Impact statement
This review article sheds light on the crucial role of 5-hydroxymethylcytosine (5hmC) in the understanding and future treatment of neurodevelopmental disorders (NDDs), such as autism spectrum disorder (ASD) and intellectual disability (ID). By focusing on this relatively underexplored epigenetic modification, the review highlights how environmental factors and genetic predispositions interact to influence brain development and function. By analyzing both animal and human studies, this review explains how alterations in 5hmC levels and distribution can profoundly influence neurodevelopmental outcomes. The findings have the potential to enhance diagnostic precision, enabling earlier identification and intervention for individuals at risk of these conditions. This article encourages the development of innovative therapeutic strategies aimed at modulating 5hmC dynamics, thereby offering hope for targeted effective treatments for NDDs. This paper stands to benefit a broad spectrum of stakeholders, including patients, clinicians and researchers, by offering new insights into the complex mechanisms of NDDs and fostering the development of targeted interventions. The global impact of this field is profound, offering hope for improved quality of life for individuals with NDDs and their families through earlier diagnosis, better understanding of the condition’s etiology and more effective, tailored treatments.
Introduction
Over the past three decades, there has been an unprecedented surge in global genetic association studies aimed at identifying genetic variants implicated in the etiology of mental illnesses, including neurodevelopmental disorders (NDDs). Despite the examination of tens of thousands of cases and controls in genome-wide association studies (GWAS), none of the identified genes have demonstrated an effect size exceeding 1% (Abdolmaleky et al., Reference Abdolmaleky, Zhou and Thiagalingam2015). Consequently, geneticists have turned their attention to the realm of epigenetics when investigating psychiatric disorders, including neurodevelopmental conditions such as autism spectrum disorder (ASD) and attention-deficit hyperactivity disorder (ADHD). Epigenetics, a term coined by Conrad H. Waddington, a prominent geneticist and developmental biologist, more than six decades ago (Waddington, Reference Waddington1957), refers to the factors that influence gene expression without altering the underlying DNA sequence (“on top of” genetics) (Slack, Reference Slack2002). In psychiatric research, the field of epigenetics primarily explores the interplay between environmental factors and the genome (Gottesman and Shields, Reference Gottesman and Shields1982). The epigenetic hypothesis is gaining increasing support among biological psychiatrists due to its potential to elucidate gender disparities, the clinical heterogeneity observed in presentations, and the rapid progression of psychiatric disorders (Petronis, Reference Petronis2001).
