Clinical differences in drug response in terms of beneficial and adverse effects, or no effect, are well established throughout medicine. In interpreting how this variability in outcome of drug therapy might affect an individual, many factors must be considered: the patient's health profile, prognosis and compliance with therapy; the severity of the disease; the quality of drug prescribing and dispensing; and the genetic profile of the individual.
Pharmacogenetics focuses on the study of gene–drug interactions and examines the extent to which variability in the human genetic make-up is responsible for the differences between patients in therapeutic efficacy, effectiveness and adverse reactions. The term pharmacogenomics has more recently been introduced and it encompasses pharmacogenetics. It refers to a genome-wide search for genes and their products relevant to the use of drugs in humans. The search includes genes determining disease susceptibility and those causing individual variations in drug response, and it is based on the knowledge derived from the Human Genome Project (Reference Aitchison, Gill, Plomin and DeFriesAitchison & Gill, 2002).
Pharmacogenetics and pharmacogenomics have a shared goal, in that they both seek to guide pharmacotherapy and improve outcome by providing individualised treatment decisions. A long-term aim of pharmacogenomics is to develop novel diagnostic procedures and therapeutic products for effective and safe individualised drug prescription. Both fields, therefore, hold great potential, particularly in psychiatry, where biologically based treatment guidelines are lacking.
Individual variability in drug response can often be understood as a combination of factors affecting the pharmacokinetic and pharmacodynamic effects of drugs (Fig. 1). Candidate genes for pharmacogenetic studies include polymorphic drug-metabolising enzymes, drug transporters and polymorphic drug targets that affect disease-related pathways.

Fig. 1 Pharmacokinetic and pharmacodynamic factors that affect individual patients’ responses to drugs.
Pharmacogenetics of antipsychotic treatment
Genetic variants of drug-metabolising enzymes
Antipsychotics are used to treat schizophrenia. There is increasing evidence to suggest that the CYP2D6 genotype might partially affect response to typical antipsychotics, and also drug side-effects. For example, on average, Asians develop higher plasma levels than Europeans and thus seem to have an increased sensitivity to antipsychotics, including haloperidol (Reference Aitchison, Jordan and SharmaAitchison et al, 2000b ).
Genetic variants of drug targets
Drug response studies for antipsychotics have focused largely on genes that code for the neuronal targets of these drugs. These include the genes encoding dopaminergic D2, D3 and D4 receptors (DRD2, DRD3, and DRD4); the serotonergic 5-HT2A (HTR2A), 5-HT2C (HTR2C) and 5-HT6 (HTR6) receptors; the histaminergic H1 and H2 receptors (H 1 and H 2); the muscarinic cholinergic receptors; neurotransmitter transporters; and other intracellular signalling molecules.
Of the antipsychotics, clozapine is thought to be the most efficacious in improving both positive and negative symptoms of schizophrenia. Clozapine is most useful for patients who fail to respond to typical antipsychotics, but even among this group only about two-thirds of patients respond to it. Clozapine exhibits large inter-individual variations in bioavailability, steady-state plasma concentrations and clearance. It is metabolised by several CYP enzymes, including CYP1A2, CYP3A4 and CYP2C19, but not by CYP2D6 (Reference CollierCollier, 2003). Reference Aitchison, Jann and ZhaoAitchison et al (2000a ) found that clozapine is primarily metabolised by CYP1A2, since CYP1A2 knock-out mice given clozapine demonstrated significantly lower clozapine clearance than did wild-type mice. These results suggested that CYP1A2 polymorphisms might be associated with clozapine response, but so far this has not been confirmed (Reference Basu, Tsapakis and KnightBasu et al, 2004a ).
There are only a few reports on pharmacogenetic studies based on the metabolic pathways of other atypical antipsychotics such as risperidone, olanzapine and quetiapine (Reference Staddon, Arranz and MancamaStaddon et al, 2002).
