the other OXA genes tested was present in any of the 53 strains.
MLST analysis identified 33 unique STs among the 51 Acb complex strains: 21 STs (five novel) for A. baumannii, five (one novel) for A. pittii, six (one novel) for A. nosocomialis and one novel ST for A. seifertii (Table 1). By reference to the MLST-IP scheme database, 11 STs (two A. nosocomialis and nine A. baumannii) had previously been identified in two states in Southeastern Brazil (Rio de Janeiro and São Paulo). Among the eight novel STs identified, five (A. pittii ST643, A. nosocomialis ST862, ST843, ST844 and ST848 of A. baumannii) had a hitherto unrecognised allelic arrangement; three novel allele profiles (A. seifertii ST846, ST845 and ST847 of A. baumannii) were added to the MLST-IP database (Table 1).
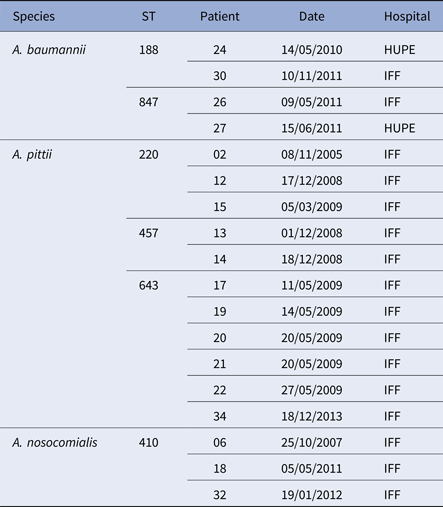
Seven paediatric patients (#6, 8, 9, 10, 18, 32 and 34) had more than one positive culture for Acinetobacter species. Two of them had persistent colonisation with the same A. baumannii ST: patient 6 by ST 411 and patient 9 by ST 228. On one occasion, patient 6 was co-colonised with A. nosocomialis (ST410). Three different Acinetobacter species were recovered from patient 10 from 2008 to 2010: A. baumannii (ST216 and ST285), A. nosocomialis (ST71) and A. variabilis (ST not determined). Moreover, two patients were sporadically colonised by different A. nosocomialis STs (patient 18: ST842 and ST410; patient 32: ST410 and ST161). Likewise, patient 34 harboured A. soli (ST not determined) and A. pittii (ST 643), and patient 8 yielded A. baumannii of two different STs, one of which was MDR corresponding to European clone I (ST1) (Table 1). Two A. baumannii, three A. pittii and one A. nosocomialis STs were also found in different patients but often separated in time and/or hospital (Table 2).
Table 2. Acinetobacter baumannii complex STs shared by different patients with CF
eBURST analysis identified STs belonging to three important A. baumannii CCs: CC1 (ST1, ST20 and ST160), CC25 (ST228) and CC79 (ST79) (Fig. 1). Moreover, comparison of the profiles of the four A. nosocomialis STs found in this study with the MLST-IP database showed that ST71, ST161 and ST410 (group founder) are SLVs of the latter (Fig. 2).