Epigenetic mechanisms, including DNA methylation and histone modifications, play pivotal roles in regulating gene function and have profound implications for behavioral and neuronal alterations observed in psychiatric disorders. These epigenetic modifications, akin to genetic mutations, can potentially influence gene expression and contribute to the pathogenesis of psychiatric disorders. Notably, individuals with ASD exhibit elevated expression of DNA methylation-promoting enzymes, such as DNMT1, DNMT3A and DNMT3B, in their cerebellum, indicating an overall increase in DNA methylation and hydroxymethylation (Keil and Lein, Reference Keil and Lein2016).
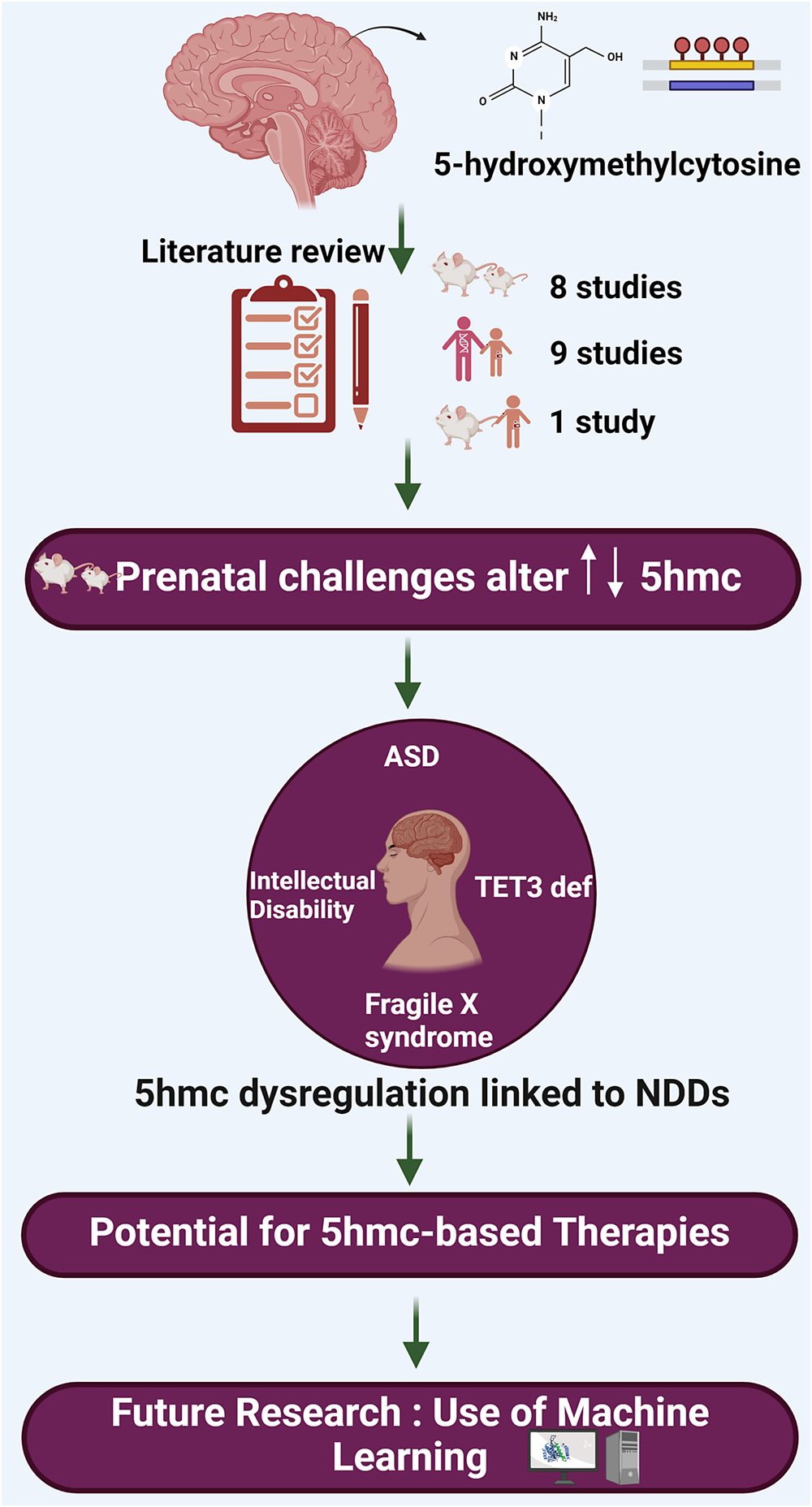
This review aims to delve specifically into the role of 5-hydroxymethylcytosine (5hmC) and its involvement in the pathogenesis of NDDs. This review is intended to bridge a critical gap in our knowledge by focusing on the epigenetic modification 5hmC, which has been relatively understudied in the context of NDDs such as ASD and intellectual developmental disorder (IDD). The scope of this review encompasses animal and human studies, focusing on how changes in 5hmC levels and distribution correlate with neurodevelopmental conditions, thereby aiming to uncover novel insights that could pave the way for innovative therapeutic strategies and a better understanding of these complex disorders.
The formation and maintenance of 5hmC
The human genome exhibits nucleotide modifications primarily through cytosine methylation and hydroxymethylation, with the latter being first observed in human Purkinje neurons in 2009 (Kriaucionis and Heintz, Reference Kriaucionis and Heintz2009). These modifications involve the addition of a methyl group to the 5′ carbon of cytosine, forming 5-methylcytosine (5mC), followed by the addition of a hydroxy group to the methyl group to generate 5hmC. The process of these modifications is illustrated in Figure 1. While 5mC is present in all cell types, current understanding suggests that 5hmC is predominantly restricted to the brain and early stages of development (Kriaucionis and Heintz, Reference Kriaucionis and Heintz2009; Tahiliani et al., Reference Tahiliani, Koh, Shen, Pastor, Bandukwala, Brudno, Agarwal, Iyer, Liu, Aravind and Rao2009; Wen and Tang, Reference Wen and Tang2014). However, ongoing research is continuously unveiling new insights into the distribution of 5hmC throughout the human body. Figure 1 also highlights two potential biological functions of 5hmC: its involvement in demethylation pathways and its ability to influence gene regulation through binding regulatory factors (Jin et al., Reference Jin, Kadam and Pfeifer2010). Both of these hypotheses represent active areas of research, with much yet to be discovered.

Figure 1. The DNA methyltransferase (DNMT) family of enzymes, DNMT1, DNMT3A and DNMT3B, catalyze the transfer of a methyl group from S-adenosyl-L-methionine (SAM) to the 5′ carbon of cytosine (C), resulting in 5-methylcytosine (5mC) (marker of gene repression). 5mC is converted back to cytosine by two possible mechanisms. One mechanism is the conversion of 5mC to 5hmC (variable marker of gene activation or repression) by the ten-eleven translocation methylcytosine dioxygenases (TET). 5hmC may then be converted to cytosine via subsequent enzymatic steps. A second possible mechanism involves a series of DNA replications or passive demethylation that, likely with decreased DNMT1 levels, fail to reproduce the methylation state of cytosine. Adapted from Khoodoruth and Khoodoruth (Reference Khoodoruth and Khoodoruth2024).
