INTRODUCTION
Cryptosporidium parvum is a protozoan parasite with a wide range of animal hosts and is a major cause of gastrointestinal illness in human populations. In immunocompetent people cryptosporidiosis is a self-limiting disease with symptoms of watery diarrhoea and abdominal cramps that may be accompanied by vomiting, low grade fever and loss of appetite, lasting for a median of 10½ days [Reference Hunter1]. In severely immunocompromised individuals, infection may result in potentially life-threatening severe and/or protracted disease [Reference Hunter and Nichols2]. The dose-dependent incubation period for cryptosporidiosis ranges from 4 to 19 days in healthy adults [Reference Chappell3].
Transmission of infective oocysts is primarily through the faecal–oral route, either through person-to-person, direct animal contact or via contaminated food, water or fomites [Reference Davies and Chalmers4]. Cryptosporidium oocysts shed in animal faeces can remain viable for long periods and are highly resistant to most commonly used chemical disinfectants, including chlorine [Reference Chalmers and Davies5]. The environmental persistence of oocysts, coupled with a low infectious dose, result in significant risk of zoonotic infection to humans in environments such as petting farms [Reference Okhuysen6].
Previous zoonotic outbreaks of C. parvum infection in humans in the UK have been linked to bottle-feeding lambs [Reference Chalmers and Giles7–Reference Gormley9], direct contact with pre-weaned calves [Reference Gait10], poor hand hygiene in farm environments [Reference McGuigan, Steven and Pollock8] and consumption of contaminated water [Reference Duke11–Reference Hoek15].
In 2009, the UK Health Protection Agency launched an independent investigation into the contributing factors of a major outbreak of verocytotoxigenic Escherichia coli O157 in visitors to an open farm in Surrey. The committee made a number of recommendations including the need for a code of practice for open farms to reduce the risk to visitors from zoonoses such as E. coli and Cryptosporidium [16]. In 2012, the Access to Farms Partnership, UK, published an industry-sponsored Code of Practice for visitor attractions offering animal contact [17].
On 17 April 2013, the local diagnostic microbiology laboratory notified Public Health England (PHE) of a cluster of eight laboratory-confirmed cases of cryptosporidiosis. Environmental health officers (EHOs) used trawling questionnaires to identify common exposures and found that all cases had visited a petting farm in the North of England over the Easter holiday period with onset of symptoms from 3 to 10 days after their reported visit date. The farm opened for the season on 29 March and over the following 2 weeks had about 600 visitors per day. The premises consisted of a farm building with animal contact areas, access to penned animals, dining and play areas. Outside there were a picnic area and further animal enclosures. Activities offered included bottle-feeding of lambs and calves, pig brushing and contact with rabbits, guinea pigs, tortoises, chickens and donkeys. When brought into the barn on 29 March, the lambs were approximately aged between 10 and 14 days, piglets between 21 and 28 days and calves were >2 months old. Lambs were brought out of the pens into an open animal contact area in the main barn to be bottle-fed while calves remained in their pen and were fed through a gate.
Following an outbreak control meeting, EHOs visited the farm on 19 April and implemented initial control measures: contact with lambs was minimized, the premises were steam cleaned and supervising staff were asked to encourage all visitors to wash their hands.
Previously in 2011, 12 confirmed cases of cryptosporidiosis had also been linked to this farm. Investigation at the time had putatively identified bottle-feeding of lambs and lamb contact near the eating area as the most likely sources but no definite conclusions were drawn at the time due to limited epidemiological and microbiological findings.
As this was the second outbreak at this establishment without an obvious aetiological source, a case-control study was conducted alongside further environmental and microbiological investigations. The objective of these investigations was to identify the source and the route of transmission of Cryptosporidium infection during this outbreak.
Due to the findings of the previous outbreak investigation at the farm in 2011 and a number of similar spring outbreaks linked to bottle-feeding lambs [Reference Chalmers and Giles7–Reference Gormley9], contact with the bottle-fed lambs as the source of the outbreak was considered the primary hypothesis. Additional exposures, including any other animal contact, hand washing behaviour, risk awareness, participation in farm activities and the consumption of food and drinks on site, were also examined to identify alternative potential transmission risks that might have contributed to the risk of cryptosporidiosis on this farm.