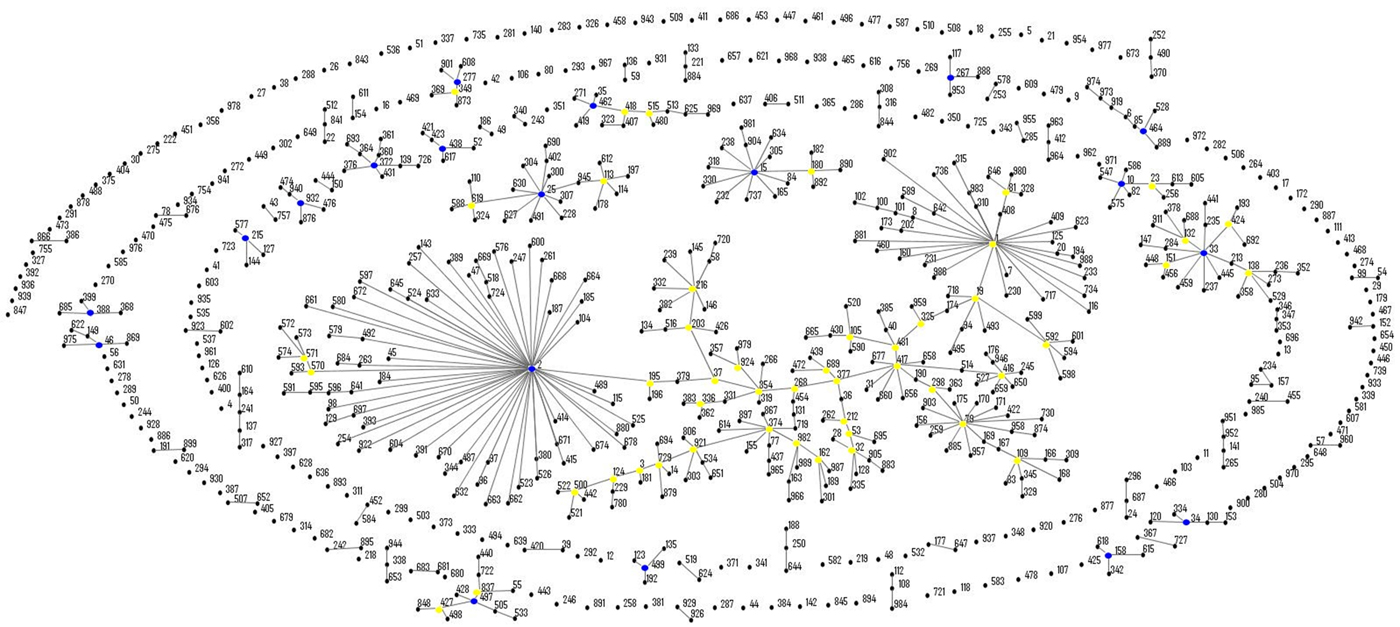
Fig. 1. Population snapshot of A. baumannii isolates in this study and existing isolates in the Institut Pasteur's MLST database by eBURST algorithm (http://pubmlst.org/abaumannii/; 24 April 2017, date last accessed). Each ST is represented by a black dot. Blue and yellow dots correspond to group and subgroup founders, respectively. SLVs are linked by lines, and clonal complexes (CC) correspond to the group of connected STs.
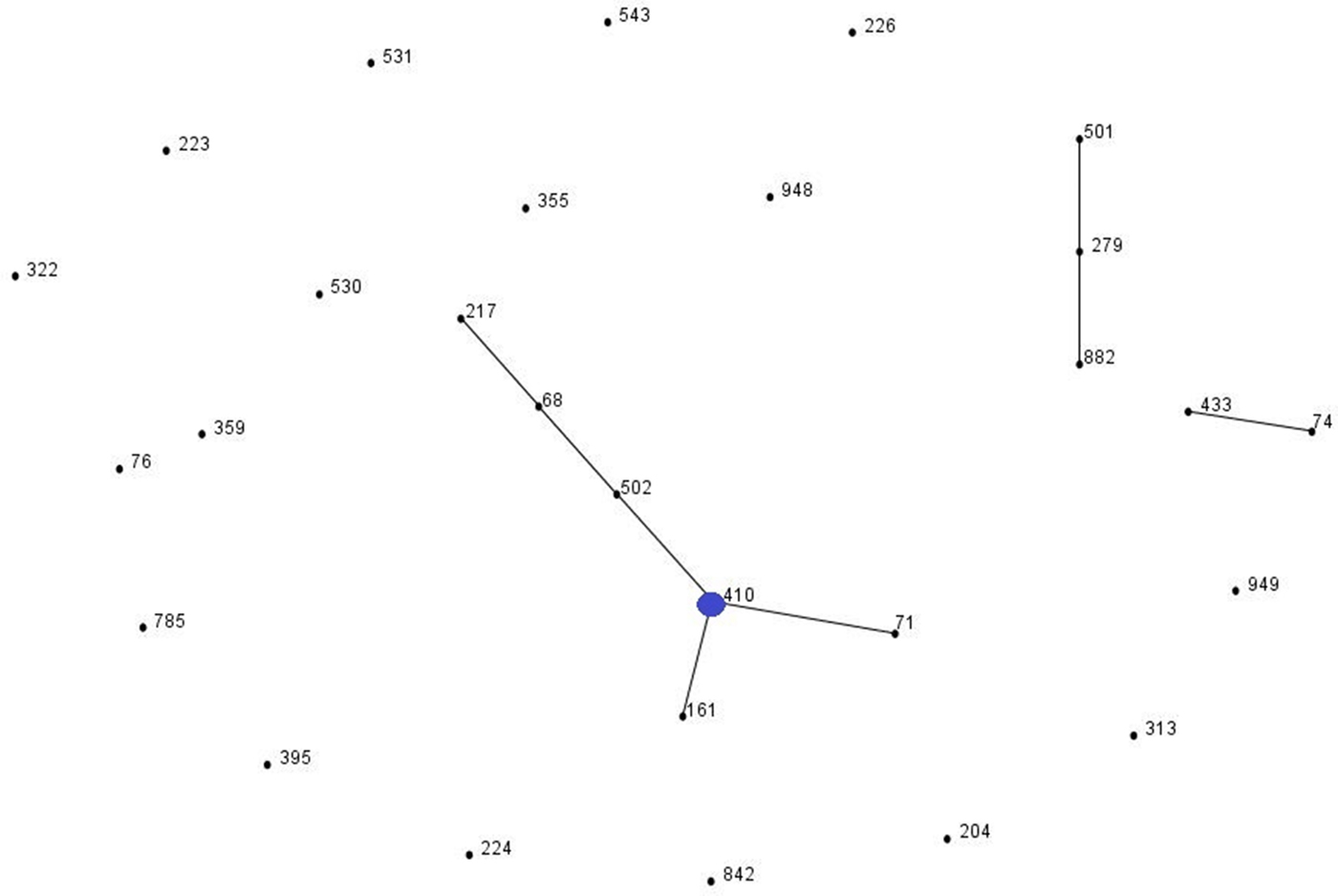
Fig. 2. Population snapshot of A. nosocomialis isolates in this study and existing isolates in the Institut Pasteur's MLST database by eBURST algorithm (http://pubmlst.org/abaumannii/; 24 April 2017, date last accessed). Each ST is represented by a black dot. Blue dot correspond to group founder. SLVs are linked by lines, and clonal complexes (CC) correspond to the group of connected STs.
Discussion
In the present study all, but two, of the strains of Acinetobacter spp. identified from CF patients were assigned to the Acb complex (96%) with A. baumannii being the most common. These results corroborate the distribution of these species found in hospitalised patients but the proportion of species other than A. baumannii normally found in the latter group of patients is usually considerably lower than the 45% of strains identified here [Reference Vasconcelos15]. Most isolates (70%) were susceptible to antimicrobials which suggests that these strains, in common with other opportunist Gram-negative bacteria, may have been acquired by CF patients from environmental sources, given their ubiquity in nature [Reference Peleg, Seifert and Paterson1, Reference Al Atrouni5].
To our knowledge, this is the first description of A. nosocomialis, A. seifertii, A. soli and A. variabilis (previously group of 15 sensu DNA Tjernberg & Ursing) [Reference Krizova16] isolated from CF patients. Although initially reported as environmental microorganisms, A. seifertii and A. soli have both been recently described from bacteraemia in hospitalised patients [Reference Kishii17, Reference Endo18].
There was marked genetic diversity among the strains investigated here which suggests that they were acquired by patients from different sources. Nevertheless, three STs accounted for 11 of 13 patients who grew A. pittii; ST 643 was identified in six patients, five of which were sampled in May 2009. This could conceivably be due to contamination of specimens, but if not, it is strongly indicative of cross-infection between these patients, or alternatively, acquisition of the strain from a single common source. Considerably less sharing of strains of the same STs between patients was evident for A. nosocomialis and A. baumannii.