Neurodevelopmental disorders
NDDs are the first chapter and represent a prominent category in the latest edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-5-TR) (American Psychiatric Association, 2022). These disorders typically emerge during the developmental period and often manifest early in life. NDDs are characterized by deficits or variations in brain processes that give rise to impairments in personal, social, academic, or occupational functioning (Khoodoruth et al., Reference Khoodoruth, Khoodoruth, Ramadan, Johnson, Gulistan, Deluvio, Alamri, Al-Abdulla, Ouanes and Khan2023b). IDD, ASD and ADHD, among others, are encompassed within the spectrum of NDDs. Notably, extensive research has focused on elucidating the regulatory factors governing genes implicated in ASD, which affects 1 in 36 children and is characterized by impairments in social interaction and communication and restricted or repetitive behavioral patterns (Centers for Disease Control and Prevention, 2021). In contrast, despite ADHD exhibiting a community prevalence of 5% and being characterized by symptoms of hyperactivity, impulsivity and inattention that are inconsistent with developmental levels, there is a scarcity of epigenetic studies exploring this specific domain (Khoodoruth et al., Reference Khoodoruth, Ouanes and Khan2022).
Search criteria
Resources were retrieved from two databases, PubMed and Scopus, for this review. Initially, 75 papers were retrieved. After removing duplicates and screening titles and abstracts, 18 papers were included that were relevant to our study (Figure 2). The keywords used to search for resources in the mentioned databases were as follows: (5-hydroxymethylcytosine OR 5-hmc OR 5hmc) AND (“neurodevelopmental disorder” OR autism OR adhd OR “intellectual disability” OR “learning disability”). The included studies are summarized in Table 1.
Table 1. Epigenetic regulation of neurodevelopmental disorders: insights from animal and human studies on 5-hydroxymethylcytosine (5hmC)

ASD, autism spectrum disorder; CB, cerebellum; CGG, cytosine–guanine–guanine; CMA, cow milk allergy; CP, cerebral palsy; CpG, 5’—C—phosphate—G—3′; DhMRs, differentially hydroxymethylated regions; DNMT3A and DNMT3B, de novo methyltransferases 3A and 3B; EN-2, engrailed-2; F, female; FC, frontal cortex; ID, intellectual disability; FGX, fragile X syndrome; FMR1, fragile X mental retardation 1 gene; FMRP, fragile X mental retardation protein; FXTAS, fragile X-associated tremor/ataxia syndrome; GAD1, glutamic acid decarboxylase 67; GAD2, glutamic acid decarboxylase 65; IDD, intellectual developmental disorder; M, male; MeCP2, methyl-CpG binding protein 2; mTOR, mammalian target of rapamycin; NDDs, neurodevelopmental disorders; OGG1, oxoguanine glycosylase 1; PBMC, peripheral mononuclear blood cells; PFC, prefrontal cortex; PM, postmortem; SVZ, subventricular zone; TET, ten-eleven translocase; RELN, reelin; WP, whey protein; 5hmC, 5-hydroxymethylcytosine; TSS, transcription start sites; 5mC, 5-methylcytosine; 8-oxo-dG, 8-oxo-deoxyguanosine.

Figure 2. Flow Chart of Included and Excluded Studies.
Results
Animal studies
Maternal infection during pregnancy has been identified as a potential risk factor for developing NDDs in offspring (Atladóttir et al., Reference Atladóttir, Thorsen, Østergaard, Schendel, Lemcke, Abdallah and Parner2010). Labouesse et al. (Reference Labouesse, Dong, Grayson, Guidotti and Meyer2015) employed a well-established mouse model of prenatal immune challenge by administering viral mimetic poly(I:C) to pregnant mice. This immune activation during pregnancy resulted in elevated levels of both 5mC and 5hmC in the prefrontal cortex, specifically in the promoter region of GAD1. Additionally, increased levels of 5mC were observed in the promoter region of GAD2 following early-life challenges. These prenatal infection-induced modifications were associated with impairments in social interaction, challenges in working memory and abnormal cognitive abilities (Labouesse et al., Reference Labouesse, Dong, Grayson, Guidotti and Meyer2015).
Papale et al. (Reference Papale, Zhang, Li, Chen, Keleş and Alisch2015) conducted a study involving three mouse models of autism (Cntnap2−/−) and three age-matched control littermates, sacrificing them at 9 weeks of age. A genome-wide sequencing approach was employed on the striatal tissue, revealing a widespread distribution of 5hmC in the Cntnap2−/− mice, particularly within genic regions and repetitive elements. Notably, a significant overlap was observed between neuronal projection morphogenesis and the known ASD genes, suggesting an enrichment of neuronal ontological functions (Papale et al., Reference Papale, Zhang, Li, Chen, Keleş and Alisch2015). Additionally, Zhang et al. (Reference Zhang, Zhang, Chen, Wang, Chen, Gao, Fan, Shi, Zhang and Li2019) highlighted a significant reduction in 5hmC and Tet proteins, global alterations in 5hmC distribution and the identification of DhMRs associated with Cerebral Palsy (CP)-related genes, such as Notch1, Slc16a2, Dmd and Grin2b, indicating disrupted epigenetic regulation and potential impacts on gene expression critical to CP pathogenesis.