Dopamine receptors
The dopamine D2 receptor is a major site of action of antipsychotics, but a functional polymorphism (−141 Ins/Del) affecting promoter activity and DRD2 expression was not shown to be associated with clinical response to clozapine or typical antipsychotics. This is in contrast to reports of missense variants (Val96Ala, Pro310Ser and Ser311Cys) determining response to various antipsychotics, including clozapine. Furthermore, the DRD3 Ser9Gly missence variant has been associated with response to typical antipsychotics, but association with response to clozapine remains controversial (for a review see Reference Shaikh and KerwinShaikh & Kerwin, 2002). Several studies on the association of a 16 amino acid repeat polymorphism in exon 3 of DRD4 have been carried out, and surprisingly, given the relatively high affinity of clozapine for the D4 receptor, suggest that this polymorphism does not seem to correlate with the degree of response to clozapine, in contrast to the positive association between this polymorphism and a rapid response to acute treatment with typical antipsychotics.
Serotonin receptors
In addition to the undoubted pivotal role of dopamine in the mechanism of action of antipsychotics, mechanisms mediated by serotonin (5-HT) may also be involved (Reference MeltzerMeltzer, 1995). Several lines of research have implicated the 5-HT2A, 5-HT2C, 5-HT5A and 5-HT6 receptors in response to treatment of schizophrenia. A strong association reported between the 102T/C polymorphism in HTR2A and clozapine response was not replicated by others, but a meta-analysis showed that the 102T/C silent polymorphism played a major role in determining clozapine response (Reference Arranz, Munro and ShamArranz et al, 1998). Similarly, a –1438G/A polymorphism in the promoter region of this gene was strongly associated with clozapine response. These polymorphisms have also been associated with response to typical antipsychotics (Reference Joober, Benkelfat and BriseboisJoober et al, 1999) and risperidone (Reference Lane, Chang and ChiuLane et al, 2002). With respect to HTR2A, positive associations were also reported between clozapine response and a structural His452Tyr variant of potential functional significance. An association has also been reported between a potentially functional Cys23Ser structural change in HTR2C and clozapine response but other studies have failed to replicate this finding. Furthermore, investigation of the 5-HT5 and the 5-HT6 receptor genes has pointed towards a minor contributing role in clozapine treatment response (Reference Yu, Tsai and LinYu et al, 1999; Reference Birkett, Arranz and MunroBirkett et al, 2000).
Pharmacogenetic prediction of antipsychotic response
Reference Arranz, Munro and BirkettArranz et al(2000) performed association studies of multiple candidate genes in an attempt to find the combination of polymorphisms that gave the best predictive value of response to clozapine in patients with schizophrenia. On the basis of clozapine-binding profiles, they studied 19 genetic polymorphisms in eight receptor subtype genes, including the α2A-adrenoceptor (ADRA2A), DRD3, HTR2A, HTR2C, HTR3A, HTR5A, H 1, H 2 and the serotonin transporter (5-HTT) genes. A combination of six polymorphisms showing the strongest association with response (HTR2A 102T/C and His452Tyr; HTR2C –330GT/–244CT and Cys23Ser; 5-HTTLPR; H 2 –1018G/A) gave a level of prediction of 76.86% (χ2 = 35.8, P=0.0001) and a sensitivity of 95.89 (s.d.=0.04) for the identification of patients who will show a satisfactory improvement with treatment. This study was the first to report on the use of combinations of pharmacodynamic factor gene polymorphisms to predict the response to antipsychotic medication. Attempts to replicate this finding have, however, been inconsistent.
Preliminary results from the same group (Reference Clark, Arranz and ArrondoClark et al, 2002), showed that, in 92 Spanish patients, a combination of polymorphisms in the genes HTR2C, HTR2A, DRD3, 5-HTT (the variable number tandem repeat (VNTR) and the serotonin-transporter-linked polymorphic region (5-HTTLPR) gene) may be used for the prediction of treatment response to olanzapine (positive predicted value = 76%, negative predicted value = 79%, sensitivity = 82%, specificity = 72%, P = 0.07). Similar studies have been performed for the response to risperidone, but in smaller groups. The hope is that if these studies are replicated, such methodology could form the basis for pharmacogenetic prediction tests for response to various antipsychotics, in a new era of treatment individualisation.
Adverse effects of antipsychotic treatment
Pharmacogenetic candidate gene studies concerning the side-effects of antipsychotic drugs have investigated the association with the relevant drug-metabolising enzymes or neurotransmitter receptors. Extrapyramidal symptoms and early-stage side-effects of antipsychotic therapy such as postural hypotension and excess sedation have been reported to be associated with overrepresentation of poor metabolisers of CYP2D6 (for a review see Reference Scordo and SpinaScordo & Spina, 2002).