METHODS
Descriptive analysis
Case-finding was primarily conducted through routine surveillance mechanisms, with neighbouring health protection teams alerted to the outbreak and asked to report any additional cases linked to the farm. Primary cases had onset of illness in the first 14 days following their visit to the farm. Diarrhoea was reported in all cases; other symptoms included vomiting, abdominal pain and fever. Confirmed cases were distinguished from probable cases through laboratory confirmation of Cryptosporidium sp. identified in a faecal sample by auramine phenol microscopy. Secondary cases may or may not have visited the farm, but had household contact with a confirmed case and had onset of symptoms >14 days after any farm visit.
Environmental investigation
The farm was visited on four separate occasions between 19 April and 27 June 2013 by EHOs, PHE and a Veterinary Investigation Officer from the Animal and Plant Health Agency (APHA), Thirsk, UK. The premises were assessed for compliance against the Industry Code of Practice [17].
Microbiological investigation
Stool samples were microscopically examined at local laboratories for Cryptosporidium oocysts [18]. Smear-positive stool samples were sent to the national Cryptosporidium Reference Unit (CRU), Swansea, UK, for species confirmation by real-time polymerase chain reaction (PCR) based on the Lib13 gene, nested PCR and sequencing part of the ssu rRNA gene and subtyping through DNA sequence analysis of the 60 kDa glycoprotein (gp60) gene [Reference Hadfield19–Reference Alves21].
During a visit to the farm on 7 May 2013, the Veterinary Investigation Officer took 43 freshly voided floor faecal samples from chickens (3 samples); horse (1); donkeys (4); sheep (1); lambs (8); calves (12); pigs (8); guinea pigs (5) and a tortoise (1). The samples were tested for presence of Cryptosporidium oocysts using the fluorescent antibody test (FAT, Cellabs Crypto Cel Antigen test, TCS Biosciences Ltd, UK) at APHA, Weybridge. Microscopy positive samples (n = 8) were sent to the national CRU for species identification and subtyping as described above.
Case-control study definitions
The at-risk population for this outbreak was defined as all farm visitors from 29 March to 14 April 2013 inclusive. Around 10 500 people visited the farm over this period.
The control group were defined as individuals aged >12 months who visited the farm over this period who did not have symptoms of diarrhoea in the following 28 days. Infants aged <12 months were not eligible as all reported cases were aged >1 year. To minimize recall error by parents, siblings of cases were excluded from the control group.
Cases were identified as any confirmed or probable case of cryptosporidiosis that visited the farm in the study period and had onset of symptoms in the following 14 days.
Recruitment of controls
A snowballing control recruitment method was used as there was no record of individuals who had visited the farm: cases were asked to provide the details of friends or family who had visited the farm over this period. These individuals were contacted by the study interviewers, invited to participate in the study and asked to provide details of further potential controls. The process was repeated until study recruitment met a 1:1 case/control ratio.
Sample size calculation
Recruitment of 32 cases and 32 controls was required in order to detect a minimum odds ratio (OR) of 5 with 80% power at the 5% significance level, where 50% of controls are exposed.
Exposures
The farm was identified as a common exposure in all primary cases through initial trawling questionnaires conducted by EHOs. A study-specific questionnaire was developed to collect further information about the farm visit; including risk awareness and information provision, individual animal contact, dry feeding of animals, bottle-feeding of lambs and calves, pig brushing, crowding during these activities, hand washing, consumption of food and/or drink on site (including food from the cafe and tap water from the farm) and contact with play facilities.
Data collection
Questionnaires were administered either through a telephone interview or, alternatively, an online survey completed directly by study participants (or the parent/guardian who visited with the child in question). Telephone interviews were conducted by seven interviewers who received training in administering the questionnaire. Responses were entered contemporaneously into the web-based survey. In order to minimize the potential for response bias, participation in the study was maximized by following-up non-responders by both telephone and e-mail and the provision of choice in preferred method of questionnaire completion. Consent to participate in the study was obtained for all interviews.
Statistical analysis
Individual risks for all 48 potential exposures measured were estimated by calculating crude ORs with corresponding 95% confidence intervals (CIs) and tested using Pearson's χ 2. Exposure effects which were significant at the 75% level were examined for inclusion in a logistic regression model (n = 16).
Due to the sparsity of data across age groups, it was not possible to adjust for the confounding effect of age completely. Adjustment was necessarily limited to comparisons between three aggregated age groups (0–5, 6–17, ⩾18 years).