Amir et al., Reference Amir, Van den Veyver, Wan, Tran, Francke and Zoghbi1999 have demonstrated that mutations in the X-linked gene methyl CpG binding protein 2 (MeCP2) lead to Rett syndrome, a severe NDD. Additionally, duplications of genomic segments of the MeCP2 gene have been associated with autistic features in humans (Mellén et al., Reference Mellén, Ayata, Dewell, Kriaucionis and Heintz2012). In a study by Cheng et al. (Reference Cheng, Chen, Wan, Tang, Tian, Liao and Qiu2017), mass spectrometry analysis was employed to identify MeCP2-associated proteins involved in mRNA splicing, utilizing samples from MeCP2-null rat brains. The functional analysis of ChIP-seq datasets provided insights into the interaction between MeCP2, 5hmC and epigenetic changes in histone markers, highlighting the role of MeCP2 in regulating mRNA splicing within the nervous system (Cheng et al., Reference Cheng, Chen, Wan, Tang, Tian, Liao and Qiu2017).
To investigate the potential influence of food allergy and inflammation on behavior and mast cell accumulation in the brain, Germundson et al. (Reference Germundson, Smith, Vendsel, Kelsch, Combs and Nagamoto-Combs2018) conducted a study utilizing mouse models of milk allergy with bovine milk whey proteins (WP) as the allergen. Male and female mice from different age groups (4 weeks and 10 months) were sensitized to the allergen for 5 weeks, followed by oral challenges for three consecutive days before sacrifice. The researchers examined various factors, including epigenetic DNA modifications in the brain. The results revealed no apparent behavioral differences in female mice. However, WP-sensitized male mice exhibited reduced digging activity compared to sham males in both age groups. Additionally, a noticeable difference in 5hmC immunoreactivity was observed in the amygdala of both age groups of WP mice, suggesting an involvement of epigenetic regulation (Germundson et al., Reference Germundson, Smith, Vendsel, Kelsch, Combs and Nagamoto-Combs2018). Similarly, Cao et al. (Reference Cao, He, Zhao, Wang, Jia, Srivastava, M-S and Li2022) suggested an association between food allergy (cow milk allergy) and an increase in ASD-like symptoms and levels of 5hmC. Results showed that mice with cow milk allergy showed autistic-like symptoms and behaviors. There was an increase in the 5hmC biomarker in the hypothalamus as well.
In a more recent paper by Papale et al. (Reference Papale, Madrid, Zhang, Chen, Sak, Keleş and Alisch2022), the brain tissue of ASD mouse models (Cntnap2) that were projected to prenatal stress was tested. The sample lacked behavioral or neuropathological abnormalities. Genomic profiling of DNA samples from hippocampal and striatal showed disruptions in 5hmC levels. The result suggests a role of 5hmC in gene regulation and possible associations with mental illness and behavioral changes.
Human studies
Nine articles, which included human subjects with ASD, FXS, TET3 deficiency and ID investigating 5hmC as an epigenetic marker in NDDs, were included in this review (Table 1). This shows the growing interest of geneticists and biological psychiatrists in exploring the role of environmental factors as epigenetic mechanisms, including DNA methylation/demethylation, in the development of NDDs.
In 2012, a study by Wang et al. (Reference Wang, Pan, Lin, Szulwach, Song, He, Wu, Warren, Jin, Duan and Li2012) unveiled significant insights into 5hmC dynamics during human cerebellum development. It demonstrated a substantial increase in 5hmC levels from fetal to adult stages, underscoring its pivotal role in brain maturation. Remarkably, 5hmC was found to be preferentially enriched in gene exons and untranslated regions (UTRs), pinpointing specific genomic areas influenced by this modification throughout development. The discovery of both fetus-specific and adult-specific DhMRs, especially in genes regulated by fragile X mental retardation protein (FMRP) and those linked with autism, suggested a widespread impact of 5hmC on neurodevelopment. These findings highlight the potential of dysregulated 5hmC in contributing to the molecular pathogenesis of NDDs. Furthermore, the observation that fetus-specific DhMRs retain epigenetic memories of embryonic stem cells implied these regions carry developmental epigenetic information crucial for cerebellum development gene expression patterns. The association of DhMRs with ASD candidate genes further supported the notion that 5-hmC dysregulation could be a contributing factor to NDDs, offering novel perspectives for understanding the epigenetic mechanisms underlying brain development and its related disorders.