Extrapyramidal side-effects
Tardive dyskinesia is an involuntary movement disorder manifested typically in the orofacial area, but frequently extending to the limbs and the trunk. Susceptibility to the development of tardive dyskinesia is currently thought to have a genetic basis, and a positive association between tardive dyskinesia and a functional polymorphism (a cytosine-to-adenine substitution, C/A) in the first intron of CYP1A2 has been reported (Reference Basile, Ozdemir and MasellisBasile et al, 2000). A meta-analysis (Reference Lerer, Segman and FangerauLerer et al, 2002) has correlated the DRD3 Ser9Gly polymorphism with tardive dyskinesia in a large sample (n=780) of patients treated with typical antipsychotics. Since both DRD3 and CYP1A2 seem to contribute to the development of tardive dyskinesia, a gene–gene interaction analysis was undertaken (Reference Basile, Masellis and PotkinBasile et al, 2002) using scores on the Abnormal Involuntary Movements Scale (AIMS) to measure symptom severity. This showed that patients who exhibited the homozygous risk genotype at both DRD3 (Gly/Gly) and CYP1A2 (C/C) had the most severe tardive dyskinesia (highest mean AIMS scores), whereas those who had none of the risk genotypes at either locus had the lowest mean scores. Following the same rationale, Reference Zhang, Zhang and HouZhang et al(2003) recently reported a possible synergistic effect of DRD3 Ser9Gly and manganese superoxide dismutase gene (MnSOD). Manganese superoxide dismutase is an enzyme that catalyses the dismutation reaction of the toxic superoxide radical to molecular oxygen and hydrogen peroxide and thus forms a crucial part of the cellular antioxidant defence mechanism. This reported interaction may affect susceptibility to tardive dyskinesia by influencing mitochondrial free radical scavenging.
Acute akathisia has also been reported to be associated with polymorphisms in DRD3 and DRD2 (Reference Basu, Tsapakis and AitchisonBasu et al, 2004b ). MnSOD alone has also been shown to be weakly associated with tardive dyskinesia. Furthermore, the contribution of ten polymorphic sites in six candidate dopaminergic and serotonergic genes to the development of tardive dyskinesia was recently examined in a small Jewish sample, with only the dopamine transporter gene (DAT) 3’-VNTR polymorphism, the serotonin transporter-linked polymorphic region (5-HTTLP) and the tryptophan hydroxylase (TPH) intron-7 polymorphism yielding trends towards a positive association (Reference Segman, Goltser and Heresco-LevySegman et al, 2003).
Hyperprolactinaemia
Antipsychotic-induced hyperprolactinaemia has been shown to be associated with the DRD2 Taq1A polymorphism (Reference Mihara, Kondo and SuzukiMihara et al, 2000) and, more recently, a significant association between DRD2 –141C and hyperprolactinaemia consistent with in vitro work was demonstrated (further details available from the authors on request). The latter association was strengthened by controlling for CYP2D6 genotypic category (P=0.023), and a trend for an association with a specific DRD2 haplotype was also shown.
Weight gain
Among antipsychotics, clozapine appears to have the greatest weight-gain liability. It is currently thought that weight gain induced by clozapine and other antipsychotics (typical and atypical) results from multiple neurotransmitter receptor interactions, leading to changes in appetite and eating behaviour. Reference Basile, Masellis and McIntyreBasile et al(2001) invesitaged ten genetic polymorphisms across nine candidate genes involved in both central hypothalamic weight regulation and peripheral thermogenic pathways. The nine candidate genes were the HTR2C, HTR2A, HTR1A, the H 1 and H 2, the CYP1A2, the β3-adrenergic receptor genes (ADRB3) and ADRA1A and the tumour necrosis factor a gene (TNFα). Only four of these (HTR2C, ADRB3, ADRA1A and TNFα) demonstrated a modest, non-significant trend towards a positive association with clozapine-induced weight gain. A positive association between a promoter polymorphism (−759C/T), thought to alter HTR2C gene expression, has also been reported by Reference Reynolds, Zhang and ZhangReynolds et al(2002), but both studies await replication.