Due to low levels of exposure individually, lamb contact risk factors (hugging lambs, having a lamb on your lap, touching or sitting on the floor in the lamb-feeding area) were combined into a single ordinal variable for inclusion in the multivariate logistic regression model. Bottle-feeding lambs, physical contact with lambs and lack of hand washing after animal contact were identified, from experience in the previous outbreak in 2011 and published literature [Reference Chalmers and Giles7–Reference Gormley9], as presumptively increasing the risk of Cryptosporidium infection. Given the limited sample size and high levels of exposures to some of these activities in the control population, the power of the study to detect a significant association would be limited to high-effect estimates (OR⩾5). For these reasons, these three primary risk factors were included in all models regardless of statistical significance.
It was not possible to include all 16 exposures in a single regression model due to related exposures and sparse data across subgroups, so forward selection of variables was performed starting from a primary model in which case-control status was modelled on age group and primary risk factors (bottle-feeding lambs, physical contact with lambs, absence of hand washing after animal contact). Additional factors were retained in the logistic regression model if they significantly improved the fit of the model to the data. Fit was assessed using likelihood ratio testing (χ 2 LR) with significance α < 0·05. Differences in medians were assessed using the non-parametric test for equality of medians.
As both laboratory-confirmed and probable cases were included in the case-control study, there may be potential for misclassification bias. In order to examine the maximum magnitude of this error, a sensitivity analysis was performed in which only laboratory-confirmed cases were included. All statistical analyses were performed using Stata v. 12.1 (StataCorp, USA).
RESULTS
Descriptive analysis
Forty-six cases of cryptosporidiosis were linked to this outbreak with onset dates ranging from 5 to 27 April 2013, corresponding to farm visits between 31 March and 10 April (Fig. 1). These cases consisted of 45 primary cases and one secondary case, a household contact of a primary case. Five cases were admitted to hospital. The median incubation period between farm visit and self-reported onset of symptoms for primary cases was 7 days (range 1–14 days). Median age for all cases where age was reported (n = 43) was 7 years (range 1–76 years).
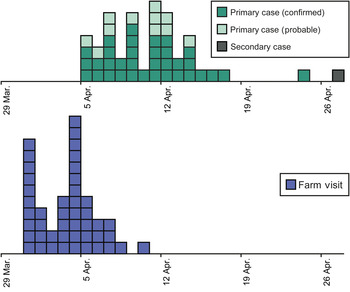
Fig. 1. Epidemiological curves showing date of farm visit and reported symptom onset for all cases (n = 46).
Environmental investigation
Inspection of the premises found that the farm was largely compliant with the Industry Code of Practice [17]. The overall environment of the farm was found to be very clean. The welfare status of the animals was deemed to be excellent. All animals were clean and no signs of ill health had been reported. Adequate hand washing facilities were available and there were multiple signs around the premises reminding visitors to wash their hands. Bottle-feeding of both lambs and calves were supervised by experienced members of staff who reminded visitors to wash their hands. No concerns were identified in the farm cafe or designated eating areas.
Microbiological investigation
Presence of Cryptosporidium oocysts was confirmed using auramine phenol microscopy in isolates from 35 cases linked to this outbreak. Of these 35 isolates, 22 were sent by local laboratories to the national CRU, for species confirmation and subtyping. All 22 available isolates were confirmed as C. parvum and identified as gp60 subtype IIaA19G1R1 which is not common in England.
Microscopic investigation by FAT of the 43 animal faecal samples identified Cryptosporidium oocysts in chicken (2/3 samples), sheep (1/1), lamb (2/8), pig (2/8) and guinea pig (1/5) samples (Table 1). Positive nested ssu rRNA gene PCR results were obtained for 3/8 samples, with an isolate from a lamb paddock identified as C. parvum and two isolates from a pig pen as C. scrofarum. Subtyping of the lamb paddock isolate confirmed the presence of the IIaA19G1R1 subtype. Representative sequences have been deposited into GenBank under accession numbers KT634306 and KT634307.
Table 1. Microbiological investigation of faecal samples from animal pens
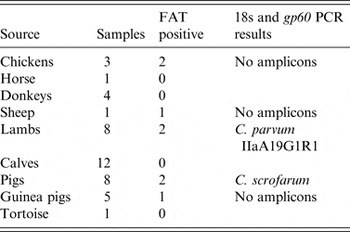
FAT, Fluorescent antibody test for presence of Cryptosporidium sp. oocysts; PCR, polymerase chain reaction.