Zhubi et al. investigated the function of 5hmC in the transcriptional regulation of ASD candidate genes, specifically GAD1 and RELN, in the cerebellum and frontal cortex of individuals with ASD (James et al., Reference James, Shpyleva, Melnyk, Pavliv and Pogribny2014; Zhubi et al., Reference Zhubi, Chen, Dong, Cook, Guidotti and Grayson2014, Reference Zhubi, Chen, Guidotti and Grayson2017). Their findings revealed an enrichment of 5hmC at the promoters of GAD1 and RELN in the cerebellar cortex of individuals with ASD. Additionally, there was a significant increase in the binding of MeCP2 and TET1 to the promoters of GAD1 and RELN (Zhubi et al., Reference Zhubi, Chen, Dong, Cook, Guidotti and Grayson2014). In fact, Mellén et al. (Reference Mellén, Ayata, Dewell, Kriaucionis and Heintz2012) characterized MeCP2 as a significant binding protein for 5-5hmC in the central nervous system (CNS). MeCP2 was found to enhance gene transcription upon binding to 5hmC, and conversely, it exhibited a repressive effect when binding to DNA containing 5mC. These results suggest that elevated levels of 5hmC, relative to 5mC, at specific gene regions enhance the binding of MeCP2 to 5hmC, thereby reducing the expression of the target genes in the cerebellum of individuals with ASD. Notably, alterations in MeCP2 levels due to gene deletions or mutations are implicated in Rett Syndrome, a syndromic NDD with prominent features of autism (American Psychiatric Association, 1994; Chahrour and Zoghbi, Reference Chahrour and Zoghbi2007). MeCP2 plays a critical role in regulating synaptic and neuronal plasticity and developing motor skills, cognitive function and social behavior (Ebert and Greenberg, Reference Ebert and Greenberg2013). Furthermore, both RELN and GAD1 have been implicated in the pathophysiology of ASD (Zhubi et al., Reference Zhubi, Chen, Guidotti and Grayson2017).
James et al. (Reference James, Shpyleva, Melnyk, Pavliv and Pogribny2014) have also shown a significant increase in 5hmC levels in the cerebellum of individuals with autism, accompanied by elevated expression of de novo methyltransferases DNMT3A and DNMT3B, ten-eleven translocase genes TET1 and TET3, and increased content of 8-oxo-deoxyguanosine (8-oxo-dG), an indicator of oxidative DNA damage. They further demonstrated a positive correlation between 5hmC content and the engrailed-2 (EN-2) gene expression within the EN-2 promoter. The EN-2 homeobox transcription factor plays a crucial role in Purkinje cell maturation, normal cerebellar patterning and development and has been implicated in ASD in multiple studies (Benayed et al., Reference Benayed, Choi, Matteson, Gharani, Kamdar, Brzustowicz and Millonig2009; Cheng et al., Reference Cheng, Sudarov, Szulc, Sgaier, Stephen, Turnbull and Joyner2010).
Cheng et al. (Reference Cheng, Li, Manupipatpong, Lin, Li, Xu, Jiang, Shu, Wu and Jin2018) conducted a study investigating 5hmC alterations in postmortem brains of individuals with ASD across different age groups. They identified differentially hydroxymethylated regions (DhMRs) specifically in the young group (age ≤ 18), while no significant DhMRs were observed in the groups over 18 years of age. These findings suggest that 5hmC alterations are associated with ASD, particularly during early development, and may contribute to the pathogenesis of the disorder. Pathway and disease association analyses further revealed that the intragenic DhMRs were enriched in genes involved in cell–cell communication and neurological disorders. Notably, many ASD risk genes play crucial roles in synapse development, which is essential for communication between brain cells (Ebert and Greenberg, Reference Ebert and Greenberg2013).
Corley et al. (Reference Corley, Vargas-Maya, Pang, Lum-Jones, Li, Khadka, Sultana, Blanchard and Maunakea2019) conducted a study focusing on the interindividual variability in DNA methylation levels among individuals with ASD by examining postmortem subventricular zone (SVZ) tissue specimens from ASD cases and control subjects. The SVZ, which is implicated in ASD pathology, serves as a significant neurogenic niche in the mammalian brain, housing neural stem cells that undergo proliferation, differentiation and migration to form the neocortex during prenatal human brain development. The study revealed significantly lower levels of DNA methylation in ASD cases than controls at differentially methylated loci (DML) in both young and middle-aged groups. Specifically, ASD brain samples exhibited hypomethylation at transcription start sites (TSS), gene bodies and canonical exons, indicating a global hypomethylation pattern in young individuals with ASD compared to typically developing individuals. In contrast, 5hmC levels were increased at TSS, gene bodies and canonical exons in ASD cases while being depleted in control samples. These findings suggest an active demethylation process in the genome associated with ASD and underscore the crucial role of 5hmC-mediated epigenetic modifications in the pathogenesis of this NDD.