Agranulocytosis
Clozapine-induced agranulocytosis has been associated with a dominant gene within the major histocompatibility complex region marked by heat shock protein 70–1 and 70–2 variants. This finding was, however, reported in two studies with small power of Jewish samples (Reference Valevski, Klein and GazitValevski et al, 1998; Reference Meged, Stein and SitrotaMeged et al, 1999). In a more recent study, of a non-Jewish Caucasian sample, clozapine-induced agranulocytosis was significantly associated with some human leukocyte antigen (HLA) polymorphisms, and age seemed to be a further major risk factor for clozapine-induced agranulocytosis (Reference Dettling, Cascorbi and RootsDettling et al, 2001). Thus, HLA loci may serve as genetic markers to identify individuals of different ethnic subgroups prone to this severe idiosyncratic drug reaction.
Pharmacogenetics of antidepressant treatment
Pharmacotherapy in depression results in effective treatment for the majority of patients, but it can take up to 6 weeks before response is seen and up to 40% do not respond sufficiently to the initial treatment. It would therefore be highly desirable to identify those who will not respond before therapy is started, thus avoiding a relatively long period of trial and error. The impact on health care costs of a test identifying these patients could be substantial, owing to the large burden of disease exerted by depression.
Polymorphisms of drug-metabolising enzymes
Most interest has focused on the CYP2D6 gene, which encodes debrisoquine hydroxylase, the enzyme that metabolises many antidepressants, including the tricyclics (TCAs), selective serotonin reuptake inhibitors (SSRIs) and venlafaxine. Studies have identified genetic polymorphisms with over 70 allelic variants; these result in clinically important functional metabolic changes, notwithstanding ethnic differences in the prevalence of both ‘poor metabolisers’ and ‘ultrarapid metabolisers’. About 7–10% of Caucasians are poor metabolisers of CYP2D6. These individuals have no functional CYP2D6 because they have no functional copy of a CYP2D6 gene (e.g. they are homozygous for null alleles, a mutation that leads to a prematurely truncated protein and hence lack of debrisoquine hydroxylase activity). Such people are likely to have increased concentrations of metabolised drugs at conventional doses (Reference BrøsenBrøsen, 1996; Reference Sachse, Brockmoller and BauerSachse et al, 1997). At the other extreme of the metabolic spectrum, in ultrarapid metabolisers (with CYP2D6 gene duplications, which result in excess metabolic activity of debrisoquine hydroxylase), who comprise 3–8% of Caucasians and up to 30% of some other ethnic groups, drugs at standard doses often do not reach therapeutic concentrations and an increased dose may be required to achieve therapeutic response. For example, nortriptyline CYP2D6 poor metabolisers require only 50% of the average effective antidepressant dose, but ultrarapid metabolisers may require up to 230% of this dose (Reference Johansson, Lundqvist and BertilssonJohansson et al, 1993; Reference Kirchheiner, Brøsen and DahlKirchheiner et al, 2001).
Pharmacodynamic variability
There is considerable evidence supporting the hypothesis that alterations in serotonergic neuronal function are involved in the pathophysiology of depression (Reference Owens and NemeroffOwens & Nemeroff, 1994). The serotonin transporter (5-HTT) protein acts as the primary mechanism for removing 5-HT from the synaptic cleft. Two polymorphisms have been identified within the human 5-HTT, a 44 bp insertion/deletion polymorphism in the promoter region (5-HTTLPR), giving rise to a short (s) and a long (l) variant, and a VNTR polymorphism in intron 2. Since the serotonin transporter is the target for serotonin reuptake inhibitors, including SSRIs, the effect of 5-HTT variants on clinical response to these drugs has been intensively studied.
The effects of 5-HTTLPR on clinical response to fluvoxamine and paroxetine in two Italian samples revealed that the presence of at least one l allele was significantly associated with greater improvement in scores on the Hamilton Rating Scale for Depression. This finding was replicated by a study of Spanish Caucasians treated with citalopram, where the s/s genotype was shown to be more frequent in the group that did not respond to SSRI treatment. Moreover, homozygosity of the l allele was associated with a faster response to paroxetine in elderly American patients with depression, and Taiwanese patients with depression were found to respond better to fluoxetine in the presence of the l/l genotype.