Case-control study
Out of the 46 cases linked to this outbreak, 45 people were eligible for inclusion in the study as primary cases of cryptosporidiosis. Forty-two (93·3%) were successfully contacted by telephone, all of whom agreed to take part in the study. Thirty-eight (84·4%) cases completed the questionnaire, either online or via a telephone interview. The remaining four cases were sent a link to the online survey but did not complete the questionnaire. Through the snowballing method, a total of 69 potentially eligible controls were identified. Of these, five could not be contacted, 64 were contacted by telephone or e-mail and 55 (79·7%) agreed to take part in the study. A total of 42 (60·9%) controls completed the online questionnaire, three of whom were excluded from the study as they reported symptoms within 28 days of their farm visit. Thirty-eight cases and 39 controls were included in the statistical analysis.
Case and control groups did not statistically differ by age, gender, length of farm visit, arrival time, day or date period of visit (Table 2). However, due to the recruitment process, the reporting delay between visit date and completion of the study questionnaire was significantly longer in controls than in cases (control median reporting delay 42 days, case median reporting delay 31·5 days, χ 2 = 36·5, P < 0·001). Reporting delay ranged from a minimum of 22 to a maximum of 51 days.
Table 2. Characteristics of case-control study participants
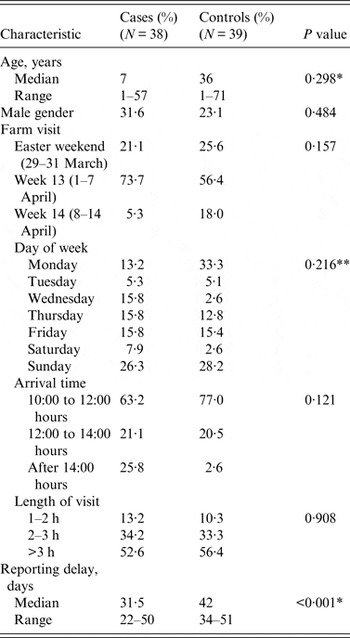
Reporting delay, period between farm visit and completion of study questionnaire.
P value, for Pearson's χ 2 test unless otherwise specified.
* Non-parametric test for the equality of medians.
** Fisher's exact test.
Out of 48 potential exposures examined through the questionnaire, 16 achieved the threshold statistical significance (75%) in univariate analysis for inclusion in a logistic regression model (Tables 3 and 4). No single risk factor could account for all cases of illness. As expected, bottle-feeding of lambs was very popular in both cases and controls. Cases were significantly more likely to report touching the tortoise and to have eaten without washing their hands. Cases were also significantly less likely to report being verbally advised about the risk of illness on the farm and how to avoid becoming ill.
Table 3. Single variable analysis for all exposures examined for inclusion in a logistic regression model (P < 0·25)
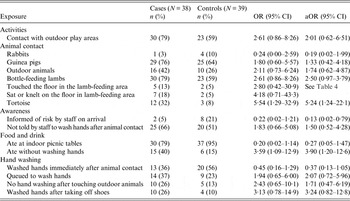
OR, Odds ratio; CI, confidence interval; aOR, odds ratio adjusted for age group, bottle-feeding lambs and not washing hands after animal contact.
Table 4. Lamb contact risk factors included in logistic regression as a combined ordinal variable (P < 0·25)
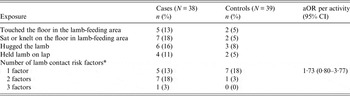
aOR, Odds ratio adjusted for age group, bottle-feeding lambs and not washing hands after animal contact; CI, confidence interval.
* Combined ordinal variable for number of lamb risk factors reported.
Only 45% of cases reported contact with calves (unadjusted OR 1·16, 95% CI 0·43–3·16) and 37% bottle-feeding calves (unadjusted OR 1·17, 95% CI 0·41–3·31). Neither of these exposures reached the threshold statistical significance necessary to be examined in the multivariate model. A substantial proportion of both cases (61%) and controls (72%) ate food from the farm cafe, but this was not associated with a significant risk of illness (unadjusted OR 0·60, 95% CI 0·21–1·73) and therefore was also not examined in the multivariable model. Only one case reported drinking tap water from the farm.