Beck et al. (Reference Beck, Petracovici, He, Moore, Louie, Ansar, Douzgou, Sithambaram, Cottrell, Santos-Cortez, Prijoles, Bend, Keren, Mignot, Nougues, Õunap, Reimand, Pajusalu, Zahid, Saqib, Buratti, Seaby, McWalter, Telegrafi, Baldridge, Shinawi, Leal, Schaefer, Stevenson, Banka, Bonasio and Fahrner2020) identified TET3 deficiency as a novel Mendelian disorder of DNA demethylation, affecting 11 patients across eight families, characterized by intellectual disability (ID), developmental delay and growth anomalies. Through analysis of mono-allelic and bi-allelic pathogenic variants, primarily within TET3’s catalytic domain, the study elucidated the enzyme’s vital role in 5hmC production and neurodevelopment, while underscoring the phenotypic commonalities with other epigenetic disorders, thereby advancing our understanding of the epigenetic regulation mechanisms involved in human development and disease.
Esanov et al. (Reference Esanov, Andrade, Bennison, Wahlestedt and Zeier2016) further elucidated the role of 5hmC in the epigenetic regulation of the FMR1 gene in FXS, revealing significant increases in 5hmC levels at the FMR1 promoter in full-mutation FXS patient brains compared to pre-mutation carriers and controls, with this enrichment being neuron-specific. Cellular models, including fibroblasts, lymphocytes and induced pluripotent stem cell (iPSC)-derived neurons, failed to recapitulate the 5hmC enrichment observed in primary neurons, suggesting they do not fully mimic the epigenetic landscape of FXS. Additionally, 5hmC and 5mC distributions were distinct between neuronal and non-neuronal DNA fractions, indicating cell-type-specific epigenetic regulation. The study further demonstrates that neither cell-cycle progression nor neuronal maturity affects 5hmC levels at the FMR1 promoter, and TET enzyme expression levels do not differ significantly between FXS-derived neuronal models and primary neurons, suggesting that the lack of 5hmC enrichment in cellular models is not due to TET dysregulation. These findings highlight the critical role of 5hmC in the FMR1 gene’s epigenetic regulation in FXS and point to the need for future therapeutic strategies targeting this pathway to restore FMR1 expression while also indicating that current cellular models may not fully capture the disease’s epigenetic nuances.
Mixed models
Shpyleva et al. (Reference Shpyleva, Ivanovsky, de Conti, Melnyk, Tryndyak, Beland, James and Pogribny2014) conducted a study examining DNA from the cerebellum of BTBR T+tf/J mice, a relevant mouse model of autism and postmortem cerebellum samples from individuals with ASD. Their findings revealed elevated levels of 8-oxo-deoxyguanosine (8-oxo-dG), 5mC and 5hmC in both mouse and human samples, corroborating earlier observations by James et al. (Reference James, Shpyleva, Melnyk, Pavliv and Pogribny2014). The study further emphasized the interplay between reduced expression of 8-oxoguanine DNA-glycosylase 1 (Ogg1) and 5hmC in the pathogenesis of ASD.
Ogg1, highly expressed in the brain, protects neurons against oxidative DNA damage during development and various pathological conditions (Wong et al., Reference Wong, McCallum, Jeng and Wells2008; Liu et al., Reference Liu, Croteau, Souza-Pinto, Pitta, Tian, Wu, Jiang, Mustafa, Keijzers, Bohr and Mattson2011). The absence of Ogg1 in the brain leads to various cellular and molecular events, including increased apoptosis and abnormal neuronal connectivity (Wong et al., Reference Wong, McCallum, Jeng and Wells2008), key pathomorphological features of autism. In fact, Bhatia et al. (Reference Bhatia, Arslan, Rodriguez-Hernandez, Bonin and Wells2022) explored the involvement of OGG1, using OGG1 knockout mice, in brain development and its role in repairing DNA lesions induced by reactive oxygen species. Their findings demonstrate that young Ogg1 knockout mice displayed sex- and gene-specific DNA damage, reduced DNA methylation marks, elevated cerebellar calbindin levels, impaired hippocampal function, altered body weight and various behavioral abnormalities, highlighting the significance of OGG1 in normal brain development through its potential functions in DNA repair and epigenetic regulation, with implications for NDDs.
Discussion
Ethical concern
In the papers mentioned in the human studies section, dissection of postmortem brains (cerebellum) of children under the age of 18 was required to be done to profile genome-wide distribution of 5hmC. There have been ethical concerns about brain banks, as many controversial concerns have arisen.
Everyone has the right to autonomy over their body per the Convention on Human Rights and Biomedicine (Hendriks, Reference Hendriks1997). Therefore, pursuing informed consent for removing brain tissue postmortem requires consent for organ donation before death. In some cases, patients such as ASD patients are considered vulnerable. They often do not have autonomy over their bodies to make that decision, so such decisions are made by their caretakers, which raises questions about the adequacy of surrogate decision-making in capturing the donor’s wishes.