On the other hand, several studies have suggested an association in the opposite direction, not only in Oriental but also in American patients treated with SSRIs. A significant association between specific SSRI response and the intron 2 VNTR 12/12 genotype has also been reported (Reference Serretti, Lilli and SmeraldiSerretti et al, 2002). We have found a positive trend towards an association between the 5-HTTLPR l/l genotype and response to treatment with TCAs in a group of Caucasian patients with unipolar or bipolar affective disorder (Reference Tsapakis, Checkley and KerwinTsapakis et al, 2003).
The tryptophan hydroxylase gene (TPH) and the brain-expressed TPH (TPH2) are further candidate genes implicated in the clinical response to SSRI treatment. TPH encodes the rate-limiting enzyme in the synthesis of 5-HT from tryptophan. Various non-functional TPH polymorphisms have been detected and, in a Finnish sample, the intron-7 779A/C polymorphism has been associated with suicidality and alcoholism. A further polymorphism (218A/C), located in a potential transcription factor binding site, may influence gene expression and, consequently, response to antidepressants. In fact, associations between TPH variants and response to both fluvoxamine and paroxetine have been reported (Reference Serretti, Zanardi and CusinSerretti et al, 2001a ,Reference Serretti, Zanardi and Rossini b ).
The postsynaptic 5-HT2A receptor may also influence the efficacy of serotonergic antidepressants. Indeed, the C-containing variants of the 102T/C 5-HT 2A polymorphism were associated with response to treatment with SSRIs, tricyclics and electroconvulsive therapy (ECT) (Reference Minov, Baghai and SchuleMinov et al, 2001). Furthermore, mutations in guanine nucleotide binding proteins (G-proteins), which represent the essential regulatory components in the transmembrane system of many receptors, might affect antidepressant efficacy. Reference Zill, Baghai and ZwanzgerZill et al(2000) have described an association between TT homozygosity in G β 3 with response to SSRI, TCAs and ECT combinations.
The serotonergic and dopaminergic systems are interconnected in the brain, and serotonergic projections inhibit dopamine function in the midbrain. SSRIs enhance dopamine function in the nucleus accumbens through increased expression of postsynaptic dopamine D2 receptors, and an association between changes in the dopaminergic system and treatment response in major depression has been suggested. However, a study testing a DRD2 and a DRD4 polymorphism failed to show evidence of an association with efficacy of SSRIs (Reference Serretti, Zanardi and CusinSerretti et al, 2001c ). Clinically significant variations within the noradrenaline (norepinephrine) transporter (NET or hNAT) genes and the ADRA2A also seem to determine the side-effect profiles of antidepressant medications. Among others, the rare Ala457Pro variant of hNAT has been associated with orthostatic intolerance (a syndrome characterised by light-headedness, fatigue, altered mentation and syncope, and associated with postural tachycardia and plasma noradrenaline concentrations that are disproportionately high in relation to sympathetic outflow), and the transporter coded for by this variant has been shown to have reduced affinity for noradrenaline (Reference Shannon, Flattem and JordanShannon et al, 2000). Furthermore, Reference Aitchison, Tandon and AshworthAitchison et al(2002) have identified a complex microsatellite-like repeat region that appeared to be polymorphic, and the most common sequence variant in this region has been reported to be associated with anorexia nervosa (Reference Urwin, Bennetts and WilckenUrwin et al, 2002). The α2-adrenergic receptor has been implicated in the aetiology of sexual dysfunction induced by SSRIs and other antidepressants, and the α2 antagonist yohimbine was shown to improve this extremely common adverse effect in a small Swedish open trial (Reference JacobsenJacobsen, 1992). Two high-frequency single-nucleotide polymorphisms (787C/G and 1817G/A) have been identified in Sweden (Reference Liljedahl, Karlsson and MelhusLiljedahl et al, 2003).