Although A. baumannii is the species most commonly associated with antimicrobial resistance in hospital outbreaks [Reference Diancourt10], there was no evidence of clonal spread of resistance among the patient cohort for this species or for A. pittii and A. nosocomialis. Indeed, only two strains of the same ST (188) of A. baumannii proved to be MDR and these were recovered from two patients attending different hospitals and sampled over a year apart. Likewise, the two MDR strains of A. pittii ST 643 were separated by an interval of over 4 years (Tables 1 and 2). It is noteworthy that overall only 16 of the 53 strains were found to be MDR, and 21 strains were susceptible to all, but one, of the antimicrobial agents tested. This finding suggests that antimicrobial resistance alone does not necessarily confer a selective advantage for the establishment or dissemination of Acinetobacter spp. in respiratory infections in CF patients. There is, however, some evidence that A. pittii was relatively more successful in adhering to human bronchial epithelial cells and in inducing IL-8 expression than A. baumannii or A. nosocomialis [Reference Peleg19]. In addition, recent whole-genome sequencing data of an A. pittii ST643 MDR strain revealed the presence of genes associated with virulence, biofilm formation and antimicrobial resistance [Reference Rocha20].
Three patients grew two or three different species of Acinetobacter, and four individuals yielded strains of the same species distinguishable by ST. Studies on the lung microbiome in CF show the presence of widely diverse bacterial populations which with increasing age of the host become well adapted to the CF lung environment and compete with other resident species for dominance [Reference Cox21]. It is likely that Acinetobacter spp. are not well adapted to this relatively unique environment and this might explain their low frequency of isolation from these patients.
One patient (#32, Table 1) was colonised with two distinguishable but phylogenetically related STs of A. nosocomialis (ST161 and its SLV ST410). A. nosocomialis ST161 has been found in clinical (MDR profile) and environmental samples (non-MDR) in Rio de Janeiro [Reference Girão22]. In this study, the ST161 clone was MDR, which might indicate that it was acquired by the patient through cross-transmission in the hospital setting. This scenario might also explain the acquisition of the European clone I A. baumannii ST1 by patient #8. Although there are no reports of cross-transmission of Acinetobacter spp. between patients with CF, it is generally accepted that cross-transmission in hospitals occurs through transient colonisation of the hands of healthcare workers and the survival of the organism on inanimate surfaces [Reference D'Agata, Thayer and Schaffner23]. Patient #9 was colonised for 2 years by the same A. baumannii ST (ST228), which is a SLV of ST25 (CC25). ST25 is an emerging genotype responsible for recent epidemics worldwide that has been associated with elevated resistance to desiccation, high biofilm-forming capacity on abiotic surfaces, and adherence to pneumocytes [Reference Giannouli24].
Eight strains of MDR A. baumannii were also resistant to imipenem and meropenem, and carried the bla OXA-23-like gene. Around the world, including Brazil, this resistance determinant is widely associated with the spread of carbapenem resistance [Reference Vasconcelos15], and to the best of our knowledge, this study is the first to report its occurrence in this species from CF patients. Further, four of the MDR strains belonged to two globally distributed CCs in A. baumannii, CC1 (ST1, ST20 and ST160) and CC79 (ST79), which have been associated with antimicrobial resistance and dispersion of the bla OXA-23-like gene in Rio de Janeiro and wider Brazilian hospitals [Reference Grosso25, Reference Chagas26].
A major limitation of this retrospective study is the lack of clinical and epidemiological data to provide a context of the significance of the findings and impact on patient care. However, in our paediatric and adult CF centres, Acinetobacter spp. are rare in the patients and we have been able to document successfully the species distribution and genetic relatedness of the different species recovered from patients over a long time period. Unlike other prevalent Gram-negative species isolated from CF, we found little evidence of widespread clonal transmission which suggests that acquisition of Acinetobacter spp. in our hospitals was most often due to sporadic and widely diverse strain populations more associated with environmental rather than hospital sources. Nevertheless, our data do raise concerns regarding the risk of gene dissemination, since the MDR A. baumannii CC79 and CC1 lineages carrying the bla OXA23-like gene may be a silent source of resistance genes which could spread to other pathogens and compromise treatment and management of patients. Further prospective studies are therefore warranted to establish the clinical significance of Acinetobacter spp. in CF and distinguish between benign colonisation and increase in infectious risk.
Acknowledgements
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant number 471480/2012-6)
Declaration of interest
None.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.