Results for the final logistic regression model are presented in Figure 2. This study identified eating without washing hands as a significant risk factor for Cryptosporidium infection in the outbreak (OR 5·48, 95% CI 1·51–19·9) and being informed of risk of infection on arrival at the farm as a protective factor (OR 0·10, 95% CI 0·01–0·71). Although not statistically significant, the other risk factors included in the model based on prior knowledge were all associated with a small increase in risk (lack of hand washing and delayed hand washing after animal contact, lamb contact, bottle feeding lambs). There were no further potential associations observed with other animal groups, activities or food and drink after adjusting for age group and primary risk factors. Excluding all cases without laboratory confirmation (n = 8) from analyses resulted in no change in effect estimates.

Fig. 2. Final multivariable logistic regression model (77 observations).
DISCUSSION
This substantial outbreak of cryptosporidiosis was unusual in the notable absence of any identifiable concerns during environmental inspection of the farm premises. Young lambs were identified as a potential primary source of the outbreak through a combination of microbiological and epidemiological investigations. The exposure period for outbreak cases corresponds with the peak oocyst excretion period for the lambs used in bottle-feeding activities: the lambs were aged between 10 and 14 days on their introduction into the barn at the start of the exposure period for this outbreak and the last reported visit date by a case was when the lambs would have been aged 23–27 days. Studies have shown that between 75–100% of lambs born in an environment with asymptomatic shedding adults or other infected neonates become infected with C. parvum within the first 2 weeks of life [Reference Ortega-Mora22, Reference De Graaf23]. In one study, C. parvum oocyst excretion was reported to peak at 8 days post-infection followed by a consistent decline over 11–15 days associated with increasing antibody levels [Reference Bukhari and Smith24]. No signs of clinical infection were present in the lambs although C. parvum was found in faecal samples. This is consistent with previous studies which have found significant oocyst shedding in asymptomatic neonatal lambs [Reference Pritchard25].
The case-control study identified a small but not statistically significant risk associated with lamb contact and bottle-feeding lambs. As none of these effect estimates exceeded an OR of 5, it is not expected that this study would have the necessary power to detect a statistical association with infection risk. Additionally, contact with lambs did not account for all cases of illness, with only 61% of cases reporting direct contact with a lamb. This finding suggests that transmission also occurred through environmental contamination of fomites with Cryptosporidium oocysts.
Although previous outbreaks of C. parvum have been linked to contact with unweaned calves [Reference Gait10], in this outbreak investigation no epidemiological association was found between reported exposure to calves and illness. Studies in UK cattle populations have demonstrated that the risk of Cryptosporidium infection in cattle is highest in calves aged between 8 and 21 days [Reference Brook26] with high levels of oocyst shedding in unweaned calves aged <1 month [Reference Smith27]. Calves at the farm were aged 30 days on their introduction to the barn. No calf faecal samples were found to be positive for Cryptosporidium oocysts at the time of microbiological sampling, but as this occurred 3 weeks after the outbreak it may not accurately reflect carriage during the period of outbreak exposure. There was no evidence to suggest that direct contact with calves was the source of this outbreak but it remains plausible that transmission could have occurred indirectly from infected calves through contamination of the environment or cross-infection of other livestock.
Microbiological investigation linked all 22 subtyped cases and a faecal sample from a lamb enclosure as the rare gp60 subtype IIaA19G1R1. Although the animal faecal sample was collected 3 weeks after the outbreak and only represents a snapshot of carriage at that time, it does indicate a persistence of C. parvum in the livestock at the farm. IIaA19G1R1 has a sparse global distribution and has been documented sporadically in a range of cross-sectional surveys including calves (n = 2) in Cheshire, UK [Reference Brook28], one soay sheep on St Kilda, UK [Reference Connelly29], one dairy calf in Buenos Aires, Argentina [Reference Del Coco30], a cow in The Netherlands [Reference Wielinga31] and in sporadic human cryptosporidiosis in Slovenia [Reference Soba and Logar32]. This subtype was found to be the causative organism in two separate outbreaks of cryptosporidiosis in school children linked to farm visits in the East Midlands in 2013 (n = 3) and 2014 (n = 3) (PHE and CRU data). IIaA19G1R1 was identified in a recurring outbreak in visitors to a Norwegian holiday farm linked to lambs and goat kids [Reference Lange33]. These authors were able to link two outbreaks 3 years apart to the persistence of the IIaA19G1R1 subtype on the farm. By contrast, this outbreak investigation identified a different subtype to that isolated in the earlier outbreak in 2011 (IIaA17G1R1) (CRU data). Increasing application of subtyping and other genetic technologies will present the opportunity to examine trends over longer time periods and identify potentially persistent outbreak subtypes.