Pharmacogenetics of treatment with lithium
Lithium response has long been thought to have a genetic component, and changes in the serotonergic system have been implicated in the mechanism of action of lithium. In a review of the literature, Reference Ikeda and KatoIkeda & Kato (2003) note that a positive association between the presence of the l allele of 5-HTTLPR and good response to lithium has been reported in a North Italian sample, but an earlier study from the South of Italy reported the l/l genotype to be associated with the non-responder phenotype. Furthermore, no association between response to lithium prophylaxis and the 102T/C or the 141C/T 5-HT 2A polymorphisms was found in another Italian sample. A significant association between TPH and prophylactic efficacy of lithium has also been reported. At therapeutic doses, lithium inhibits the activity of enzymes involved in the phosphatidylinositol and phospholipase C intracellular pathways, and a higher frequency of a phospholipase C γ-1 gene (PLCG1) repeat has been shown to be positively associated with lithium response in the treatment of bipolar disorder. In addition, a PLCG1-8 repeat was reported to be more frequent among lithium responders than normal controls, and it was also suggested that the 973C/A polymorphism in the inositol polyphosphate 1-phosphatase gene was an indication of positive lithium response. Reference Washizuka, Ikeda and KatoWashizuka et al(2003) have reported on the association of the mitochondrial DNA (mtDNA) 10398 polymorphism and maintenance lithium treatment response in a small Japanese sample of patients with bipolar affective disorder.
Alzheimer's disease
Although the aetiology of Alzheimer's disease remains largely unknown, three genes have been identified whose mutations cause the early-onset familial Alzheimer's disease and show nearly 100% penetrance with autosomal dominant inheritance. These genes encode β-amyloid precursor protein (APP), presenilin-1 (PSEN1) and presenilin-2 (PSEN2). Moreover, the risk of occurrence of Alzheimer's disease has been associated with apolipoprotein E (ApoE) and its three common isoforms Apo E2, E3 and E4. It has further been established that the ε 4 allele confers a significant risk for late-onset, sporadic Alzheimer's disease, the most common form of the illness. As a result, E4 has served as the primary target for all pharmacogenetic studies related to Alzheimer's disease conducted to date, and it has been demonstrated that the therapeutic response to drugs in Alzheimer's disease is ApoE genotype-specific. Some studies have suggested that individuals carrying ε 4 respond poorly to the acetylcholinesterase inhibitor tacrine, compared with people with other genotypes, but other studies have produced conflicting results.
Patients with Alzheimer's disease may benefit from S12024, a drug that increases vasopressinergic activity. A multifactorial therapy combining three different drugs yielded positive results during 6–12 months’ treatment in about 60% of patients. With this therapeutic strategy, APOE-4/4 carriers were the worst responders, and patients with the APOE-3/4 genotype were the best responders. It was also possible to differentiate the influential effect of PSEN1 and PSEN2 polymorphic variants on mental performance in response to multifactorial therapy (Reference CacabelosCacabelos, 2003).
Genetic profiling of drug response in Alzheimer's disease was recently extended for tacrine and S12024, and Variagenics, a pharmacogenomics biotechnology company have patented an association with a polymorphism in the butyrylcholinesterase gene (Reference AmouyelAmouyel, 2002).
Pharmacogenetics of attention-deficit hyperactivity disorder
Family, twin and adoption studies in attention-deficit hyperactivity disorder (ADHD) have shown both an increased risk in the relatives of probands and a moderate to high heritability; transmission is believed to be polygenic, each gene having a small effect on phenotype. As regards the pharmacogenetics of ADHD, homozygosity of the 10-repeat allele at DAT1 was associated with poor response to methylphenidate, and this finding has been replicated in a Brazilian study. A positive association between the 10-repeat allele at DAT1 and response to methylphenidate has, however, been reported, and homozygosity of the 9-repeat allele at DAT1 was associated with a poor response to methylphenidate. Further, it was shown that individuals with the 7-repeat allele at DRD4 achieved less normalisation of symptoms and required 1.5 times more methylphenidate to achieve improvement than did those without the 7-repeat allele. Individuals who both presented the 7-repeat allele at DRD4 and were homozygous for the l allele of 5-HTTLPR showed a reduced improvement in general functioning during methylphenidate treatment (Reference Rohde, Roman and HutzRohde et al, 2003).
Newer developments
Besides drug-metabolising enzymes and neurotransmitter receptors and transporters, other important candidates that warrant further investigation have now emerged, mostly focusing on elucidating mechanisms responsible for the slow onset of therapeutic effect of psychotropics (particularly antidepressants). Such mechanisms may include the evoking of adaptive changes in intracellular signal transduction and synaptic connectivity. There has been an extensive search for genes to explain this phenomenon, and current candidates include the following: brain-derived neurotrophic factor (BDNF), G-proteins, the association between mood stabilisers (e.g. lithium and sodium valproate) and human B-cell lymphoma protein-2 (Bcl-2), and other related genes such as Bcl-2 associated athanogene-1 protein (Bag-1).