Additionally, there are no international laws about postmortem body organs and tissue donation to legalize and regulate it worldwide in a harmonized manner (Huitinga et al., Reference Huitinga, de Goeij and Klioueva2019). To mitigate these ethical dilemmas and enhance the ethical integrity of research involving postmortem brain dissection, international collaboration should advocate for and participate in international efforts to develop harmonized legal and ethical standards and laws for postmortem donation and research. Moreover, more transparency and public engagement are needed in the future to educate people who are considering donating to brain banks on the importance of postmortem brain tissues in research, explain the ethical aspects and make sure consent forms reflect all those aspects to ensure the informed decision of donors and caretakers of vulnerable donors.
Future perspectives
Studies showed that 5hmC is critical in brain development and neurological disorders. 5hmC-mediated pathways are essential for gene regulation (Cheng et al., Reference Cheng, Bernstein, Chen and Jin2015). Moreover, it is an intermediate in the DNA demethylation process. This finding is crucial as it will help researchers and professionals better understand the role of 5hmC on neurodevelopment disorders, and it could accelerate the research to find novel treatments or even come up with preventative measures to minimize the effect of neurodevelopment disorders.
Most studies included in this review, if not all, utilized ASD as the prototypical NDD. However, other NDDs, such as ADHD, IDD, communication disorders, learning disorders and tic disorders, warrant further epigenetic investigations. These disorders also have a profound negative impact on children’s social, psychological and academic functioning.
Moreover, most investigations focused on analyzing genome-wide DNA hydroxymethylation changes in brain samples from the cerebellum and frontal cortex. This choice of regions was motivated by the observed positive correlation between levels of 5hmC and cerebellum development in previous studies. It would be interesting to explore the enrichment of 5hmC in other brain regions, such as the parietal, temporal and occipital lobes.
Animal studies that model ASD-like symptoms offer crucial insights, yet it is imperative to distinguish between the simulation of specific behaviors and the broader scope of NDD research. This differentiation is essential for advancing our comprehension and treatment strategies for ASD and other NDDs within a comprehensive neurodevelopmental context. While rodents, particularly mice, are frequently utilized for ASD modeling due to their cost-effectiveness and sophisticated genetic manipulation capabilities, they lack the sulcus and gyrus structures present in the human brain, posing a significant limitation to their translational relevance (Li et al., Reference Li, Zhu, Gu and Cheng2021). Emerging technologies, including single-cell sequencing, multiphoton in vivo imaging and gene editing, represent promising advancements in unraveling the pathogenesis of NDDs (Li et al., Reference Li, Zhu, Gu and Cheng2021). These technologies hold the potential to overcome existing limitations by providing deeper insights into the cellular and molecular complexities underlying these disorders.
Increasing evidence is shedding light on the connection between inflammation and neuropsychiatric disorders, highlighting the significant impact of immunological responses on neurological health. Notably, the association of Pediatric Autoimmune Neuropsychiatric Disorders Associated with Streptococcal Infections (PANDAS) with conditions such as Tourette Syndrome and obsessive-compulsive disorder is well-known (Khoodoruth et al., Reference Khoodoruth, Ahammad, Khan and Mohammad2023a). Furthermore, research has revealed the intricate interactions among the redox system, autophagy and the Nrf2 pathway in the pathogenesis and progression of neuropsychiatric disorders (Calabrese et al., Reference Calabrese, Mancuso, Calvani, Rizzarelli, Butterfield and Giuffrida Stella2007, Reference Calabrese, Cornelius, Dinkova-Kostova, Calabrese and Mattson2010, Reference Calabrese, Giordano, Signorile, Laura Ontario, Castorina, De Pasquale, Eckert and Calabrese2016; Scuto et al., Reference Scuto, Mancuso, Tomasello, Ontario, Cavallaro, Frasca, Maiolino, Salinaro, Calabrese and Calabrese2019). These findings underscore the pivotal role of oxidative stress and inflammation in NDDs and point to the promising therapeutic potential of targeting these pathways.
Given the limited options for pharmacological intervention in managing challenging behaviors in NDDs—where only risperidone and aripiprazole have been approved for autism-related irritability (Cohen et al., Reference Cohen, Raffin, Canitano, Bodeau, Bonnot, Périsse, Consoli and Laurent2013)—there is a compelling need for innovative therapeutic approaches. In this context, epigenetic therapies, such as DNMT modifiers, emerge as promising candidates. Drawing on their successful application in oncology (Leone et al., Reference Leone, Voso, Teofili and Lübbert2003), these therapies hold significant promise for precision medicine in NDDs, offering new hope for addressing these complex conditions with more targeted and effective interventions.