In response to these findings, the farm implemented a number of changes to practice. These included bringing in lambs at an older age, after their peak oocyst excretion period, minimizing physical contact with visitors by feeding the lambs through a fence and increasing the ratio of supervisors to visitors at bottle-feeding sessions.
The results of the epidemiological study identified not being informed of risk and the absence of hand washing before eating as statistically significant risk factors for infection. Cases were 5½ times more likely than controls to report eating without washing their hands and 90% less likely to have been informed of infection risk on arrival to the farm. Risk awareness was a significant issue across the study population with 51% of those interviewed reporting that they were not adequately aware of the risk of infection on an open farm and how to avoid such risks, despite industry-standard recommended levels of signage and written information [17]. Although hand washing facilities were sufficient, hand hygiene recommendations were not complied with by large numbers of visitors and several participants reported the use of hand gels in preference to hand washing. As the farm was very clean in appearance we conclude that this limited the immediate perception of risk in visitors, and in combination with the limited effectiveness of signs in communicating risk, resulted in poor hand hygiene compliance in visitors.
To address the identified issues, the farm has gone beyond industry-standard recommendations and applied a number of additional processes to improve the communication of risk to their visitors; maximizing verbal communication of information, increasing staff supervision, improving staff training and adding interactive information to entrance tickets and staff uniforms. Communicating risk of infection from animal contact activities and importance of hand washing effectively to the public remains a substantial challenge.
This study has a number of additional limitations. The snowballing sampling technique employed to recruit controls resulted in a high response rate and adequate control recruitment but as a non-random sampling technique is subject to a number of potential issues. Immediate friends and family of cases may not be representative of the wider target population and any similarity in exposure status between cases and controls resulting from this technique would result in an underestimation of any risk association. This would be compounded if any of the control population had been misclassified and in fact had become infected on their visit. In order to minimize this misclassification bias, respondents reporting diarrhoea or vomiting in the 28 days after their farm visit were excluded from the control group. Despite this, there may potentially be asymptomatic carriers in the control group [Reference Davies34] whose inclusion would underestimate the effect of true risk exposures.
Case-finding in this investigation was limited only to those identified through routine surveillance methods. A study by Tam et al. in 2012 estimated that the true population incidence of cryptosporidiosis in England and Wales between 2008 and 2009 was 8·2 times greater than that identified through routine surveillance [Reference Tam35]. It is likely that we have underestimated the actual size of this outbreak, as the self-limiting course of infection will restrict the number of cases seeking healthcare as well as through under-ascertainment through standard sample submission and testing processes. However, the cases of highest clinical significance will have been successfully captured through this process and included in the investigation.
Due to the statistically significant differential reporting delay between cases and controls there is increased potential for recall bias in participants’ responses where controls may be less likely to remember an exposure. The median delays between farm visit and completion of study questionnaire were long for both controls (42 days) and cases (31½ days).
Cryptosporidiosis is a widespread infection with 3520 confirmed cases in England and Wales in 2013 (PHE data). There were 12 petting-farm associated outbreaks of cryptosporidiosis across England between January and May 2013 alone [36]. The outbreak described in this investigation was the largest of these and affected a wide range of ages. Combined epidemiological, environmental and microbiological investigations identified asymptomatic lambs as the most likely source of infection. This is the third documented outbreak of cryptosporidiosis in European farm visitors caused by the rare C. parvum gp60 subtype IIaA19G1R1 [Reference Lange33]. This study also highlights the limited effectiveness of signs in communicating risk, particularly on farms where the immediate risk of contamination is not obvious to visitors. Improving risk awareness and hand hygiene compliance in farm visitors is a substantial challenge which must be addressed in order to prevent future outbreaks of not only cryptosporidiosis, but other zoonotic infections of significant public health importance such as Vero cytotoxin-producing E. coli.
ACKNOWLEDGEMENTS
We thank our PHE colleagues in Yorkshire and the Humber PHE Centre and the Field Epidemiology Service, Leeds, as well as the staff of Craven, Bradford, Leeds, Calderdale, Kirklees and Wakefield District Councils, the APHA, Thirsk and the national Cryptosporidium Reference Unit, Swansea, especially Kristin Elwin and Guy Robinson for gp60 subtype analysis.
This research received no specific grant from any funding agency, commercial or non-for-profit sectors.
DECLARATION OF INTEREST
None.