Ethical considerations
The emergence of pharmacogenetics is about to mark a new era in clinical psychiatry, in which genotype or other biomarkers may influence choice of therapy, increasing the safety and efficacy of commonly used medications. The ethical issues that arise from pharmacogenetic research and its clinical applications (Box 2) should, however, be addressed before the benefits of treatment individualisation can be realised (Reference Buchanan, Califano and KahnBuchanan et al, 2002).
Box 2 Ethical issues in pharmacogenetics
Pharmacogenetic tests would need to be approved and overseen by major national regulatory bodies such as the UK Medicines Control Agency (MCA)
-
• Over 500 pharmaceutical company and government representatives from around the world recently discussed the Food and Drug Administration's draft guidance for industry on pharmacogenomics data submission (Reference AbbottAbbott, 2003)
Appropriate protection for privacy and confidentiality is crucial
-
• Pharmacogenetic tests can carry several types of potentially psychosocially harmful secondary information
-
• Genotype-based information about drug response may inform prognosis
Information on poor drug response could easily affect an individual in terms of:
-
• employment opportunities
-
• insurance status
-
• their relatives, who might be genetically predisposed to the same mental illness and therefore might also take the drug.
Participation in pharmacogenetic research requires informed consent, and participants should be fully informed of:
-
• the need to obtain DNA for the research
-
• the protections of confidentiality and privacy (including how the sample and test results will be stored and their privacy and confidentiality maintained)
-
• whether further research may be done using their DNA in the future
-
• the risks and benefits of providing DNA for participation in the trial
-
• whether research results will be disclosed to the participants
At present, it is unclear whether, or to what extent, pharmacogenetic tests will be included in standard packages of care. Much will depend on whether pharmacogenetic testing appears to be cost-effective in the long-run, when the costs of integrating the methodology into the health care delivery system are taken into account. However, if a test's positive predictive value and utility in determining whether to prescribe a drug, in what dosage and in what combinations with other drugs are well established, then physicians would probably have a duty to offer it if the benefits outweigh the risks and costs of doing so. Consequently, physicians are likely to have to learn the role that pharmacogenomic tests may play in choosing therapy and to respond accordingly. The pace at which these tests will be integrated into psychiatric practice will, no doubt, differ across diseases and between specialists and generalists. Without major changes in the teaching of genetics psychiatric problem include random errors, confounding and bias, and effect modifications and interactions (Box 3). The rigorous application of basic epidemiological principles to avoid confounding, misclassification and bias may not be sufficient. Matching according to ethnicity and/or the ethnic homogeneity of the study population become important, although these are often difficult to achieve in practice. Hence, testing for hidden population stratification is mandatory and can be performed at tolerable expense by use of a limited number of multi-allelic unlinked marker loci (Reference Pritchard and RosenbergPritchard & Rosenberg, 1999). This strategy should allow for the detection of moderate or even minor differences in ethnic composition, and therefore the question of statistical power is crucial. For research into disease susceptibility genes, most investigators nowadays consider studies with family controls (e.g. so-called trios, where the genotype of an individual is studied with that of two of his or her siblings) as the best option. This approach, however, is not usually feasible for the study of genes that modify drug response or the risk of adverse drug effects. Such studies require families in which several members have the same disease and are exposed to the same drug; this is only rarely seen (even retrospectively). Thus, genetic modifiers of drug response and adverse drug reactions usually have to be studied in unrelated controls, and population stratification may be excluded in these instances. Multiple testing is another major concern in genetic association studies. Sample sizes should be large enough to allow all predefined hypotheses to be tested at the desired confidence level, and all other tests should be identified as explorative.