Research has shown that machine learning can offer new insights into the intricate ways epigenetic changes affect our genes, as it has great potential to advance our understanding of the complex relationships between epigenetic modifications and gene expression. Some research highlights the importance of artificial intelligence in advancing our understanding of epigenetic modifications and their potential implications in disease diagnosis and therapy. Liu et al. (Reference Liu, Chen, Su, Chen and Wei2020) discuss the important role of RNA 5hmC modification in various biological processes, including NDDs. However, identifying 5hmC sites in RNA is expensive and challenging. Therefore, the authors developed a computational protocol called iRNA5hmC, which uses machine learning to predict RNA 5hmC sites. The researchers found that the proposed feature representations were more accurate at distinguishing true 5hmC sites from non-5hmC sites compared to existing feature descriptors. This could potentially increase the efficiency of exploring 5hmC sites and their association with NDDs for scientists when exploring connection between 5hmC sites and NDDs such as ASD, encouraging more opportunities for research in this area.
In another study, Pavlovic et al. (Reference Pavlovic, Ray, Pavlovic, Kotamarti, Chen and Zhang2017) developed a machine learning framework called DIRECTION to predict and characterize DNA methylation and hydroxymethylation in mammalian genomes, providing a cost-effective and efficient alternative to existing methods. The paper presents a machine-learning framework that predicts and characterizes DNA methylation and hydroxymethylation in mammalian genomes. The framework was tested on human embryonic stem cells and neural progenitor cells, and it accurately predicted DNA methylation and hydroxymethylation, demonstrating the potential for large-scale reconstruction of epigenetic maps in mammalian model systems.
As we look to the future, enhancing machine learning algorithms to better predict 5hmC patterns—and possibly other epigenetic modifications—becomes crucial. Such advancements could provide deep insights into the role of epigenetics in NDDs. Importantly, machine learning models that accurately predict epigenetic changes hold the promise for clinical application, offering new avenues for diagnosis and treatment. By identifying specific epigenetic markers linked to disorders such as ASD, these models could pave the way for personalized medicine, enabling tailored treatment plans based on an individual’s epigenetic makeup for more effective outcomes.
Conclusion
In summary, this review article has provided an overview of the current literature investigating the involvement of 5hmC in the development of NDDs, underscoring its diagnostic and therapeutic importance. This emphasis is particularly critical given the current absence of definitive clinical laboratory tests for the diagnosis and management of NDDs. The identification of 5hmC as a prominent epigenetic mark in the brain has expanded our understanding of the intricate mechanisms governing neurogenesis and the emergence of complex behavioral disorders. NDDs entail a multifaceted molecular pathogenesis encompassing a range of genomic, epigenomic, proteomic, metabolic and physiological alterations. Therefore, gaining comprehensive insights into the molecular processes of NDDs using advanced machine learning algorithms, whenever feasible, is crucial for effective clinical and personalized management and prevention strategies.
Open peer review
To view the open peer review materials for this article, please visit http://doi.org/10.1017/pcm.2024.2.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article.
Acknowledgements
We are grateful for the support received from the College of Health and Life Sciences of Hamad Bin Khalifa University and the Child and Adolescent Mental Health Service of Hamad Medical Corporation. Figures were created with BioRender.com.
Author contribution
M.A.S.K.: conceptualization, methodology, project administration, writing- original draft preparation, writing- reviewing and editing. W.N.C.K.: visualization, writing-reviewing and editing. R.A.: conceptualization, methodology, writing- original draft preparation, writing- reviewing and editing.
Financial support
Open Access funding provided by the Qatar National Library.
Competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.






Comments
Dr. Mohamed Adil Shah Khoodoruth
Royal College of Psychiatrists (UK) Member
Child and Adolescent Psychiatry Clinical Fellow
Hamad Medical Corporation
Qatar
Division of Genomics and Precision Medicine
College of Health and Life Sciences
Hamad Bin Khalifa University
Qatar
29 November 2023
Editor-in-Chief
Cambridge Prisms: Precision Medicine
Professor Dame Anna Dominiczak
University of Glasgow | UK
Dear Professor Dominiczak
I hope that this letter finds you in good health.
Please find attached our review entitled “ 5-hydroxymethylcytosine: A new player in neurodevelopmental disorders” for consideration for publication in Cambridge Prisms: Precision Medicine. This review delves into the role of 5hmC in neurodevelopmental disorders, particularly autism spectrum disorder, highlighting its significance in epigenetic control, behavioral disorders, and precision medicine. By examining both mice models and human samples, the review explores elevated 5hmC levels in the brain, connecting it with autism-like symptoms and other neurodevelopmental disorders.
This review would greatly interest the readers of Cambridge Prisms: Precision Medicine as it provides an overview of the current state of knowledge in this field. The broad range of topics covered in this review would interest the readership of Cambridge Prisms: Precision Medicine, and would provide a useful resource for researchers and clinicians working in this field.
Thank you in advance for your consideration, and we look forward to your response.
Sincerely,
(on behalf of co-authors)
Adil