Box 3 Causes of discrepant results among different association studies on the same problem (adapted from Reference Meisel, Gerloff and Kirchheiner Meisel et al, 2003 )
Random errors
-
• multiple testing
-
• inappropriate sample size
-
• inappropriate selection of genetic variants
-
• errors in genotyping
Confounding and bias
-
• genetic heterogeneity (the phenotype is affected by variants in numerous genes)
-
• aetiological heterogeneity (several causes lead to the same phenotype)
-
• differences in ethnic background within the population studied
-
• population stratification (multiple subgroups, each with different allele frequencies)
-
• inappropriate or missing consideration of haplotypes
-
• intermixing of incident and prevalent cases of the same phenotype
-
• inappropriate control groups
Effect modifications and interactions
-
• undefined gene–gene interactions
-
• undefined gene–environment interactions
Future directions
In view of the large numbers of conflicting studies and the need for careful data collection and statistical, epidemiological and genetic analysis during data generation, future pharmacogenomic analysis should be integrated much more widely into clinical psychiatric research. If study participants were stratified or even selected according to their pharmacogenetic traits, variabilities in pharmacokinetics and drug response could be reduced considerably. Sample sizes could then be smaller and the studies less expensive, although selection biases would have to be carefully avoided as they could materially compromise study results. According to accepted criteria of evidence-based medicine, the value of pharmacogenetic testing for routine clinical practice should be verified prospectively in clinical trials. Proven benefits and pharmaco-economic considerations (e.g. the number of tests necessary to avoid one case of toxicity), including the costs of pharmacogenomics test kits, are among the factors that will determine whether or not such tests will become part of routine clinical practice.
MCQs
-
1 With regard to clozapine:
-
a response to treatment with clozapine is subject to interindividual variations
-
b it is primarily metabolised by CYP1A2
-
c it is unlikely to be metabolised by CYP2D6
-
d dopamine D4 receptor polymorphisms have been shown to be strongly associated with clozapine response
-
e combinations of pharmacodynamic gene polymorphisms have been shown to predict response to clozapine.
-
-
2 Regarding tardive dyskinesia:
-
a susceptibility to the development of tardive dyskinesia is thought to have a genetic basis
-
b the AIMS is a commonly used scale for rating the presence of tardive dyskinesia
-
c the DRD3 Ser9Gly polymorphism has been significantly correlated with the development of tardive dyskinesia
-
d the development of tardive dyskinesia has not been linked with CYP1A2
-
e no association has been shown between the development of tardive dyskinesia and superoxide dismutase genes.
-
-
3 The serotonin transporter-linked polymorphic region (5-HTTLPR):
-
a is an insertion/deletion polymorphism
-
b has been identified in intron 2 of the serotonin transporter gene
-
c has been associated with clinical response to SSRIs
-
d has been associated with clinical response to TCAs
-
e has not been associated with response to lithium treatment.
-
-
4 The following are genes thought to be associated with the corresponding phenotypes:
-
a the 5-HT 2A gene and response to treatment with electroconvulsive therapy
-
b the HLA gene family and clozapine-induced agranulocytosis
-
c the phospholipase C γ-1 gene and lithium response in the treatment of bipolar disorder
-
d the apolipoprotein ε 4 gene and poor response to tacrine in the treatment of Alzheimer's disease
-
e the dopamine transporter gene and response to methylphenidate in the treatment of ADHD.
-
-
5 Rate the following statements as true or false:
-
a haloperidol is metabolised via CYP2D6
-
b CYP2D6 polymorphisms do not affect the metabolism of nortriptyline
-
c poor CYP2D6 metabolisers have multiple functional copies of a CYP2D6 gene
-
d up to 10% of Caucasians may be CYP2D6 ultrarapid metabolisers
-
e the P450 enzymes are Phase II drug-metabolising enzymes.
-
MCQ answers
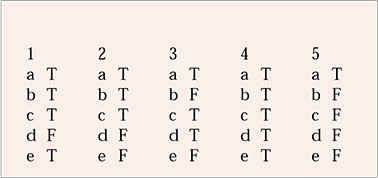
| 1 | 2 | 3 | 4 | 5 | |||||
|---|---|---|---|---|---|---|---|---|---|
| a | T | a | T | a | T | a | T | a | T |
| b | T | b | T | b | F | b | T | b | F |
| c | T | c | T | c | T | c | T | c | F |
| d | F | d | F | d | T | d | T | d | F |
| e | T | e | F | e | F | e | T | e | F |




eLetters
No eLetters have been published for this article.